Abstract
This study aims to evaluate the feasibility of a label-free nanobiosensor based on blood plasma surface-enhanced Raman spectroscopy (SERS) method for exploring variability of different tumor (T) stages in nasopharyngeal cancer (NPC). Au nanoparticles as the SERS-active nanostructures were directly mixed with human blood plasma to enhance the Raman scattering signals. High quality SERS spectra can be acquired from blood plasma samples belong to 60 healthy volunteers, 25 NPC patients with T1 stage and 75 NPC patients with T2–T4 stage. A diagnostic accuracy of 83.5% and 93.3%, respectively, can be achieved for classification between early T (T1) stage cancer and normal; and advanced T (T2–T4) stage cancer and normal blood groups. This exploratory study demonstrates that the nanobiosensor based on SERS technique in conjunction with PCA-LDA has great potential as a clinical complement for different T stages detection in nasopharyngeal cancer.
Nasopharyngeal cancer (NPC) is a malignant neoplasm arising in the nasopharyngeal epithelial lining presenting a distinct racial and geographical distribution. According to the Global Cancer Statistics, NPC has extremely high incidence and mortality in southeast Asia and southern China, accounting for 20–30 new cases per 100,000 Chinese populations per year, occurring 10–30 times more frequently than in the United States and other western countries1,2. The most important prognostic factor for NPC is the tumor (T), nodes (N), and metastases (M) staging,which also has high directive value in making therapy plan. Particularly, T stages show the variation of tumor volume and express independent prognostic factor of local control for primary tumor volume. It has been confirmed that the risk of local failure is increased by estimated 1% for every 1 cm3 increase in primary tumor volume3. Recent report has shown that 5-year overall survival (OS) is approximately 100% for T1, however that will be tremendously decreased to 74.1% for T2, 63.9% for T3 and 18.7% for T4, suggesting the significant role of T stage classification for predicting 5-year OS of the NPC patients4. Hence, early detection and analysis of different T stages coupled with timely and standard treatment (e.g. radiotherapy or chemoradiotherapy) is critical to improving patients' survival. However, it is challenging for clinicians to detect T1 stage cancer by conventional nasopharyngoscope examine, since the insidious nature of NPC and the relative anatomical inaccessibility of the nasopharynx, leading to a limited accuracy5. Although magnetic resonance imaging (MRI) is more sensitive than nasopharyngoscope for detecting subtle and infiltrating lesions, performing MRI on all suspect patients is not practical. In this regard, plasma Epstein-Barr virus (EBV) analysis base on immunofluorescence assay would be more useful for high-risk individuals screening, but it suffers the disadvantage of low positive predictive value, high individual variation, lack of a standardized method and time-consuming procedure6. Excisional nasopharyngeal suspicious lesions for histopathological examinations currently remains the gold standard diagnosis, though this approach contains the subjective methodology in the procedures and is invasive and unsuitable for mass screening of high-risk patients who may have multiple suspicious lesions7. Therefore, development of a sensitive, convenient, non-invasive and biopsy-free diagnostic technique would be of imperative clinical value to identify and analyze different T stages, especially for early stage (T1) in NPC.
Raman spectroscopy (RS) technique base on inelastic light scattering process is capable of capturing ‘fingerprints' of specific biomolecular structures and conformation, since the scattered photon will shifted to another frequency correlated with specific molecular vibration respect to the incident excitation light7,8. Recently, RS technique has been widely investigated for biomolecular alterations associated with diseases and particularly shown promise for human cancer-related detection, including cancer cells, tissue and others7,9,10. Nevertheless, there are some limitations associated with conventional RS technique. For instance, the Raman scattering efficiency is extremely weak resulted from inherently small cross-section (10−30 per molecule) of biological samples11. To overcome this drawback, it is obliged to increase laser power and/or data collection time to acquire good quality spectra, which may change and even damage the biological sample. Beyond that, a strong background autofluorescence from biological samples makes it difficult to extract the pure Raman signals. These main disadvantages of conventional RS technique hinder its further clinical applications in medical cancer diagnosis12.
The drawbacks of conventional RS technique can be overcome by the development of surface-enhanced Raman spectroscopy (SERS) based on nanotechnology. Due to the intense electromagnetic field near the metallic nanostructures, Raman signals are dramatically enhanced when the Ag or Au nanoparticles (NP) are mixed with analyte molecules in SERS measurements. Besides, the strong background autofluorescence can be greatly reduced at the same time11,13. Recently, SERS technique has attracted considerable interest as a promising tool in cancer detection. Although currently polymerase chain reaction (PCR) and immunoassays can be used for testing those cancer biomarkers, they suffer the disadvantage of expensive reagents and time-consuming sample preparation processes compared with SERS. In addition, SERS can provide further advantages over conventional fluorescence-based assays: (1) narrower spectral widths; (2) multiple labels detection under single source; and (3) no photobleaching issues for SERS labels14. Most currently approaches are focused on immunoassays (cancer biomarker) with antibodies that are conjugated to metal NPs13,15. For example, Wang. et al.16 developed an antibody-labeled Au NPs based SERS immunoassay that relies on the recognition of biomarkers to monitor mucin protein MUC4 levels in patient sera for the early stage diagnosis of pancreatic cancer. Additionally, a rapid and reproducible SERS-based immunoassay technique combining magnetic beads with antibody-labeled Au NPs was reported for sensitive target marker (CEA) detection in lung cancer17. In the latter, a simultaneous detection of dual cancer markers in blood serum has been achieved using SERS-based immunoassay technique18. Additionally, there is a series of studies in which SERS has been used to discriminate tumor cells from benign cells19,20,21. In brief, different from conventional RS that captures the biomolecular information on the whole samples, SERS technique can achieve single molecule detection and identification, proving a unique opportunity to enabling earlier cancer detection13.
Human blood-based assays are the most used tests for noninvasive cancer detection in clinical practice, due to the key role of blood markers in early diagnosis, prognosis, and monitoring responses to therapy16. Compared with other biological sample such as tissue and cell, blood sample has significant advantages in conveniently and even repeatedly collection for high-risk patients. Additionally, as tumor formation, the content of biomolecules such as protein and DNA included in human blood will alter due to apoptosis and necrosis of cells11. Recently, substantial progress has also been achieved in label-free SERS technology. Our group has applied ultra-sensitive SERS technique into human blood detection to develop a label-free nanobiosensor for cancer screening11,12,22,23. However, the potential of SERS technique for identification and analysis of different T stages especially for early stage (T1) has not been assessed in detail for cancer detection.
The aim of this study thus was to assess the feasibility of Au NP based SERS for biomolecular analysis of blood plasma samples belong to normal, T1 stage cancer and T2–T4 stage cancer for NPC detection. Multivariate statistical techniques, including principal component analysis combined with linear discriminant analysis (PCA-LDA), were employed to analyze and differentiate the blood plasma SERS spectra obtained from the three groups. To the best of our knowledge, this is the first report on blood SERS for NPC detection at different T stages which is crucial for clinician to evaluate the patient's condition and make optimal treatment decisions. This exploratory work may further promote this label-free blood SERS technique into practical clinical applications.
Results
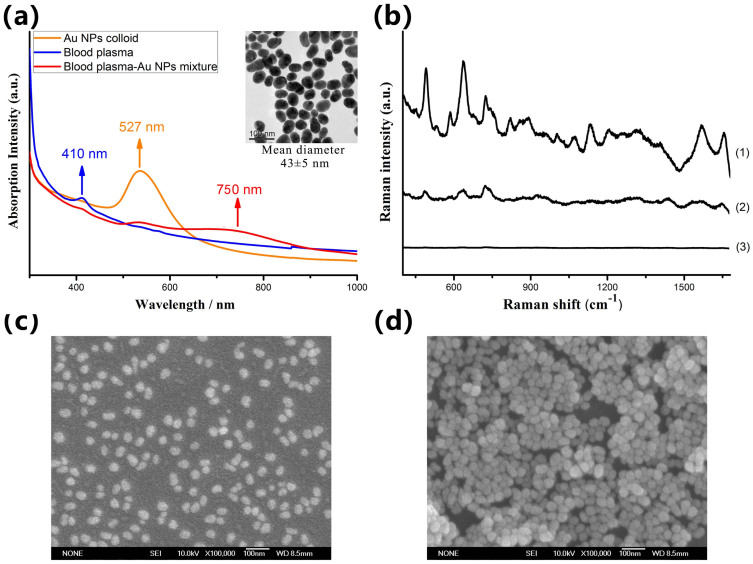
Using Au-NPs as substrate, we have successfully acquired blood plasma SERS spectra from 160 subjects. In these samples, 60 were histopathological normal and 100 were NPC. According to TNM classification, 25 cancers were of T1 stage and 75 T2–T4 stages. As shown in Fig. 1a, when the Au NPs were mixed with blood plasma, a new broad plasmon resonance absorption band around 750 nm wavelength region appeared, while the intense blood plasma and Au NPs colloid absorption bands (410 and 527 nm) were reduced to some extent. This change is usually believed to originate from the localized surface plasmons resonance of deposited Au NPs. The new plasmon resonance absorption band largely overlaps with the excitation laser, which will lead to a tremendous enhancement in the intensity of the Raman scattering. Figure 1b shows a comparison of blood plasma SERS spectrum and regular Raman spectrum of the same plasma sample without the Au NPs, demonstrating that the intensity of many dominant vibration bands increased dramatically due to a strong localized surface plasmons resonance of Au NPs on the surface of biomolecules in the blood plasma. Figure 1c and 1d show the SEM image of Au NPs, and the mixture of plasma-Au NPs, respectively, which confirmed that biochemical substances in the blood plasma were conjugated to the surfaces of Au NPs leading to the obvious aggregation.
Figure 1.
(a) The UV/visible absorption spectra of the Au NPs colloid, blood plasma and blood plasma-Au NPs mixture. The insert picture shows the TEM micrograph of Au NPs. (b) Comparison of (1) SERS spectrum of blood plasma-Au NPs mixture, (2) regular Raman spectrum of the same plasma sample without the Au NPs and (3) background Raman signal of the anticoagulant mixed with Au colloid. (c–d) The SEM images of Au NPs, and blood plasma-Au NPs mixture, respectively.
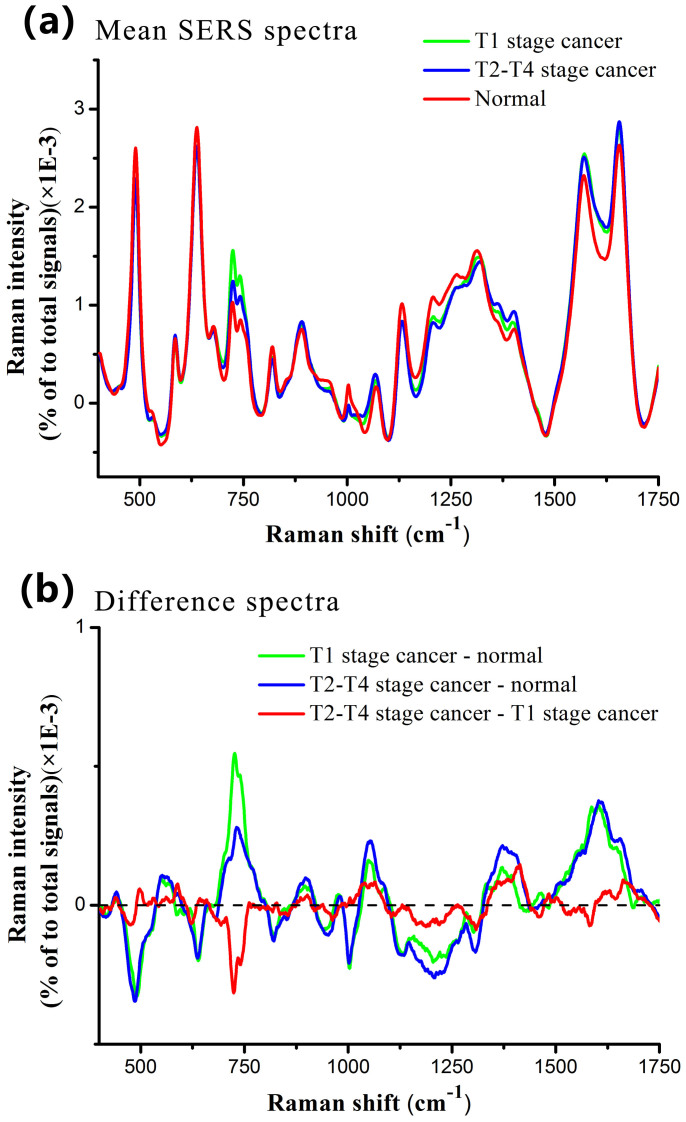
Figure 2a shows the normalized mean SERS spectra from normal, T1 stage and T2–T4 stage NPC blood plasma samples. The prominent SERS peaks located at 492, 638, 725, 743, 818, 890, 1003, 1068, 1131, 1208, 1402, 1573 and 1655 cm−1 can be consistently observed in both normal and cancer plasma, with the strongest signals at 492, 638, 1131, 1573 and 1655 cm−1. To better understand the molecular basis for the observed SERS spectra of blood plasma, Table 1 lists tentative assignments for the observed SERS bands, according to the literatures11,12,22,23,24,25,26,27,28,29. A comparison of blood SERS spectra of normal and different T stages of NPC reveals distinctive SERS spectral features and intensity differences in the spectral ranges of 700–800, 950–1100, 1200–1350, 1550–1675 cm−1 which primarily contain signals related to DNA/RNA bases, proteins and lipids. These significant Raman spectral changes can be viewed more clearly in the corresponding difference spectra between the three groups in Fig. 2b.
Figure 2.
(a) Comparison of normalized mean SERS spectra from 60 normal, 25 T1 stage NPC and 75 T2–T4 stage NPC blood plasma samples. (b) Difference spectra calculated from the mean SERS spectra among the three groups.
Table 1. The peak positions and tentative assignment of major vibrational bands observed in plasma samples11,12,22,23,24,25,26,27,28.
| Peak positions (cm−1) | Vibrational mode | Major assignment |
|---|---|---|
| 492 | Ring vibration | L-arginine |
| 638 | C-C twisting | Tyrosine |
| 725 | Hypoxanthine | |
| 743 | Ring vibration | Thymine |
| 818 | C-C stretching | Collagen |
| 890 | C-O-H bending | D-galactosamine |
| 1003 | C-C symmetric stretch ring breathing | Phenylalanine |
| 1068 | C-C stretching | Lipids |
| 1131 | D-mannos | |
| 1208 | C-C6H5 stretching | Tryptophan |
| 1402 | Phospholipids | |
| 1573 | DNA/RNA bases | |
| 1655 | C = O stretching | AmideI, α-helix |
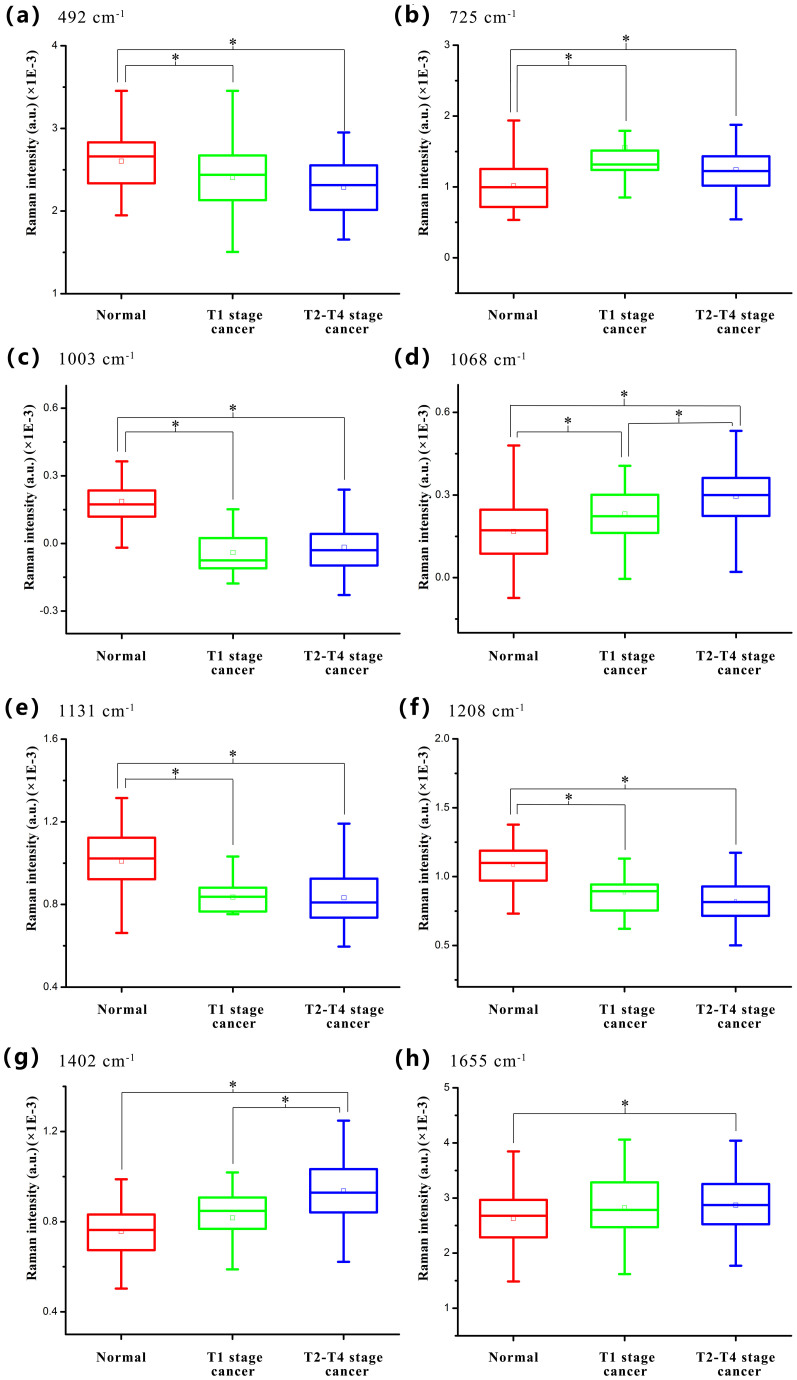
Particularly, the eight important SERS peaks at around 492, 725, 1003, 1068, 1131, 1208, 1402 and 1655 cm−1 were identified among the three blood groups. Figure 3 shows the comparison of these SERS spectral intensities in the box plots. It can be found that cancer groups show lower intensities at 492, 1003, 1131 and 1208 cm−1 but exhibit much increased SERS signals at 725, 1068, 1402 and 1655 cm−1 as compared to normal group. In addition, the intensities of most SERS peaks except 725 and 1003 cm−1 SERS peaks, represent linear changes as T stages development compared to normal, indicating special changes of biomolecules in blood plasma samples between different T stages of NPC patients and normal subjects. Moreover, T-test represents that different spectral intensities were largely associated with different degrees of diagnostic utility for discriminating different blood groups (normal, T1 stage cancer and T2–T4 stage cancer). For instance, spectral intensity of 492, 725, 1003, 1131 and 1208 cm−1 is optimal in discriminating normal from T1 stage cancer and T2–T4 stage cancer; spectral intensity of 1068 cm−1 shows efficacy in classification of the three different blood types; spectral intensity of 1402 cm−1 can be used for differentiating T2–T4 stage cancer from normal and T1 stage cancer; and spectral intensity of 1655 cm−1 can be used to separate normal from T2–T4 stage cancer. The detailed diagnostic results were presented in the Supplementary Table S2 online. From the results, spectral intensity of 1003 cm−1 achieved the most optimal classification effect between different T stage cancer and normal. Also, it can be seen that T1 stage cancer and T2–T4 stage cancer can not be significantly discriminated using most of SERS spectral intensities. More detailed analysis about the changes of biomolecules with the assignment of SERS bands was presented in next discussion section. These results indicate that there are significant changes in the percentage of biomolecules in different blood groups, suggesting a potential role of blood SERS for different T stages detection in NPC.
Figure 3. Box plots of the eight significant SERS peak intensities for the three blood sample types (normal, T1 stage cancer and T2–T4 stage cancer): (a) 492 cm−1, (b) 725 cm−1, (c) 1003 cm−1, (d) 1068 cm−1, (e) 1131 cm−1, (f) 1208 cm−1, (g) 1402 cm−1 and (h) 1655 cm−1.
The line within each notch box represents the median, and the lower and upper boundaries of the box indicate first and third quartiles respectively. Error bars (whiskers) represent the 1.5-fold interquartile range. *p < 0.05 (pairwise comparison of blood groups with T-test).
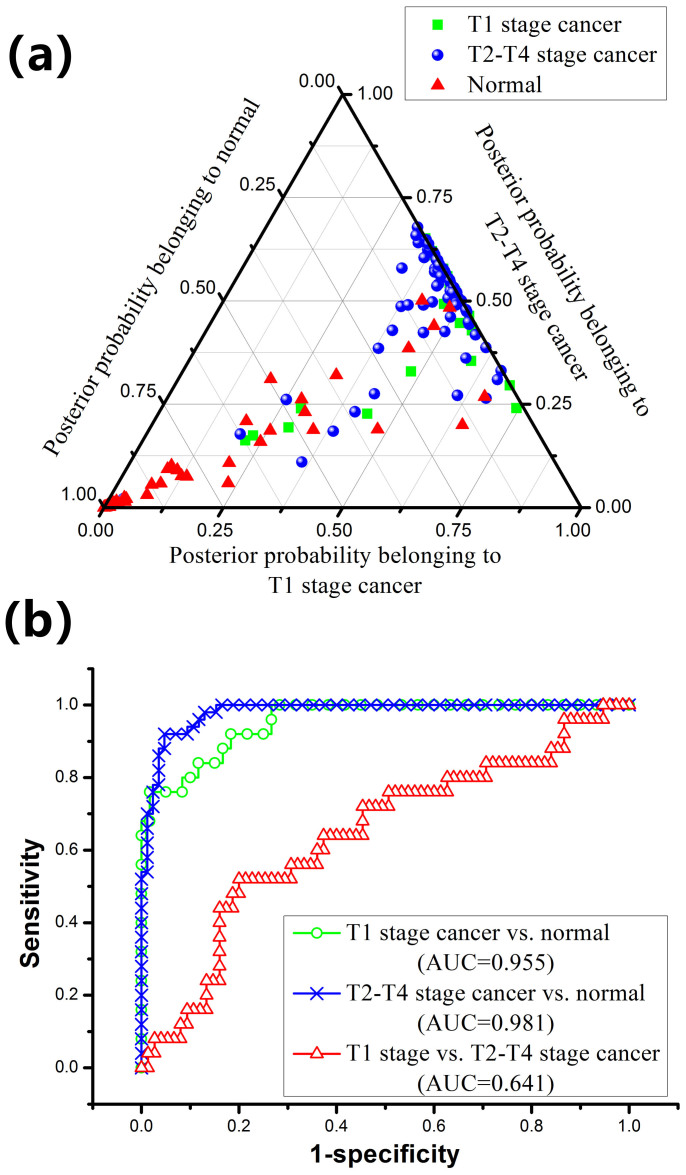
To further develop diagnostic algorithms for improving blood SERS analysis and differentiation between normal, T1 stage cancer and T2–T4 stage cancer samples, the normalized whole SERS spectrum data set was fed into the SPSS software package (SPSS Inc., Chicago) for PCA-LDA analysis after the removal of fluorescence background. Three PCs (PC1, PC2 and PC3) accounting for 63% of the variance were found to be the most diagnostically significant (p < 0.05) as defined by T-test on all the PC scores for discriminating the three groups. Then, all the three PCs were loaded into the LDA with the leave-one-out, cross-validation method for generating effective diagnostic model for blood sample classification. Figure 4a displays a ternary plot of the posterior probabilities belonging to normal, T1 stage cancer and T2–T4 stage cancer blood plasma groups. Table 2 summarizes the diagnostic results for SERS spectra using PCA-LDA method in classifying the three blood groups. Although there are some degree of overlap in plots showed in Fig. 4a, high diagnostic sensitivities of 84% and 92%, specificities of 83.3% and 95%, and accuracies of 83.5% and 93.3%, respectively, can be achieved for classification of T1 stage cancer and normal, and for T2–T4 stage cancer and normal blood groups. However, the diagnostic sensitivity, specificity and accuracy are only 64%, 62.7% and 63%, respectively, for classification of T1 stage cancer and T2–T4 stage cancer. The differences between samples belonging to one pathology type have also been assessed using PCA-LDA method (see Supplementary Table S3 online). The statistical results show that the differences between samples from the same group are not diagnostically significant.
Figure 4.
(a) Two-dimensional ternary plot of the posterior probabilities belonging to normal, T1 stage cancer and T2–T4 stage cancer blood plasma samples, illustrating the good clustering of the three distinctive groups achieved by the PCA-LDA diagnostic algorithms. (b) Receiver operating characteristic (ROC) curves of classification results for different T stages of NPC generated from PCA-LDA analysis. The integration areas under the ROC curves (AUC) are 0.641, 0.955 and 0.981, respectively, for the three groups' classification.
Table 2. Classification results of blood SERS prediction of the three pathology groups using PCA-LDA method.
| Predicted parameters | |||||
|---|---|---|---|---|---|
| Diagnostic combinations | Sensitivity | Specificity | Accuracy | ||
| T1 stage cancer | vs. | Normal | 84% (21/25) | 83.3% (50/60) | 83.5% (71/85) |
| T2–T4 stage cancer | vs. | Normal | 92% (69/75) | 95% (57/60) | 93.3% (126/135) |
| T1 stage cancer | vs. | T2–T4 stage cancer | 64% (16/25) | 62.7% (47/75) | 63% (63/100) |
To further evaluate and the performance of the blood SERS together with PCA-LDA for different T stages of NPC detection, receiver operating characteristic (ROC) curves were generated (Fig. 4b) at different threshold levels. The integration areas under the ROC curve (AUC) are 0.641, 0.955 and 0.981, respectively, for T1 stage cancer vs. T2–T4 stage cancer; T1 stage cancer vs. normal; and T2–T4 stage cancer vs. normal classification, respectively. These results demonstrate that PCA-LDA-based SERS technique has potential capacity for different T stages of NPC detection.
Discussion
T stage is confirmed as an independent prognostic factor and plays a significant role for predicting 5-year OS of the NPC patients. Particularly, the 5-year OS of T1 stage NPC is greatly higher than that of other stages. In this work, we investigate the blood plasma SERS spectral properties of T1 stage cancer and T2–T4 stage cancer samples of NPC. To further characterize different stages of NPC blood samples, we also compared with SERS spectra of normal blood samples. Specific differences in SERS spectra between normal, T1 stage cancer and T2–T4 stage cancer samples can be observed (Fig. 2), suggesting a promising potential of blood SERS technique for NPC detection and analysis applications.
The most important detection mechanism of blood SERS is based on the enhancement in electromagnetic field due to strong localized surface plasmons resonance between the aggregated Au NPs. When the Au colloid was mixed with the blood plasma sample, the biochemical substances in the blood plasma were nonspecifically absorbed to the surfaces of Au NPs leading to this aggregation. Another mechanism involves chemical enhancement as a result of charge transfer between NP surface and the blood plasma molecules30,31. Due to the two major mechanisms, the Raman signals of blood plasma can be greatly enhanced (Fig. 1b), which provides a unique opportunity to explore subtle changes in blood plasma from different T stages of cancers at the molecular level.
For example, the SERS peak intensities at 492, 638, 725, 743, 818, 890, 1003, 1068, 1131, 1208, 1402, 1573 and 1655 cm−1 appear to be unique with a certain degree of similar alterations (increase/decrease) of blood SERS signals in T1 stage cancer and T2–T4 stage cancer as compared to normal. This indicates that T1 stage cancer and T2–T4 stage cancer samples still contain some similar constituents. The variations in protein-related SERS peaks at 1003, 1208 and 1665 cm−1 generally associated with T stage development likely reflect special content and conformation changes. A decrease in SERS signals at 1003 and 1208 cm−1 respectively was found in cancer samples as compared to normal, indicating a decrease in the percentage of phenylalanine and tryptophan relative to the total SERS-active constituents in cancer blood. This is consistent with Huang group's report on lower Raman signals of phenylalanine and tryptophan associated with malignancies in gastric and tissues10. The Raman peak at 1003 cm−1 is a prominent and stable Raman signal reflecting the changes of phenylalanine in tissue, cell, blood and other samples. Previous results (see Supplementary Table S2 online) confirm this peak has great potential as a biomarker for classification between different T stage cancer and normal.
The relative peak intensities at 1655 cm−1 due to the amide I band exhibited higher signal, indicating that cancer blood may be associated with an increase in the relative amount of proteins in the a-helix conformation. Similar change could also be observed in cervical cancer blood samples11. Besides, this kind of α-helical proteins, such as histones which are main protein component that makes up the chromatin, also excited a higher content in gastric tissues samples32. The changes of these proteins may reflect more chemical interaction between the proteins (gene) and the microenvironment occurring in the cells, which is probably due to the disordered mitotic activity and other stimulatory effect, including DNA damage and oncogene activation24,33. For example, NPC transformation is consistently associated with the Epstein-Barr virus (EBV) infection, resulting in the high expression of EBV transforming proteins in NPC patients. Besides, high levels of p53 (nuclear protein) and low levels of p16 (cyclin-dependent kinase inhibitory protein) proteins are frequently found in NPC patients, which could result in rapid progression to advanced cancer3.
In addition, there is also a relative increase of DNA/RNA bases-related bands (725, 743 and 1573 cm−1) in intensity, suggesting that cancer blood may be associated with an increase in the relative amount of nucleic acid bases relative to the whole SERS-active constituents. The appearance of these nucleic acid bases may signify an abnormal metabolism of DNA or RNA bases in the blood of NPC subjects, resulting from the apoptosis and necrosis, or release of intact cells in the bloodstream and their subsequent lysis. Recently, investigation of circulating RNA in human blood has received much interest for its newly tumor-specific markers role for cancer detection and diagnosis. Cancer patients had elevated cell-free RNA levels in their peripheral blood34. Similarly, a report35 shows that blood DNA level is significantly increased in esophageal cancer patients and has also been regarded as an important molecular marker for monitoring the response to adjuvant therapy in ovary, lung, and lymphoma cancer patients. In our previous studies on nasopharyngeal and colorectal cancer blood detection, similar changes at 725 cm−1, an important ‘fingerprint' was observed22,23. Despite our report on nasopharyngeal cancer detection has shown an increase of nucleic acid bases in cancer subjects, the change tendency of these DNA/RNA bases was not explicit in various risk levels of cancer. In this study, it can be found that the content maximum of DNA/RNA bases-related (725 cm−1) appears in T1 stage NPC, higher than normal and T2–T4 stage NPC subjects (Fig. 3). This could be another evidence that the SERS band of 725 cm−1 has promising potential as a biomarker for early cancer detection. Besides, the SERS peak intensity at 818 cm−1 due to collagen showed lower percentage signals than those of normal samples, indicating a decrease in the percentage of collagen contents in cancer blood. This is explainable. The collagen was cleaved probably due to the cytoplasmic mucin depletion and the elevated concentration of metalloproteinase, thereby resulting in an overall decrease of SERS intensity at 818 cm−1 in cancer blood plasma. On top of all these, a decrease in the SERS bands of L-arginine (492 cm−1), tyrosine (638 cm−1), and D-mannos (1131 cm−1) and a increase in the SERS bands of D-galactosamine (890 cm−1) and lipid (1068 cm−1) were found in plasma of cancer patients, suggesting a special molecular changes in quantity or structure in the NPC patients. The abnormal metabolism associated with cancer transformation, may lead to these changes, which is in agreement with biochemical analysis results of colorectal cancer blood detection23. Therefore, the differences in SERS spectra between T1 stage cancer and T2–T4 stage cancer samples further confirm that SERS method can be used to reveal biomolecular changes associated with different T stages development in NPC.
It should be noted that the simplistic peak intensities analysis above only uses limited SERS peak information, and there are significant variations and overlapping intensities of SERS spectra of normal and cancer blood plasma among inter-subjects. Thus, a multivariate statistical analysis based on PCA-LDA was employed to incorporate the entire spectrum and automatically determine the most diagnostically significant features for improving the efficiency for plasma analysis and differentiation. This powerful procedure has been widely investigated by researchers for Raman spectral analysis in cancer tissue, cell and blood discrimination10,11,25. The present study showed that the diagnostic accuracy of 93.3% for identifying T2–T4 stage cancer from normal can be achieved using the PCA-LDA model, which had almost a 10% improvement in diagnostic accuracy compared with the discrimination between T1 stage cancer and normal groups. For advanced T stage of NPC, the abnormal metabolism is greater than that of early stage (T1), besides, advanced T stage cancer is probably with distant metastasis, thus resulting in more complex constitute in blood compared to the normal. On the other hand, only 63% diagnostic accuracy can be achieved for identifying T1 stage cancer and T2–T4 stage cancer. This result reflects that there are similar alterations of blood components in T1 stage cancer and T2–T4 stage cancer as compared to normal, indicating that different stages of cancer samples still contain some similar constituents. Therefore, it is difficult to differentiate different T stages of NPC subjects without control group. Further work combining quantitative analysis of tumor biomarker with PCA-LDA diagnostic algorithm may be a robust method to resolve this problem. In addition, Supplementary Table S4 online represents the diagnostic results for classification of SERS spectra between negative control (10 rhinitis patients' blood samples) and previous three groups (normal, T1 stage cancer and T2–T4 stage cancer) using PCA-LDA method. We found that the negative control can not be distinguished from normal group, whereas high classification accuracies can be achieved for negative control and T1 stage cancer, and for negative control and T2–T4 stage cancer, respectively. This preliminary result suggests that rhinitis disease has almost no impact on the NPC detection. Finally, receiver operating characteristic analysis (Fig. 4b) further confirms that blood SERS together with PCA-LDA diagnostic algorithm employing the entire SERS spectral features is powerful for classification between different stages cancer and normal groups. Meanwhile, we recognize that it is still a challenge to apply this spectroscopic staging approach for clinical application so far. The current spectroscopic staging approach is focused on the methodology development and a pilot application for nasopharyngeal cancer detection. Next step, we will collect more samples including benign tumor, rhinitis and other relative disease patients to verify the reliability of this method and develop more powerful diagnosis algorithm to improve this novel spectroscopic approach for accurate cancer staging.
In conclusion, the Au NPs based SERS technology is capable of characterizing biomolecular differences in NPC blood plasma samples from different T stages, and good classification between early stage (T1) cancer and normal can be achieved by PCA-LDA diagnostic algorithm, demonstrating the potential of SERS technique to be a clinical complement for early T stage detection in NPC.
Methods
Preparation of Au colloids
Stable Au colloid solutions were produced using the method reported by Grabar et. al36. In brief, a total of 500 ml HAuCL4 (1 mM) was added to a rolling boil with stirring. Then, quickly bring 50 ml sodium citrate (38.8 mM) to the vortex of the solution resulting in a color change from pale yellow to ruby-red. Boiling process was continued for 10 min and stirring was continued for an additional 15 min after the remove of the heating mantle. Figure 1a shows an absorption spectrum and a transmission electron microscopy (TEM) photograph of prepared Au colloid NPs. There is a significant absorption at 527 nm, and the particle sizes of Au colloid follow a normal distribution with a mean diameter of 43 nm and standard deviation of 5 nm. Lastly, the Au colloidal solution was concentrated by centrifugation at 15000 rpm for 10 min, and the final concentration was obtained for use.
Collection of human blood plasma samples
Human blood experiments were performed in agreement with the ethical committee at our institution (Fujian Provincial Cancer Hospital, Fujian, China), and the informed consent was obtained from all subjects. Three subject blood plasma samples were prepared in this study including 60 blood plasma samples from healthy volunteers as the control group, 25 blood plasma samples from NPC patients with T1 stage and 75 blood plasma samples from NPC patients with T2–T4 stage defined by histopathological diagnosis. Supplementary Table S1 online represents more clinical detailed information on these patients. All samples were provided by the Fujian Tumor Hospital. After 12 hours of overnight fasting, a single 3 ml blood samples were collected from the study subjects between 7:00–8:00 A.M. with the use of anticoagulant. Prior to SERS measurement, 10 μl blood plasma was mixed with 10 μl gold colloid yielding a color change from ruby-red of gold colloid to dark purple of mixture upon colloid aggregation. Figure 1a shows the absorption spectra of the Au NPs colloid, blood plasma and blood plasma-Au NPs mixture. And the prepared mixture was incubated for 2 h at 4°C. Then, a drop of this plasma-Au NPs mixture was transferred onto a rectangle aluminum plate for SERS analysis.
SERS measurement
A Renishaw Raman micro-spectrometer (Great Britain) was employed for blood plasma SERS measurement under a 785 nm laser excitation source. The system acquires SERS spectra in the wavelength region of 400–1750 cm−1 within 10 s integration time with ×20 objective, 2 cm−1 resolution using Peltier cooled charge-coupled devise camera. The software package WIRE 2.0 was used for spectral acquisition and analysis.
Data processing and analysis
Fluorescence background removal and normalization
The measured blood plasma SERS spectra represented a combination of blood Raman scattering, autofluorescence and noise. To remove the autofluorescence background of the original SERS data and yield the blood SERS spectrum alone, a Vancouver Raman Algorithm based on fifth-order polynomial fitting method was performed for raw spectra preprocess24. Then all background-subtracted SERS spectra were normalized to the area under curve in the 400–1750 cm−1 wavenumber range of each spectrum to reduce the influence of inter – and/or intra-subject spectral intensity variability for making a better comparison of the spectral characteristics (spectral shapes) among different samples for multivariate data analysis.
Multivariate statistical analysis
To reduce the high dimension of the spectral space (each SERS spectrum ranging from 400–1750 cm−1 with a set of 1433 intensity variables), PCA was employed to generate few principal components (PCs) that account for the most of the whole variance in original spectra while retaining the most diagnostically significant information for plasma differentiation. Normalized spectral data sets were assembled into data matrices with wavenumber columns and individual case rows. Thus, PCA was performed on the data matrices to generate PCs comprising a reduced number of orthogonal variables. Each loading vector is related to the original spectrum by a variable called the PC score, which represents the weight of that particular component against the basis spectrum, and is capable of reflecting the differences between different groups. An independent sample T-test was used to identify diagnostically significant PC scores calculated by PCA for each case using an alpha of 5%. The collected statistically significant PC scores (p < 0.05) were lastly retained and input into an LDA model for correctly predicting the blood plasma samples obtained from the three groups (i.e. T1 stage cancer vs. normal; T2–T4 stage cancer vs. normal; T1 stage cancer vs. T2–T4 stage cancer). Here, LDA was applied in an unbiased manner with the leave-one-out and cross-validation method on all spectral data. In this method, one sample (i.e., one spectrum) was left out from the data set, and the algorithm based on PCA -LDA was redeveloped using the remaining blood spectra. The algorithm was then used to classify the withheld spectrum. The process was repeated until all withheld spectra were classified7,37.
To compare the performance of the PCA-LDA based multivariate statistical method for blood plasma discrimination using the SERS spectral data, receiver operating characteristic (ROC) curves were generated by successively changing the thresholds to determine discrimination sensitivity and specificity for all samples11.
Author Contributions
D.L., J.P., S.F. and R.C. designed the experiments. S.Q. collected human blood samples. D.L. and H.H. performed the experiments. G.C., H.S., W.C. and Y.Y. analyzed the data. D.L. wrote the paper. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary information
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. 11104030, 61178090, 81101110, and 11274065), the major projects of international cooperation (No. 61210016), the program for Changjiang Scholars and Innovative Research Team in University (No. IRT1115), the Science and Technology Project of Fujian Province (No. 2012J05004; No.WKJ-FJ-01), the Project of the Educational Office of Fujian Province (No. JA11055), and Natural Science Foundation of Fujian Province (No. 2013J01330). Finally, we thank Prof. Haishan Zeng's meaningful discussions.
References
- Ji X. et al. Nasopharyngeal carcinoma risk by histologic type in central China: Impact of smoking, alcohol and family history. Int. J. Cancer 129, 724–732 (2011). [DOI] [PubMed] [Google Scholar]
- Jemal A. et al. Global cancer statistics. CA. Cancer. J. Clin. 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Yoshizaki T. et al. Current understanding and management of nasopharyngeal carcinoma. Auris Nasus Larynx 39, 137–144 (2012). [DOI] [PubMed] [Google Scholar]
- El-Sherbieny E., Rashwan H., Lubis S. H. & Choi V. J. Prognostic factors in patients with nasopharyngeal carcinoma treated in hospital Kuala Lumpur. Asian. Pac. J. Cancer. Prev. 12, 1739–1743 (2011). [PubMed] [Google Scholar]
- Kwok A. W. et al. Nasopharyngeal Carcinoma: a Cantonese Cumber. J. AMSA. 2, 1–20 (2013). [Google Scholar]
- Chan K. et al. Early detection of nasopharyngeal carcinoma by plasma Epstein-Barr virus DNA analysis in a surveillance program. Cancer 119, 1838–1844 (2013). [DOI] [PubMed] [Google Scholar]
- Teh S. et al. Diagnostic potential of near-infrared Raman spectroscopy in the stomach: differentiating dysplasia from normal tissue. Brit. J. Cancer 98, 457–465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Lee A. M., Lui H., McLean D. I. & Zeng H. A Method for accurate in vivo micro-Raman spectroscopic measurements under guidance of advanced microscopy imaging. Sci. Rep. 3, 1–5 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q. & Chang C. Diagnostic applications of Raman spectroscopy. Nanomedicine 8, 545–558 (2012). [DOI] [PubMed] [Google Scholar]
- Teh S. K. et al. Near-infrared Raman spectroscopy for gastric precancer diagnosis. J. Raman Spectrosc. 40, 908–914 (2009). [Google Scholar]
- Feng S. et al. Blood plasma surface-enhanced Raman spectroscopy for non-invasive optical detection of cervical cancer. Analyst 138, 3967–3974 (2013). [DOI] [PubMed] [Google Scholar]
- Feng S. et al. Gastric cancer detection based on blood plasma surface-enhanced Raman spectroscopy excited by polarized laser light. Biosens. Bioelectron. 26, 3167–3174 (2011). [DOI] [PubMed] [Google Scholar]
- Srajer J., Schwaighofer A. & Nowak C. Surface-enhanced Raman spectroscopy for biomedical diagnostics and imaging. Biomed. Spectrosc. Imaging. 2, 51–71 (2013). [Google Scholar]
- Bantz K. C. et al. Recent progress in SERS biosensing. Phys. Chem. Chem. Phys. 13, 11551–11567 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrell M., Maiti K. K., Dhaliwal K. & Chang Y.-T. Surface-enhanced Raman scattering in cancer detection and imaging. Trends Biotechnol. 31, 249–257 (2013). [DOI] [PubMed] [Google Scholar]
- Wang G. et al. Detection of the potential pancreatic cancer marker MUC4 in serum using surface-enhanced Raman scattering. Anal. Chem. 83, 2554–2561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chon H., Lee S., Son S. W., Oh C. H. & Choo J. Highly sensitive immunoassay of lung cancer marker carcinoembryonic antigen using surface-enhanced Raman scattering of hollow gold nanospheres. Anal. Chem. 81, 3029–3034 (2009). [DOI] [PubMed] [Google Scholar]
- Chon H. et al. Simultaneous immunoassay for the detection of two lung cancer markers using functionalized SERS nanoprobes. Chem. Commun. 47, 12515–12517 (2011). [DOI] [PubMed] [Google Scholar]
- Kim J.-H. et al. Nanoparticle probes with surface enhanced Raman spectroscopic tags for cellular cancer targeting. Anal. Chem. 78, 6967–6973 (2006). [DOI] [PubMed] [Google Scholar]
- Lee S. et al. Biological imaging of HEK293 cells expressing PLCγ1 using surface-enhanced Raman microscopy. Anal. Chem. 79, 916–922 (2007). [DOI] [PubMed] [Google Scholar]
- Scaffidi J. P., Gregas M. K., Seewaldt V. & Vo-Dinh T. SERS-based plasmonic nanobiosensing in single living cells. Anal. Bioanal. Chem. 393, 1135–1141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S. et al. Nasopharyngeal cancer detection based on blood plasma surface-enhanced Raman spectroscopy and multivariate analysis. Biosens. Bioelectron. 25, 2414–2419 (2010). [DOI] [PubMed] [Google Scholar]
- Lin D. et al. Colorectal cancer detection by gold nanoparticle based surface-enhanced Raman spectroscopy of blood serum and statistical analysis. Opt. Express 19, 13565–13577 (2011). [DOI] [PubMed] [Google Scholar]
- Huang Z. et al. Near-infrared Raman spectroscopy for optical diagnosis of lung cancer. Int. J. Cancer 107, 1047–1052 (2003). [DOI] [PubMed] [Google Scholar]
- Chan J. W. et al. Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells. Biophys. J. 90, 648–656 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. et al. A novel blood plasma analysis technique combining membrane electrophoresis with silver nanoparticle-based SERS spectroscopy for potential applications in noninvasive cancer detection. Nanomedicine 7, 655–663 (2011). [DOI] [PubMed] [Google Scholar]
- Li Y. et al. Research on the Raman spectral character and diagnostic value of squamous cell carcinoma of oral mucosa. J. Raman Spectrosc. 41, 142–147 (2010). [PubMed] [Google Scholar]
- Wang H. et al. Depth-resolved in vivo micro-Raman spectroscopy of a murine skin tumor model reveals cancer-specific spectral biomarkers. J. Raman Spectrosc. 42, 160–166 (2011). [Google Scholar]
- Premasiri W., Lee J. & Ziegler L. Surface-Enhanced Raman Scattering of Whole Human Blood, Blood Plasma, and Red Blood Cells: Cellular Processes and Bioanalytical Sensing. J. Phys. Chem. B. 116, 9376–9386 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M. et al. Gold nanostructures: engineering their plasmonic properties for biomedical applications. Chem. Soc. Rev. 35, 1084–1094 (2006). [DOI] [PubMed] [Google Scholar]
- Kattumuri V. et al. Agarose-stabilized gold nanoparticles for surface-enhanced Raman spectroscopic detection of DNA nucleosides. Appl. Phys. Lett. 88, 153114 (2006). [Google Scholar]
- Bergholt M. S. et al. Characterizing variability in in vivo Raman spectra of different anatomical locations in the upper gastrointestinal tract toward cancer detection. J. Biomed. Opt. 16, 037003–037010 (2011). [DOI] [PubMed] [Google Scholar]
- Ryan K. M., Phillips A. C. & Vousden K. H. Regulation and function of the p53 tumor suppressor protein. Curr. Opin. Cell Biol. 13, 332–337 (2001). [DOI] [PubMed] [Google Scholar]
- Tsui N. B., Ng E. K. & Lo Y. D. in Clinical Applications of PCR 123–134 (Springer, 2006). [DOI] [PubMed]
- Banki F. et al. Plasma DNA is more reliable than carcinoembryonic antigen for diagnosis of recurrent esophageal cancer. J. Am. Coll. Surgeons. 207, 30–35 (2008). [DOI] [PubMed] [Google Scholar]
- Grabar K. C., Freeman R. G., Hommer M. B. & Natan M. J. Preparation and characterization of Au colloid monolayers. Anal. Chem. 67, 735–743 (1995). [Google Scholar]
- Lin K., Cheng D. L. P. & Huang Z. Optical diagnosis of laryngeal cancer using high wavenumber Raman spectroscopy. Biosens. Bioelectron. 35, 213–217 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information