Abstract
DNA methylation is one of the best characterized epigenetic modifications. In mammals it is involved in various biological processes including the silencing of transposable elements, regulation of gene expression, genomic imprinting, and X-chromosome inactivation. This article describes how DNA methylation serves as a cellular memory system and how it is dynamically regulated through the action of the DNA methyltransferase (DNMT) and ten eleven translocation (TET) enzymes. Its role in the regulation of gene expression, through its interplay with histone modifications, is also described, and its implication in human diseases discussed. The exciting areas of investigation that will likely become the focus of research in the coming years are outlined in the summary.
DNA methylation serves as a cellular memory system that is dynamically regulated through the action of DNA methyltransferases (DNMTs) and TET proteins.
OVERVIEW
The DNA of vertebrate animals can be covalently modified by methylation of the cytosine base in the dinucleotide sequence 5′CpG3′. CpG is an abbreviation for cytosine and guanine separated by a phosphate, which links the two nucleotides together in DNA. In mammals, DNA methylation patterns are established during embryonic development by de novo methylating enzymes called Dnmt3a and Dnmt3b. They are maintained by a Dnmt1-mediated copying mechanism when cells divide. The heritability of DNA methylation patterns provides an epigenetic marking of the genome that is stable through multiple cell divisions and therefore constitutes a form of cellular memory. For this reason, historically, DNA methylation has represented the archetypal mechanism of epigenetic inheritance.
DNA methylation is found in some lower eukaryotes such as Neurospora and invertebrates (discussed in Aramayo and Selker 2013 and Elgin and Reuter 2013). There is also quite an elaborate DNA methylation system in plants, involving many enzymes and specific binding proteins covered in depth in Pikaard and Mittelsten Scheid (2014).
Molecular and genetic studies in mammals have shown that DNA cytosine methylation (abbreviated to 5mC, for 5-methyl cytosine) is associated with gene silencing. It also plays an important role in developmental processes such as X-chromosome inactivation and genomic imprinting. The methyl moiety of methyl cytosine resides in the major groove of the DNA helix where many DNA-binding proteins make contact with DNA. The methylation, therefore, likely exerts its effect by attracting or repelling various DNA-binding proteins. A family of proteins, known as methyl-CpG binding domain proteins (or MBDs), are attracted to and bind DNA-containing methylated CpG dinucleotides and have been shown to recruit repressor complexes to methylated promoter regions, thereby contributing to transcriptional silencing. Conversely, regions of CpG methylation are known to prevent protein binding of certain transcription factors, thereby preventing transcription.
Certain regions of the genome contain clusters of CpG sequences, termed CpG islands, and are mostly found directly upstream of gene promoters. In general, CpG islands are DNA methylation-free. Certain transcription factors have been discovered to bind to nonmethylated CpG-containing DNA sequences via a CXXC binding domain motif, and contribute to creating a transcriptionally competent chromatin configuration, preventing DNA methylation from occurring at these regions.
Although DNA methylation patterns can be transmitted from cell to cell, they are not permanent. In fact, changes in DNA methylation patterns can occur throughout the life of an individual. Some changes can be a physiological response to environmental changes, whereas others might be associated with a pathological process such as oncogenic transformation or cellular aging. DNA methylation marks can be removed by either an active demethylation mechanism involving a family of DNA hydroxylases called Tet proteins or a passive demethylation process by inhibition of the maintenance methyltransferase, Dnmt1, during cell divisions. DNA methylation patterns fit into an epigenetic framework directly, but also indirectly through their intimate link to other epigenetic mechanisms such as histone lysine methylation and acetylation.
The clinical relevance of DNA methylation first became apparent in relation to cancer. Reduced levels of DNA methylation led to the suppression of some forms of tumors in mouse models of cancer through genetic manipulation or treatment with DNA methyltransferase inhibitors. Conversely, low levels of DNA methylation (referred to as DNA hypomethylation) can enhance the formation of certain tumor types as well. Several other human diseases have been linked to mutations of genes that encode critical components of the DNA methylation machinery. Mutations of the DNA methyltransferase Dnmt3b leads to immune deficiency, whereas mutations of the methyl-CpG binding protein, MeCP2, causes a severe neurological disorder known as Rett syndrome. It is apparent that the integrity of the DNA methylation system is of paramount importance for the health of mammals. Thus, the study of DNA methylation in human disease represents an important frontier in medicine and will contribute to our understanding of the impact of epigenetic modification on human life.
1. A MECHANISM OF CELL MEMORY
1.1. The Hypothesis
Cytosine methylation in mammalian cells occurs predominantly in CpG dinucleotides (Fig. 1). The idea that DNA methylation in animals could represent a mechanism of cell memory arose independently in two laboratories (Holliday and Pugh 1975; Riggs 1975). Recognizing that the CpG dinucleotide is self-complementary, both groups reasoned that patterns of methylated and nonmethylated CpG could be copied when cells divide. Immediately after replication of DNA, the parental DNA strand would maintain its pattern of modified cytosines, but the newly synthesized strand would be unmodified. To insure copying of the parental pattern onto the progeny strand, they postulated a “maintenance methyltransferase” that would exclusively methylate CpGs base-paired with a methylated parental CpG. Nonmethylated CpGs would not be substrates for the maintenance methyltransferase (see Fig. 2). The consequence of this simple mechanism is that patterns of DNA methylation would be replicated semiconservatively like the base sequence of DNA itself.
Figure 1.
Cytosine methylation in DNA. (A) Addition of a methyl group, CH3 (red), at the five position of the cytosine pyrimidine ring (black arrow) does not sterically interfere with GC base pairing (blue lines). DNA methyltransferases associate covalently with the carbon 6 position (straight green arrow) during methyl group transfer. (B) A model of B-form DNA methylated at cytosines in two self-complementary CpG sequences. The paired methyl moieties (magenta and yellow) lie in the major groove of the double helix.
Figure 2.
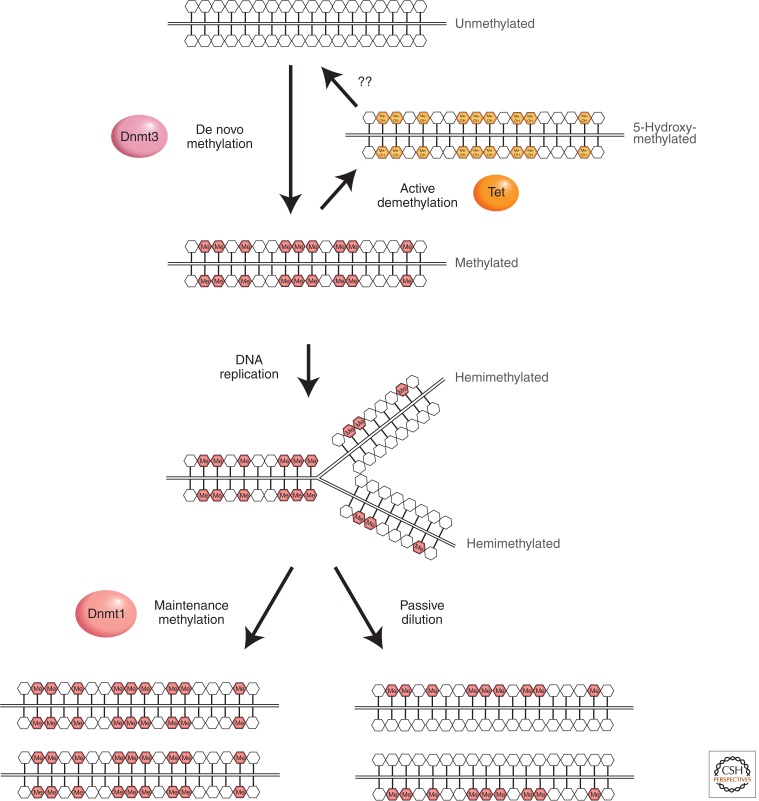
De novo methylation and maintenance methylation of DNA. A stretch of genomic DNA is shown as a line with self-complementary CpG pairs marked as vertical strokes. Unmethylated DNA (top) becomes methylated “de novo” by Dnmt3a and Dnmt3b to give symmetrical methylation at certain CpG pairs. On semiconservative DNA replication, a progeny DNA strand is base-paired with one of the methylated parental strands (the other replication product is not shown). Symmetry is restored by the maintenance DNA methyltransferase, Dnmt1, which completes half-methylated sites, but does not methylate unmodified CpGs.
1.2. Maintenance of DNA Methylation Patterns
The mammalian DNA methyltransferase activity responsible for semiconservatively replicating DNA methylation patterns was detected early on in crude cellular extracts. It was finally purified as a 200-kDa protein (Bestor and Ingram 1983). This enzyme, called Dnmt1 for DNA methyltransferase 1, is specific to CpG and has significant activity against nonmethylated DNA in biochemical assays. Its preferred DNA substrate, however, is DNA methylated at CpG on one of the two strand (so-called hemimethylated DNA; see Fig. 2). Subsequent genetic studies indicated that inactivation of Dnmt1 in mouse embryonic stem cells (Table 1) led to a genome-wide loss of CpG methylation (Li et al. 1992). This evidence fit with the view that Dnmt1 maintains DNA methylation at CpGs by completing hemimethylated sites as originally postulated by Riggs (1975) and Holliday and Pugh (1975) (Fig. 2).
Table 1.
Function of mammalian DNA methyltransferases
| DNA methyltransferase | Species | Major activity | Major phenotypes of loss of function |
|---|---|---|---|
| Dnmt1 | Mouse | Maintenance methylation of CpG | Genome-wide loss of DNA methylation, embryonic lethality at embryonic day 9.5 (E9.5), abnormal expression of imprinted genes, ectopic X-chromosome inactivation, activation of silent retrotransposon. In cancer cell lines, it leads to cell cycle arrest and mitotic defects. |
| Dnmt3a | Mouse | De novo methylation of CpG | Postnatal lethality at 4–8 wk, male sterility, and failure to establish methylation imprints in both male and female germ cells |
| Dnmt3b | Mouse | De novo methylation of CpG | Demethylation of minor satellite DNA, embryonic lethality around E14.5 days with vascular and liver defects. (Embryos lacking both Dnmt3a and Dnmt3b fail to initiate de novo methylation after implantation and die at E9.5.) |
| DNMT3B | Human | De novo methylation of CpG | ICF syndrome: immunodeficiency, centromeric instability, and facial anomalies. Loss of methylation in repetitive elements and pericentromeric heterochromatin. |
2. The ESTABLISHMENT OF DNA METHYLATION PATTERNS
The discovery of the maintenance DNA methyltransferase, Dnmt1, provided a mechanism by which DNA methylation patterns could be maintained through cell generations. This left the problem of determining when in development new DNA methylation patterns are established in an individual and how de novo methylation occurs. This is addressed in this section, as well as describing important work mapping the distribution and methylation patterns of CpG sequences in the mammalian genome and their functional significance.
2.1. De Novo Methylation of DNA in Early Embryos
In the life cycle of an individual, the genome undergoes dynamic changes in DNA methylation during early development. After fertilization, genome-wide demethylation takes place. De novo DNA methylation then occurs around the time of implantation when the inner cell mass cells start to differentiate to form the embryonic ectoderm (Fig. 3).
Figure 3.
Dynamics of 5mC/5hmC/5fC/5caC in paternal and maternal genomes during preimplantation development. DNA demethylation of the zygote, gauged by 5mC levels, occurs by a passive mechanism in the female pronucleus, diluting the marks with the passage of every cell cycle. The male pronuclear genome becomes demethylated actively by the action of the Tet enzymes. Tet3 is expressed in the oocyte and zygote. After fertilization, Tet3 is relocated from the cytoplasm to the paternal nucleus to convert 5mC to 5hmC/5fC/5caC. Subsequently, paternal and maternal genomes undergo replication-dependent dilution of 5hmC/5fC/5caC in males and 5mC in females. It is possible that replication-independent active DNA demethylation may occur in a loci-specific manner in zygotes, but the exact mechanism is currently unclear. DNA methylation patterns in ICM are reestablished by de novo DNA methyltransferases Dnmt3a and Dnmt3b at the blastocyst stage.
De novo DNA methylation was first detected when foreign DNA was introduced into a preimplantation embryo in an unmethylated state, which then became methylated. Reports of both retroviral DNA from infected mouse preimplantation embryos and DNA injected into mouse zygotes showed that the DNA became stably methylated in cells of the animal (Jahner et al. 1982). However, retroviral DNA did not become methylated if infection occurred in embryos at the later stage of gastrulation. This suggested that the process of de novo methylation of DNA is confined to pluripotent cells of early embryos. The hypothesis was further tested using mouse embryonal carcinoma cells and then embryonic stem (ES) cells. When these cells were infected by retroviruses, retroviral DNA became completely methylated and viral genes were consequently silenced (Stewart et al. 1982). Even when the maintenance Dnmt1 gene was deleted, de novo methylation of retroviral DNA in ES cells still occurred proving that other DNA methyltransferases must be at work (Lei et al. 1996). Somatic cells that are infected with viral DNA, however, did not become methylated, again indicating that it is a process that occurs in early development (Stewart et al. 1982).
2.2. Discovery of De Novo Methyltransferases
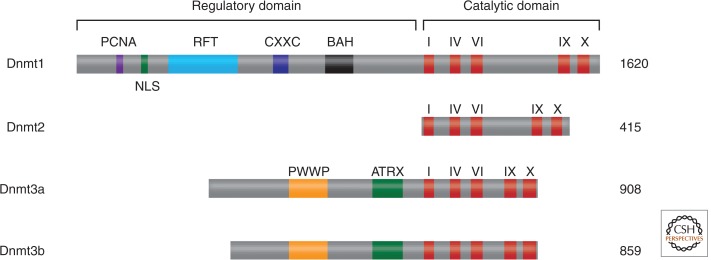
De novo methyltransferases were discovered by searching for sequence homology with prokaryotic cytosine DNA methyltransferases using expressed sequence tag databases. Prokaryotic cytosine DNA methyltransferases share a set of conserved protein motifs (Posfai et al. 1989), and these features were also found in the mammalian maintenance DNA methyltransferase, Dnmt1. The homology search identified three genes that could potentially encode novel DNA methyltransferases (Fig. 4). One candidate protein, Dnmt2, has minimal DNA methyltransferase activity in vitro and its absence has no discernible effect on de novo or maintenance methylation of DNA (Okano et al. 1998b). The other two genes, Dnmt3a and Dnmt3b, encoded related catalytically active polypeptides that showed no preference for methylating hemimethylated DNA in vitro, unlike Dnmt1 (Okano et al. 1998a). Inactivation of both Dnmt3a and Dnmt3b by gene targeting in ES cells confirmed that these genes constituted the missing de novo DNA methyltransferases (Table 1)—ES cells and embryos lacking both proteins were unable to de novo methylate proviral genomes and repetitive elements (Okano et al. 1999). Moreover, Dnmt3a and an associated regulatory factor Dnmt3L were shown to be required for the establishment of distinct DNA methylation patterns found at imprinted genes (Hata et al. 2002; Table 1 in Kaneda et al. 2004; see also Barlow and Bartolomei 2014).
Figure 4.
Mammalian DNA methyltransferases. The catalytic domains of Dnmt1, Dnmt2, and the Dnmt3 family members are conserved (the signature motifs, I, IV, VI, IX, and X, are most conserved in all cytosine methyltransferases), but there is little similarity among their amino-terminal regulatory domains. Domain abbreviations: PCNA, PCNA-interacting domain; NLS, nuclear localization signal; RFT, replication foci-targeting domain; CXXC, a cysteine-rich domain implicated in binding DNA sequences containing CpG dinucleotides; BAH, bromo-adjacent homology domain implicated in protein–protein interactions; PWWP, a domain containing a highly conserved “proline-tryptophan-tryptophan-proline” motif involved in heterochromatin association; ATRX, an ATRX-related cysteine-rich region containing a C2-C2 zinc finger and an atypical PHD domain implicated in protein–protein interactions.
2.3. CpG Islands and Patterns of DNA Methylation across the Genome
DNA across the genome of mammalian somatic tissues is methylated at 70%–80% of all CpG sites. With the continual advances in technology there has been a constant refinement in the level of detail with which we can measure DNA methylation at single CpG site resolution in any given cell type. Mapping studies (see Box 1) indicate that highly methylated sequences include satellite DNAs, repetitive elements (including transposons and their inert relics), nonrepetitive intergenic DNA, and exons of genes. Most sequences are methylated according to their frequency of CpG dinucleotides. Key exceptions to this global methylation of the mammalian genome are the CpG islands (CGIs).
BOX 1. MAPPING DNA METHYLATION.
To understand the functions of DNA methylation, it is first necessary to map methyl CpG in the genome and its dynamic changes during cell proliferation and differentiation or in development and disease. Several methods have been developed for quantitative analysis of DNA methylation at the genome scale as well as in a gene locus-specific manner.
Bisulfite-sequencing (Frommer et al. 1992) This is the most reliable method for testing all cytosines within a region of the genome. It involves the “bisulfite modification” of single-stranded DNA, which leads to deamination of unmodified cytosines, whereas 5-methylcytosine is protected. As a result, cytosines that survive bisulfite treatment are identified as methylated. Because of its high resolution and positive identification of methylated cytosine, this is the method of choice for analyzing DNA methylation patterns. In combination with next-generation sequencing, the method is widely used for methylome analysis at the whole-genome scale. Several polymerase chain reaction (PCR)-based methods that depend on prior bisulfite treatment of DNA have also been developed for rapid analysis of methylation of genes of interest (Herman et al. 1996).
MeDIP (Weber et al. 2005) Methylated DNA immunoprecipitation (MeDIP) is a versatile approach for unbiased detection of methylated DNA and can be applied to generate comprehensive DNA methylation profiles on a genome-wide scale. This method uses a monoclonal antibody that specifically recognizes 5-methylcytidine to enrich methylated genomic DNA fragments by immunoprecipitation. The methylation status is then determined by PCR for specific regions or by using DNA microarrays for the whole genome.
Pyrosequencing (Tost and Gut 2007) Analysis of DNA methylation patterns by pyrosequencing yields reproducible and accurate measures of the degree of methylation at several CpGs in close proximity with high resolution. The method is highly sensitive and quantitative, and it is often applied to methylation analysis of specific regions.
CHARM DNA methylation analysis (Irizarry et al. 2009) Comprehensive high-throughput array-based relative methylation (CHARM) analysis is a microarray-based method. It can be applied to custom-designed microarray covering the whole genome (usually nonrepetitive sequences) or specific regions (e.g., all CGIs). The method is quantitative and data analysis is straightforward. It has an advantage over other methods when it comes to comparing DNA methylation patterns from a large number of samples.
-
A few commercially available technologies
Illumina methylation chip http://www.illumina.com/products/methylation_450_beadchipkits.ilmn
Sequenom EpiTYPER http://www.sequenom.com/Sites/Genetic-Analysis/Applications/DNA-Methylation
CGIs are GC-rich sequences of ∼1 kb in length that are nonmethylated in germ cells, the early embryo, and most somatic tissues (Fig. 5) (Bird et al. 1985). It was in early mapping studies of individual gene promoters that these GC-rich regions were identified (McKeon et al. 1982). It is now evident that most (if not all) CGIs mark the promoters and 5′ regions of genes. In fact, ∼60% of human genes have CGI promoters.
Figure 5.
CpG islands. (A) CGIs are regions of high CpG density (>50%), usually 200 bp–2 kb in length that lack CpG methylation, found at promoters of most human genes. Long-term silencing of the gene can be insured by methylation of the CGI region. For example, genes on the inactive X chromosome and certain imprinted genes are silenced in this way. Also, in cancer cells certain genes are aberrantly silenced by CGI methylation. Shores are regions of the genome that reside up to 2 kb from CGIs, whereas shelves are found 2–4 kb away from CGIs. (B) Chromatin immunoprecipation (IP) analysis of Kdm2b binding sites shows that Kdm2b is enriched at the CpGs of the Hox locus in which the unmethylated CGIs (green bars) are located. (C) Cfp1, Kdm2a, and Kdm2b proteins share a common CXXC domain that binds specifically to unmethylated CpG sites. Protein length is indicated to the right of each protein. Abbreviations for other domains include PHD, plant homeodomain; A, acidic domain; B, basic domain; S, Set1 interacting domain; C, coiled coil domain; LRR, leucine-rich repeat domain.
Genes with CGI promoters that are expressed in a tissue-specific manner are usually expressed in early embryos and then in the soma. A distinct pattern of DNA methylation is found on the inactive X chromosome in females where, contrary to the norm, CGIs become de novo methylated in large numbers during the embryonic process of X-chromosome inactivation in female placental mammals (Wolf et al. 1984). This process is essential for the leak-proof silencing of genes on the inactivated chromosome necessary for dosage compensation, as DNA methylation-deficient mice or cells show frequent transcriptional reactivation of X-linked genes (see also Brockdorff and Turner 2014).
Studies of DNA methylation patterns have focused on the question of how gene expression is regulated by DNA methylation. CGIs normally remain unmethylated and multiple mechanisms are involved in protecting CGIs from de novo methylation by Dnmt3a and Dnmt3b. The discovery that two proteins containing a DNA-binding CXXC domain—Cfp1 and Kdm2a (Fig. 5C)—are able to bind specifically to unmethylated CpGs within CGIs (Blackledge et al. 2010, 2013; Thomson et al. 2010) has provided an inroad to understanding how unmethylated DNA can contribute to creating a transcriptionally competent chromatin environment as well as protecting it from de novo DNA methylation (see Section 5.3 and Deaton and Bird 2011; Cheng 2014).
The increasing resolution with which the methylome of particular cell types is being analyzed is beginning to reveal patterns of DNA methylation associated with normal development versus disease states such as cancer and aging. The detail has resulted in the categorization of different types of CGIs—that is, CGIs containing a transcriptional start site versus orphan CGIs that can be located intra- or intergenically. Studies have also revealed other differentially methylated regions termed shores (up to 2 kb away from a CGI) and shelves (located within 2–4 kb of a CGI; Fig. 5A) (Irizarry et al. 2009). One study proposes that the DNA methylation status of its first exon is a better indicator of transcriptional repression than its CGI (Brenet et al. 2011). The relevance these categories of CpG-containing regions of the genome have with the chromatin environment, transcriptional control and repression, and disease development or association requires further analyses.
3. DNA DEMETHYLATION
Developmental biologists have described the waves of genome-wide DNA demethylation that occur in the germline and in early embryogenesis (Fig. 3), however, the process by which DNA methylation is erased has been elusive. Recent discoveries, described in Secs. 3.2 and 3.3, have advanced our understanding of the DNA demethylation process.
3.1. Active and Passive Demethylation during Development
The mammalian genome is reprogrammed during early development by both active and passive DNA demethylation processes (Wu and Zhang 2010). Active DNA demethylation refers to an enzymatic process that results in the removal of the methyl-group from 5mC. In contrast, passive DNA demethylation refers to the lack of maintenance methylation during successive rounds of DNA replication either in the absence of Dnmt1 or because of its inhibition (Fig. 2). Immunostaining using an anti-5mC antibody initially showed that the 5mC levels of the maternal genome went through a replication-dependent dilution process (i.e., passive demethylation) during preimplantation development (Rougier et al. 1998). In contrast, the 5mC levels of the paternal genome dramatically decrease a few hours after fertilization (Fig. 3) (Mayer et al. 2000). Bisulfite sequencing confirmed that some of the repeat sequences, but not imprinted genes, of the paternal genomes were indeed demethylated (Oswald et al. 2000). Given that no DNA replication occurs during this period, the loss of 5mC in the paternal genome is considered “active.”
Another place where global loss of 5mC is observed is in the primordial germ cells (PGCs). At embryonic stage E7.5, a subset of posterior epiblast cells is specified to become PGCs. At the beginning of their specification as well as during migration toward the genital ridge, PGCs are believed to have the same epigenetic marks as other epiblast cells. However, by the time they arrive at the genital ridge at E11.5 many of the epigenetic marks including DNA methylation have been erased (Hajkova et al. 2002; Yamazaki et al. 2003). Because PGCs have undergone several cell cycles in the presence of Dnmt1 during this process, the loss of DNA methylation is likely to be active. Also, because DNA methylation of the imprinted genes is also erased during this period, it is believed that DNA methylation patterns are reset during germ cell development (Sasaki and Matsui 2008).
Active DNA demethylation has been reported in somatic cells in a locus-specific manner. For example, activated T lymphocytes undergo active demethylation at the interleukin-2 promoter-enhancer region in the absence of DNA replication within 20 min of stimulation (Bruniquel and Schwartz 2003). In addition, locus-specific demethylation occurs at the promoter of brain-derived neurotrophic factor (Bdnf) in depolarized neurons (Martinowich et al. 2003), and for other loci during nuclear hormone regulated gene activation (Kangaspeska et al. 2008; Metivier et al. 2008). Furthermore, locus-specific DNA demethylation occurs at the Oct4 and Nanog promoters when mouse ES cells are fused with human fibroblasts (Bhutani et al. 2010). Because no DNA replication takes place in the processes described above, active DNA demethylation is believed to be responsible for the loss of DNA methylation.
3.2. Tet-Mediated 5mC Oxidation
The observations described above have prompted the hunt for putative DNA demethylases. Most of the early studies were inconclusive in identifying the putative enzyme(s) or elucidating the mechanism of demethylation (Ooi and Bestor 2008; Wu and Zhang 2010). This situation changed with the identification of 5-hydroxymethylcytosine (5hmC) as a bona fide base of mammalian genomic DNA (Kriaucionis and Heintz 2009; Tahiliani et al. 2009) and the demonstration that Tet proteins are responsible for the conversion of 5mC to 5hmC (Fig. 6) (Tahiliani et al. 2009; Ito et al. 2010). This initial discovery is described in the article by Skirmantas and Tahiliana (this collection).
Figure 6.
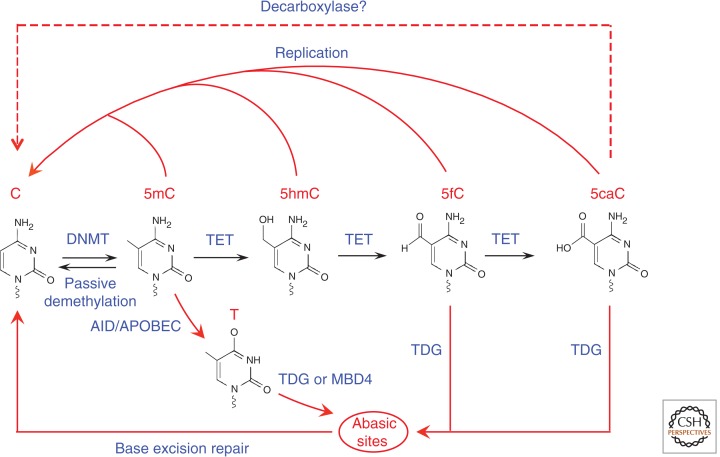
Model of Tet-initiated DNA demethylation pathways. DNA methylation (5mC) is established and maintained by DNMT. 5mC can by oxidized by Tet family of dioxygenases to generate 5hmC, 5fC, and 5caC. Because the oxidized 5mC derivatives cannot serve as substrates for DNMT1, they can be lost by replication-dependent passive demethylation. 5hmC can be deaminated by AID/APOBEC to become 5hmU, which together with 5fC and 5caC can be excised by glycosylases such as TDG, followed by DNA repair to generate C. Alternatively, a putative decarboxylase may convert 5caC to C.
Using thin layer chromatography and mass spectrometry assays, it was found that 5hmC is relatively abundant in the Purkinje neurons and mouse ES cells (Kriaucionis and Heintz 2009; Tahiliani et al. 2009). Importantly, the human TET1 protein was shown to be able to convert 5mC to 5hmC in an iron and 2-oxoglutarate-dependent manner (Tahiliani et al. 2009). This enzymatic activity is conserved in all three Tet family proteins (Fig. 7) (Ito et al. 2010). Based on the similar chemistry between Tet-catalyzed 5mC oxidation and thymine hydroxylase-catalyzed thymine oxidation, it was proposed that Tet-mediated 5mC oxidation should be able to proceed further to generate 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC) (Fig. 6) (Wu and Zhang 2010). This proposed activity was experimentally shown both in vitro and in vivo (He et al. 2011; Ito et al. 2011). Mass spectrometry analysis has indicated that 5hmC and 5fC are broadly present in the genomic DNA of various tissues and cell types (Ito et al. 2011; Pfaffeneder et al. 2011), whereas the presence of 5caC appears to be more limited (He et al. 2011; Ito et al. 2011). Collectively, Tet proteins can oxidize 5mC not only to 5hmC, but also to 5fC and 5caC.
Figure 7.
Domain structure of the mouse Tet family proteins. Schematic diagrams of predicted conserved domain structures in the three mouse Tet proteins. The conserved domains include CXXC zinc-binding domain, the cysteine-rich domain, and the double-stranded β-helix (DSBH) found in members of the dioxygenase superfamily proteins. Both the Cys-rich and the DSBH domains have been shown to be critical for enzymatic activity. Numbers indicate amino acid numbers.
3.3. Tet-Mediated DNA Demethylation
Tet-mediated iterative oxidation of 5mC described in Sec. 3.2 (Fig. 6), may contribute to the dynamic changes in global or locus-specific levels of 5mC (Wu and Zhang 2011). The conversion of 5mC to 5hmC is expected to prohibit maintenance of existing DNA methylation patterns given that 5hmC is not recognized by Dnmt1 during DNA replication (Valinluck and Sowers 2007), leading to passive DNA demethylation during cell division. In addition, the 5mC oxidation products 5fC and 5caC may serve as intermediates for active DNA demethylation as both can be cleaved by the thymine-DNA glycosylase (TDG) (He et al. 2011; Maiti and Drohat 2011). These studies have raised the possibility that Tet-mediated 5mC oxidation followed by TDG-mediated excision of 5fC/5caC and base excision repair (BER) might be one of the pathways for active DNA demethylation (Fig. 6). Consistent with this, disruption of the Tdg gene in mouse results in increased DNA methylation at certain genomic loci (Cortazar et al. 2011; Cortellino et al. 2011). In plants, a similar glycosylase-initiated BER mechanism is responsible for DNA demethylation through members of the repressor of silencing 1 (ROS1)/Demeter family of 5mC glycosylases (Zhu 2009). More studies are required to ascertain which in vivo mechanisms of active or passive DNA demethylation are at work at the different loci and genomic regions, in different cell types, and at different stages of development. A start has been made in establishing the players involved in early embryonic DNA demethylation as described in Sec. 4.4.
3.4. Tet-Mediated Demethylation in Zygotes and Preimplantation Embryos
The discovery that Tet proteins are biochemically able to oxidize 5mC made it possible to test which protein may be responsible for the specific loss of 5mC in the paternal genome in zygotes. Immunostaining revealed that the decrease in 5mC staining in the paternal pronucleus in zygotes coincides with appearance of 5hmC/5fC/5caC (Gu et al. 2011; Inoue et al. 2011; Iqbal et al. 2011; Wossidlo et al. 2011). Studies showed that Tet3 is highly expressed in zygotes, and its small interfering RNA-mediated knockdown or targeted deletion abolishes 5mC oxidation, supporting the notion that Tet3 is responsible for 5mC oxidation in the zygotes (Gu et al. 2011; Wossidlo et al. 2011).
The asymmetric appearance of 5hmC in the sperm-derived chromosomes persists into two-cell-stage embryos, and gradually decreases until the morula stage (Inoue and Zhang 2011). This suggests that 5hmC is not rapidly removed at a genome-wide scale, but rather lost by “passive” replication-dependent dilution. Similar results have also been obtained for 5fC and 5caC (Inoue et al. 2011). Therefore, the dynamic changes in 5mC during preimplantation appear to involve both “passive” and “active” processes: DNA methylation in the maternal genome is “passively” diluted through replication, and 5mC in the paternal genome first goes through a Tet3-mediated oxidation process in zygotes followed by a “passive” replication-dependent dilution process (Fig. 3). The biological significance of the Tet3-catalyzed oxidation in zygotes is currently unknown, and it remains to be seen whether individual loci conform to this general trend.
4. REGULATION OF GENE EXPRESSION BY DNA METHYLATION
DNA methylation of gene promoter regions is associated with transcription repression. Studies of the effects of DNA methylation in mammalian cells became possible with the discovery that the nucleoside analog 5-azacytidine could inhibit DNA methylation in living cells (Jones and Taylor 1980). 5-azacytidine is incorporated into DNA in place of cytidine and forms a covalent adduct with DNA methyltransferases preventing further DNA methylation. Silencing of several genes, including viral genomes (Harbers et al. 1981) and genes on the inactive X chromosome (Wolf et al. 1984), had previously been shown to correlate with their methylation. The ability of 5-azacytidine treatment to restore their expression (Mohandas et al. 1981) argued that DNA methylation played a causal role in their repression. This was later corroborated by genetic analysis using Dnmt1 knockout mice, demonstrating that inactivation of Dnmt1 resulted in genome-wide loss of DNA methylation and activation of inactive X silenced genes, viral genes, and imprinted genes such as H19 and IGF2 (Li et al. 1993).
4.1. Interference with Transcription Factor Binding
How does DNA methylation interfere with gene expression? One obvious possibility is that the presence of methyl groups in the major groove (see Fig. 1) interferes with the binding of transcription factors that activate transcription from a specific gene. A number of transcription factors recognize GC-rich sequence motifs that can contain CpG sequences. Several of these are unable to bind DNA when the CpG sequence is methylated (Watt and Molloy 1988). Proof that this mechanism operates in gene regulation comes from studies of the role of the CTCF protein in imprinting at the H19/Igf2 locus in mice (Bell and Felsenfeld 2000). CTCF is associated with transcriptional domain boundaries (Bell et al. 1999) and can insulate a promoter from the influence of remote enhancers. The maternally derived copy of the Igf2 gene is silent owing to the binding of CTCF between its promoter and a downstream enhancer. At the paternal locus, however, the CpG-rich CTCF binding sites are methylated, preventing CTCF binding and thereby allowing the downstream enhancer to activate Igf2 expression. Although there is evidence that H19/Igf2 imprinting involves additional processes, the role of CTCF represents a clear example of transcriptional regulation by DNA methylation (for more details, see Barlow and Bartolomei 2014).
A recent study revealed that DNA-binding factors can also shape DNA methylation patterns. Whole genome bisulphite sequencing analysis of embryonic stem cells and neuronal progenitors revealed the existence of low-methylated regions (LMRs) at CpG-poor distal regulatory regions (Stadler et al. 2011). Interestingly, LMRs are occupied by transcription factors and their binding is both necessary and sufficient to generate LMRs, indicating transcription factors can influence local DNA methylation.
4.2. Recruitment of Methyl-CpG Binding Proteins and Repressor Complexes
The second mode of repression is opposite to the first as it involves proteins that are attracted to rather than repelled by methyl-CpG (Fig. 8). Evidence for such a mechanism came initially from the identification of the methyl-CpG binding protein complex, MeCP1, and subsequent purification and cloning of MeCP2 (Meehan et al. 1989). Proteins with DNA-binding motifs related to that of MeCP2 were identified using database searches, and designated the methyl-CpG binding domain (MBD) family comprising MeCP2, MBD1, MBD2, MBD3, and MBD4 (Bird and Wolffe 1999). Three of the MBD proteins, MBD1, MBD2, and MeCP2, have been implicated in methylation-dependent repression of transcription (Table 2) (Bird and Wolffe 1999). An unrelated protein, Kaiso, has also been shown to bind methylated DNA and bring about methylation-dependent repression in model systems (Table 2) (Prokhortchouk et al. 2001; Yoon et al. 2003).
Figure 8.
Proteins that bind methyl-CpG. Five members of the MBD protein family are aligned at their MBD domains (purple). Other domains are labeled and include TRD; CXXC domains, which are zinc fingers, some of which are implicated in binding to nonmethylated CpG; GR repeats that may bind; a T:G mismatch glycosylase domain that is involved in repair of 5mC deamination. Kaiso lacks the MBD domain, but binds methylated DNA via zinc fingers (orange) and possesses a POB/BTB domain that is shared with other transcriptional repressors. Domain abbreviations: MBD, methyl-CpG binding domain; TRD, transcriptional repression domain; POZ, poxvirus and zinc finger, a protein–protein interacting domain.
Table 2.
Functions of methyl-CpG binding proteins
| MBP | Major activity | Species | Major phenotypes of loss-of-function mutations |
|---|---|---|---|
| MeCP2 | Binds mCpG with adjacent run AT-rich run Transcriptional repressor | Mouse | Delayed onset neurological defects including inertia, hindlimb clasping, nonrhythmic breathing, and abnormal gait. Postnatal survival ∼10 wk. |
| MECP2 | Binds mCpG with adjacent AT run Transcriptional repressor | Human | Heterozygotes suffer from Rett syndrome, a profound neurological disorder characterized by apraxia, loss of purposeful hand use, breathing irregularities, and microcephaly |
| Mbd1 | Binds mCpG via MBD; a major splice form is also able to bind CpG via a CxxC domain | Mouse | No overt phenotype, but subtle defects in neurogenesis detected |
| Mbd2 | Binds mCpG Transcriptional repressor | Mouse | Viable and fertile, but show reduced maternal nurturing behavior. Defective gene regulation in T-helper cell differentiation leading to altered response to infection. Highly resistant to intestinal tumorigenesis. |
| Mbd3 | Core component of NuRD corepressor complex Does not show strong binding to mCpG | Mouse | Early embryonic lethal |
| Mbd4 | DNA repair protein that binds mCpG and T:G mismatches at mCpG sitesThymine DNA glycosylase that excises T from T:G mismatches | Mouse | Viable and fertile. three- to fourfold increase in mutations at CpG sites. Increased susceptibility to intestinal cancer correlates with C to T transitions within the Apc gene. Mbd4 functions to minimize the mutability of 5-methylcytosine. |
| Kaiso | Binds mCGmCG and CTGCNA Transcriptional repressor | Mouse | No overt phenotype. Small but significant delay in tumorigenesis on Min background. |
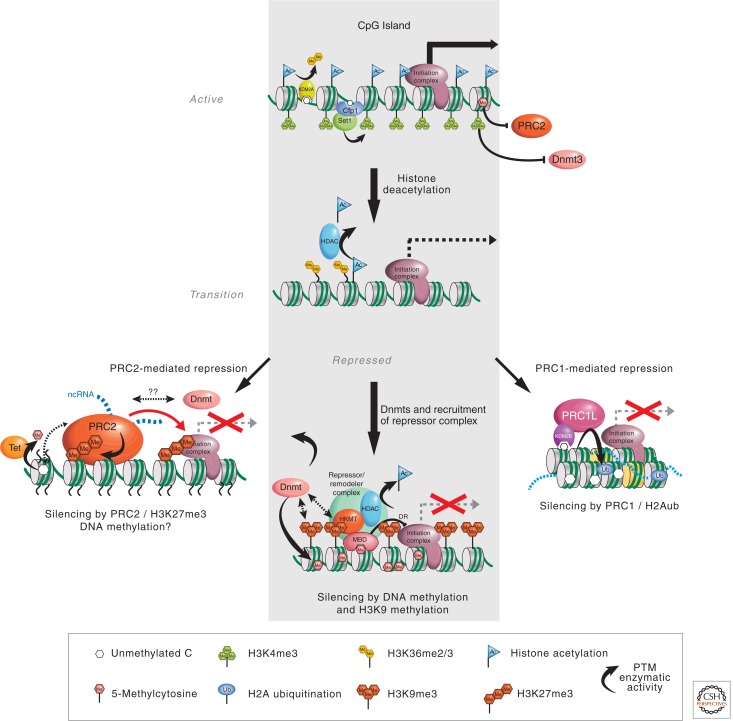
MeCP2 associates with the mSin3a corepressor complex and depends on histone deacetylation for its action (Jones et al. 1998; Nan et al. 1998). This finding showed that DNA methylation can be read by MeCP2, and provides a signal to alter chromatin structure (Fig. 9). Each of the four methyl-CpG binding proteins has since been shown to associate with different corepressor complexes. Of particular interest is MBD1, which associates with the histone lysine methyltransferase SETDB1 only during DNA replication (Sarraf and Stancheva 2004). This may ensure continued histone H3K9 methylation at chromosomal MBD1 target sequences for stable silencing of the associated genes.
Figure 9.
Recruitment of corepressors by methyl-CpG binding proteins. A hypothetical transition between an active, nonmethylated gene promoter and a repressed promoter whose silence is attributable to DNA methylation, as mediated by complexes containing an MBD protein such as MeCP2 (gray shading). The transition phase represents an intermediate step during which transcription is silenced and DNA methylation occurs. MeCP2 is envisaged to recruit the NCoR histone deacetylase (HDAC) complex and histone lysine methyltransferase (HKMT) activity to the methylated sites. In addition, there is some evidence that MeCP2 can directly repress (DR) transcription by contact with the transcription initiation complex. Other methyl-CpG binding proteins can also interact with and potentially recruit distinct corepressor complexes that include HKMT and/or HDAC activity. PRC1 and PRC2 are also involved in silencing gene expression through histone H3K27 methylation, catalyzed by PRC2 (left outcome). One of the mechanisms by which they function together to regulate the same set of target genes is through the recognition of the H3K27me3 mark by the chromodomain protein in the PRC1 complex. PRC1 can also be recruited by the CxxC domain–containing protein Kdm2b to effect gene silencing (right outcome). Proteins with known histone posttranslational modifying (PTM) activity are indicated.
MBD2 is the DNA-binding component of MeCP1, which was initially implicated as a transcriptional repressor in cellular extracts (Meehan et al. 1989; Boyes and Bird 1991). MeCP1 is a large multiprotein complex that includes the NuRD (or Mi-2) corepressor complex and MBD2 (Wade et al. 1999; Feng and Zhang 2001). NuRD comprises histone deacetylases (HDAC) and a large chromatin remodeling protein (Mi-2) (Zhang et al. 1998). NuRD can be recruited to DNA by several DNA-binding proteins other than MBD2. Cells that lack MBD2 are unable to effectively repress methylated reporter constructs, despite the presence of other methyl-CpG binding proteins in these cells, arguing that it is an important component of the repression system (Hendrich et al. 2001). MBD2-deficient mice are viable and fertile, although they have a defect in maternal behavior (Hendrich et al. 2001), and careful examination has revealed aberrations in tissue-specific gene expression. For example, expression of the interleukin-4 and interferon-γ genes during T-helper cell differentiation is significantly disrupted (Hutchins et al. 2002).
5. INTERACTIONS BETWEEN DNA METHYLATION AND HISTONE MODIFICATIONS
One of the important questions in the field is how DNA methylation coordinates with other modifications in chromatin to regulate gene expression and other chromatin-based processes. Given that DNA and histones are integral components of chromatin, it is not surprising that DNA methylation interacts with histone modifications, as introduced in Section 4. Recent studies have revealed that interactions between DNA methylation and histone modifications play an important role in regulating chromatin dynamics and various biological processes (Cedar and Bergman 2009). Because histone modifications are highly conserved in evolution from yeast to mammals, whereas DNA methylation is not, the interaction between histone modifications, chromatin-associating factors, and DNA methylation may reveal new epigenetic mechanisms that are unique to vertebrates or mammals.
5.1. DNA Methylation and Histone Deacetylation
Previous studies have established that promoter DNA methylation is generally linked to transcriptional repression whereas histone acetylation is generally linked to transcriptional activation. Consequently, DNA methylation inversely correlates with histone acetylation. Consistent with this general trend, MBD proteins that recognize and bind to methylated DNA sequences are mostly associated with HDACs (Fig. 9) (Bird and Wolffe 1999). For example, the first identified MBD protein, MeCP2 associates with the Sin3A HDAC complex and is believed to be responsible for mediating transcriptional silencing by DNA methylation (Jones et al. 1998; Nan et al. 1998). In addition, the MBD protein, MBD2, associates with the nucleosome remodeling and histone deacetylase (NuRD) corepressor complex (Feng and Zhang 2001), and is believed to be the MeCP1 transcriptional repressor initially identified in cellular extracts (Meehan et al. 1989; Boyes and Bird 1991). Another example comes from a report demonstrating that interaction between MBD1 and HDAC3 mediates the establishment and maintenance of a silenced chromatin state directed by the chimeric PML-RARa gene, which functions as a constitutive transcriptional repressor in cancer (Villa et al. 2006).
5.2. DNA Methylation and H3K9 Methylation
Both DNA methylation and H3K9 methylation are associated with stable gene silencing, which is observed mainly in heterochromatin. Heterochromatin encompasses transcriptionally inert genomic regions that are enriched with repeat sequences and retrotransposons. It is believed that one of the major functions of DNA methylation is the silencing retrotransposons to maintain genome integrity (Bestor and Bourc’his 2004). Given that both H3K9 methylation and DNA methylation are features of heterochromatin, several studies have focused on the relationship between DNA methylation and H3K9 methylation. The first evidence that links DNA methylation to H3K9 methylation came from genetic studies in the fungus Neurospora crassa, in which deletion of the H3K9 methyltransferase Dim5 resulted in a complete loss of DNA methylation (Tamaru and Selker 2001; Aramayo and Selker 2013). However, the relationship between DNA methylation and H3K9 methylation is more complex in mammals. Absence of the H3K9me3 methyltransferase, Suv39h1/Suv39h2, does not lead to a complete loss of DNA methylation, but results in reduction of DNA methylation in heterochromatic repeat sequences (Lehnertz et al. 2003). Other studies showed that the H3K9me2-specific methyltransferase G9a/GLP interacts directly with Dnmt1 (Esteve et al. 2006) and is required for de novo DNA methylation of retrotransposons in ES cells (Dong et al. 2008). The UHRF1 (ubiquitin-like PHD and RING finger domain-containing 1) protein further links DNA methylation to H3K9 methylation by binding directly to methylated H3K9 to maintain the stability of DNMT1 (Rothbart et al. 2012). Furthermore, MPP8, the H3K9me-binding protein, can mediate the interaction between G9a/GLP and Dnmt3a (Chang et al. 2011). Finally, a third H3K9 methyltransferase SETDB1 was reported to interact with Dnmt3a (Li et al. 2006). Therefore, multiple H3K9 methyltransferases can directly or indirectly associate with DNA methyltransferases to coordinate H3K9 methylation with DNA methylation.
5.3. H3K4 Methylation Inhibits Promoter CGI Methylation
A key question in the DNA methylation field has been how a CGI is protected from DNA methylation when most CpGs are methylated. The discovery of two proteins—Cfp1 and Kdm2a—that bind specifically to nonmethylated CpGs via the CXXC domain (Fig. 5) has brought us closer to understanding the mechanism. A feature of active gene promoters is its enrichment of the H3K4me3 mark (Guenther et al. 2007). An enzyme that writes this mark, Setd1, a member of the MLL family of H3K4 methyltransferases, has been shown to be recruited to CGIs in neuronal progenitor cells, through its interaction with the CpG binding protein Cfp1 (Thomson et al. 2010). Cfp1 binds only unmethylated CpG sequences found in CGIs through its CXXC domain. The enrichment of H3K4me3 at the CGIs, through the action of Setd1, is likely to be one of the reasons that prevent CGIs from being methylated. Conversely, unmethylated H3K4 enhances the access of the de novo DNA methylation enzymes, Dnm3a/Dnmt3b, to CGIs (Fig. 9). As Dnmt3a and Dnmt3b function in complex with Dnmt3L (Ooi et al. 2007), it was interesting to note that Dnmt3L selectively recognizes nucleosomes that lack H3K4 methylation (elaborated in Cheng 2014). Another factor contributing to the protection of CGIs from DNA methylation is an enrichment of Tet1 (Williams et al. 2011; Wu et al. 2011). Given that Tet1 has the capacity to oxidize 5mC to 5hmC and 5fC/5caC, and the latter can be processed by TDG followed by base excision repair (Fig. 6), any “accidental” methylation at CGIs could thus be potentially removed.
5.4. DNA Methylation and H3K27 Methylation
The first link between DNA methylation and H3K27 methylation was suggested by the demonstration that EZH2 can interact with all three DNMTs in vitro (Vire et al. 2006). However, subsequent studies in cancer cells revealed that gene silencing by H3K27me3 is independent of promoter DNA methylation (Kondo et al. 2008). Genome-wide location studies in ES cells have revealed that CpG-rich sequences are indeed enriched for the H3K27 methyltransferase PRC2 (Mendenhall et al. 2010) indicating that H3K27me3 enriched promoters are usually devoid of DNA methylation. However, many promoters marked by H3K27me3 in ES cells become silenced and DNA methylated during differentiation, suggesting a cell type–dependent cross talk between H3K27 methylation and DNA methylation (Fig. 9) (Mohn et al. 2008). In addition to cell types, the relationship between H3K27 methylation and DNA methylation can also be affected by the location of the modifications (Wu et al. 2010).
5.5. DNA Methylation and ATP-Dependent Chromatin Remodeling
In addition to histone modifications, ATP-dependent nucleosome remodelers have been linked to DNA methylation. Evidence that ATP-dependent remodeling factors are needed to ensure appropriate DNA methylation came initially from research in plants, in which the SNF2-like protein DDM1 was shown to be essential for full DNA methylation of the Arabidopsis thaliana genome (Jeddeloh et al. 1999). An equivalent dependence is seen in animals as mutations in human ATRX (Gibbons et al. 2000) and the mouse Lsh2 genes (Dennis et al. 2001), both of which encode relatives of the chromatin remodeling protein SNF2, have significant effects on global DNA methylation patterns. Loss of the LSH2 protein, in particular, matches the phenotype of the DDM1 mutation in Arabidopsis, in which methylation of highly repetitive DNA sequences is lost, although some DNA methylation is retained elsewhere in the genome. Perhaps efficient global DNA methylation of the genome requires conformational changes in chromatin structure by these chromatin remodeling factors so that DNMTs can gain access to the DNA. Collaboration between DNMTs and chromatin remodeling factors that allow them access may be particularly important in regions that are “heterochromatic” and inaccessible (Fig. 9).
6. DNA METHYLATION AND DISEASES
DNA methylation plays an important role in regulating tissue-specific gene expression and the repression of viral gene expression. It is also involved in the establishment and maintenance of genomic imprinting and X inactivation, and in the regulation of chromosome stability. Alterations in DNA methylation have been associated with cancer and several other diseases.
6.1. Methyl CpG as Hot Spots for Mutation
The disadvantage of having DNA methylation as an epigenetic system of cellular memory is the mutability of 5mC. Cytosine (C) deaminates spontaneously to give uracil (U), which is then mispaired with guanine. This potential mutation is recognized by uracil DNA glycosylases, which efficiently remove the inappropriate base and initiate repair to restore C in place of U. When 5mC deaminates, however, thymine (T) is formed. This also results in a mismatch, but the fact that T, unlike U, is a natural DNA base appears to interfere with the efficient repair of the lesion. As a result, the mutant thymine base can persist through DNA replication and is passed on to progeny cells as a C to T transition mutation. Mutations of this kind appear to be one of the most frequent single causes of genetic disease in humans, as approximately one third of all point mutations are C to T transitions at CpG sequences (Cooper and Youssoufian 1988). The instability of CpG over evolutionary time is further shown by the four- to fivefold underrepresentation of CpG in the mammalian genome (Bird 1980). The only exceptions are CGIs, within which CpGs are nonmethylated and therefore stable.
MBD4 is so far unique among methyl-CpG binding proteins in that it has enzymatic activity. The MBD4 carboxy-terminal domain is a thymine DNA glycosylase that can selectively remove T from a T-G mismatch in vitro (Hendrich et al. 1999). This activity would be expected of a DNA repair system that corrects 5mC deamination. Confirming this hypothesis, mice lacking MBD4 show enhanced mutability of methylated cytosine residues at a chromosomal reporter sequence (Millar et al. 2002). In addition, Mbd4-null mice acquire C to T transition mutations within the adenomatous polyposis coli gene and have an increased frequency of intestinal tumorigenesis (Table 2). It is noteworthy that, in spite of the existence of a dedicated repair system, sites of cytosine methylation persist as hot spots for mutation.
6.2. Methylation of Promoter CGIs
Aberrant methylation of promoter CGIs can lead to silencing of gene expression and disease. Inactivation of growth inhibitory genes (and pathways) by methylation of promoter CGIs is the most common epigenetic mechanism contributing to cancer. What triggers aberrant methylation of promoter CGIs remains largely unknown (for further discussion, see Jones and Baylin 2014).
Methylation of the CGI at the 5′ region of the FMR1 gene is shown to be the cause of fragile X syndrome (FXS). FXS is associated with triplet repeat expansion in the 5′ region of the FMR1 gene. Normal individuals have less than 50 CGG repeats, whereas the FXS patients have more than 200 CGG repeats. The genetic change caused by repeat expansion to more than 200 copies leads to de novo methylation of the CGIs and silencing of the FMR1 gene (Peprah 2012). In rare cases, individuals with more than 200 triplet repeats but an unmethylated CGI are completely normal. This indicates that promoter CGI methylation induced by CGG repeat expansion causes the disease and can be stably inherited (Peprah 2012).
6.3. DNA Hypomethylation and Chromosome Instability
Although DNA methylation is clearly mutagenic, there is evidence that its presence is beneficial with respect to chromosomal stability. Mice possessing ∼10% of normal levels of DNA methylation, caused by a hypomorphic mutation of Dnmt1, acquire aggressive T cell lymphomas that often display trisomy of chromosome 15 (Gaudet et al. 2003). Mutations of DNMT3B in patients with ICF syndrome or inactivation of Dnmt3b in mice lead to various chromosomal aberrations including chromosome fusion, breakage, and aneuploidy (Ehrlich 2003; Dodge et al. 2005). These results are of interest because cancers often display reduced levels of DNA methylation, which may contribute to tumor initiation or progression (see also Jones and Baylin 2014). One possible explanation for the result is that DNA methylation contributes to accurate chromosome segregation, and in its absence it is more frequent to have nondisjunction leading to chromosome aberrations. Alternatively, DNA methylation may suppress the expression and recombination of retrotransposons in the mammalian genome thereby protecting chromosomes from deleterious recombination. Indeed DNA methylation has been shown to play a critical role in silencing the transcription of retrotransposons during embryonic development and spermatogenesis (Bestor and Bourc’his 2004).
6.4. Rett Syndrome
The existence of multiple methyl-CpG binding proteins with repressive properties argues that these may be important mediators of the methylation signal. This is illustrated most strikingly by the finding that mutations in the human MECP2 gene are responsible for a severe neurological disorder called Rett syndrome (RTT). RTT affects females that are heterozygous for new mutations in the X-linked MECP2 gene (Table 2) (Amir et al. 1999). Because of random X-chromosome inactivation, the patients are mosaic for expression of either the mutant or the wild-type gene. Affected girls develop apparently normally for 6–18 months, at which time they enter a crisis that leaves them with greatly impaired motor skills, repetitive hand movements, abnormal breathing, microcephaly, and other symptoms (Table 2). Males who are hemizygous for comparable mutations do not survive. Interestingly, duplication of the MECP2 gene also leads to a profound autism-like syndrome, suggesting that too much of this protein is also deleterious (Lubs et al. 1999; Meins et al. 2005). Mecp2-null mice are born and develop normally for several weeks, but they acquire neurological symptoms at about 6 weeks of age, leading to death at about 12 weeks. Several features of this phenotype recall human Rett syndrome, making the mouse a convincing model for the disorder (Guy et al. 2001). There is no evidence for increased brain cell death in MeCP2-deficient human patients or mice, raising the possibility that restoration of a functional MECP2 gene could rescue the phenotype. Indeed, activation of the gene in severely affected male or female mice dramatically reverses neurological symptoms, raising the possibility that Rett syndrome in humans is curable (Guy et al. 2007).
Biochemical and immunocytochemical studies have established that MeCP2 is extremely abundant in the brain, particularly in neurons in which there are nearly 20 million molecules per nucleus (Skene et al. 2010). Accordingly, chromatin immunoprecipitation shows that MeCP2 is not targeted to specific genes, but coats the genome in a DNA methylation-dependent manner. Interestingly MeCP2 becomes phosphorylated at a specific serine residue on neuronal firing and this may play a role in modulating MeCP2 function (Zhou et al. 2006). The role of MeCP2 is not confined to neurons, however, as its presence is also required in glia (Lioy et al. 2011).
Given the potential role of MeCP2 as a transcriptional repressor, an attractive hypothesis to explain Rett syndrome is that genes in the brain needing to be silenced escape repression in its absence. Although there is evidence for derepression of transcription from retrotransposons (Muotri et al. 2005) and repetitive elements (Skene et al. 2010) in the absence of MeCP2, results of genome-wide transcription analysis are complex. MeCP2-deficient mouse brains show modest over- and underexpression of many genes. Bdnf, for example, is consistently down-regulated in the MeCP2-deficient brain (Chen et al. 2003; Martinowich et al. 2003). Interestingly, overexpression of MeCP2 leads to increased expression of many genes that are down-regulated in Mecp2-null brains and vice versa. One potential explanation for this reciprocal behavior is that MeCP2 can activate as well as repress transcription (Chahrour et al. 2008). There is evidently more to learn about the precise role of MeCP2 in the brain.
7. FUTURE DIRECTIONS
Our understanding of the biological functions of DNA methylation in mammals has been growing steadily, but is far from complete. For instance, unlike genetic mutations, we know very little about the rate of changes in CpG methylation in mammals, or the intrinsic and environmental factors that induce changes in DNA methylation patterns. Accumulating evidence indicates that changes in DNA methylation and histone modifications contribute to the pathogenesis of many diseases such as cancer, diabetes, autoimmune, and neurological disorders as well as the aging process. Modulation of epigenetic states of the genome or gene expression is thus becoming a new therapeutic approach for the treatment of these diseases. Advances in the following areas will have significant impact on our understanding of the epigenetic mechanisms in disease and development of new medicine.
7.1. Profiling the Epigenome in Disease
Many complex diseases, such as type II diabetes, schizophrenia, autoimmune diseases, and some forms of cancer often cannot be explained by simple genetic alterations. The dynamic nature of epigenetic regulation is increasingly providing an alternative explanation for some of the features of complex diseases, which include late onset, gender effects, parent-of-origin effects, discordance of monozygotic twins, and fluctuation of symptoms (Gasser and Li 2011). Although growing evidence has linked aberrant DNA methylation and histone modifications to cancer, the role of epigenetic mechanisms in the etiology of many other complex diseases is largely unknown. Comparative studies of genome-wide DNA methylation patterns between normal and disease populations may provide insight into the epigenetic basis for various complex diseases (see also Beaudet and Zoghbi 2014).
Recent advances in next generation sequencing technology has made it technically and financially possible to perform genome-wide analysis of DNA methylation patterns (i.e., the methylome) in normal and disease tissues, using bisulfite sequencing or MeDIP methods (Bibikova and Fan 2010) (Box 1). These approaches have been used to analyze DNA methylation changes, for example, during stem cell differentiation into neurons, and comparative analysis between normal and cancer tissues (Cortese et al. 2011; Hon et al. 2012). Such studies are beginning to shed light on how DNA methylation is regulated and its role in cell differentiation and disease. Two methods for the mapping of genome-wide 5hmC patterns at single base resolution have also been introduced recently (Booth et al. 2012; Yu et al. 2012). These technological breakthroughs mark a new era for understanding the role of DNA methylation and demethylation in normal development and disease.
7.2. Genetic and Environmental Factors That Induce Epigenetic Changes
It is well established that DNA methylation patterns in the mammalian genome are highly regulated during development. Genetic alterations such as CGG triplet repeat expansion in the FMR1 gene provide examples of genetic lesions that can lead to heritable changes in promoter CGI methylation and transcription repression. Mutations in genes encoding key epigenetic regulators such as MeCP2 and DNMT3B can also alter the epigenome and gene expression patterns, which lead to disease. An increasing number of other key epigenetic enzymes or chromatin-associated proteins, when mutated in cancer or other diseases, results in dramatic changes to the epigenome (You and Jones 2012).
How environmental factors may affect DNA methylation and gene expression is less well understood, although some recent studies are beginning to shed light on how they may induce epigenetic changes that can have long-lasting biological effects. One such example is the observation that rat maternal behavior produces stable alterations in DNA methylation in the offspring. Weaver and coworkers have reported that baby rats receiving different levels of maternal care have differences in DNA methylation in the promoter region of the glucocorticoid receptor (GR) gene, which are inversely correlated with GR expression, and these differences persist into adulthood (Weaver et al. 2004). Other factors, such as dietary supplements, drugs, and smoking can also impact the epigenome and lead to physiological or pathological changes.
7.3. Modulation of Epigenetic State and Reprogramming by Tet-Mediated 5mC Oxidation
One of the most exciting advances in the DNA methylation field is the demonstration that 5mC can be oxidized by Tet proteins to generate 5hmC, 5fC/5caC, and then later processed by the DNA repair mechanism enzyme TDG (Wu and Zhang 2011). Although there is clear evidence suggesting global loss of DNA methylation of the paternal genome is attributable to Tet-mediated oxidation (Wu and Zhang 2011), whether Tet proteins play a similar role in PGC reprogramming awaits to be determined (for more discussion, see Surani and Reik 2014). Given the accumulation of 5hmC in certain tissues and cell types, 5hmC has also been proposed to serve as an epigenetic mark that may mediate a specific function. In this regard, a component of the NuRD complex has been shown to recognize and bind to 5hmC (Yildirim et al. 2011). Future studies along this line should reveal whether 5hmC signals as a new epigenetic mark or simply as an intermediate for DNA demethylation. Activation of pluripotency genes, such as Oct4 and Nanog, requires demethylation of their promoters. Thus, it is not surprising that factors implicated in DNA demethylation have been reported to facilitate somatic cell reprogramming (Bhutani et al. 2010). Whether Tet proteins contribute to Oct4 and Nanog activation during reprogramming should be determined in the near future.
7.4. Modulation of Reversible Epigenetic States
Most, if not all, epigenetic modifications are reversible, which makes modulation of epigenetic states a promising new therapeutic option for cancer and other diseases. A number of agents that alter patterns of DNA methylation or inhibit HDACs are in clinical use for the treatment of cancer (Baylin and Jones 2011). The demethylating agent 5-azacytidine, for instance, has been approved by the U.S. Food and Drug Administration for the treatment of myelodysplastic syndrome, a heterogeneous disease characterized by morphologic dysplasia of hematopoietic cells in 2004. The clinical use of 5-azacytidine and other nucleoside analogs, however, is limited by their toxicity, partly because these compounds are being incorporated into DNA. This has encouraged the search for agents that can inhibit DNA methyltransferases directly or target other epigenetic regulators that can modulate DNA methyltransferase activities or CGI methylation. Because DNA methylation is just one component of the complex epigenetic regulatory network, one approach to maximize therapeutic effects and minimize toxicity is combination therapy using DNA methyltransferase or HDAC inhibitors in combination with other anticancer therapeutics (see Jones and Baylin 2014 for more detail).
ACKNOWLEDGMENTS
We wish to thank Adrian Bird for his contribution to this chapter in the first edition, and critical reading and expert comments during the writing of this article.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY 1999. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet 23: 185–188 [DOI] [PubMed] [Google Scholar]
- *.Aramayo R, Selker EU 2013. Neurospora crassa, a model system for epigenetics research. Cold Spring Harb Perspect Biol 5: a017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Barlow D, Bartolomei M 2014. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Jones PA 2011. A decade of exploring the cancer epigenome—Biological and translational implications. Nat Rev Cancer 11: 726–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Beaudet A, Zoghbi H 2014. Epigenetics and human disease. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AC, Felsenfeld G 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405: 482–485 [DOI] [PubMed] [Google Scholar]
- Bell AC, West AG, Felsenfeld G 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98: 387–396 [DOI] [PubMed] [Google Scholar]
- Bestor TH, Bourc’his D 2004. Transposon silencing and imprint establishment in mammalian germ cells. Cold Spring Harb Symp Quant Biol 69: 381–387 [DOI] [PubMed] [Google Scholar]
- Bestor TH, Ingram VM 1983. Two DNA methyltransferases from murine erythroleukemia cells: Purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci 80: 5559–5563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM 2010. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463: 1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Fan JB 2010. Genome-wide DNA methylation profiling. Wiley Interdiscip Rev Systems Biol Med 2: 210–223 [DOI] [PubMed] [Google Scholar]
- Bird AP 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8: 1499–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A, Wolffe AP 1999. Methylation-induced repression—Belts, braces and chromatin. Cell 99: 451–454 [DOI] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D 1985. A fraction of the mouse genome that is derived from islands of non-methylated, CpG-rich DNA. Cell 40: 91–99 [DOI] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ 2010. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell 38: 179–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Blackledge N, Thomson J, Skane P 2013. CpG binding proteins. Cold Spring Harb Perspect Biol 5: a018648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336: 934–937 [DOI] [PubMed] [Google Scholar]
- Boyes J, Bird A 1991. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 64: 1123–1134 [DOI] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND, Scandura JM 2011. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS One 6: e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Brockdorff N, Turner B 2014. Dosage compensation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH 2003. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol 4: 235–240 [DOI] [PubMed] [Google Scholar]
- Cedar H, Bergman Y 2009. Linking DNA methylation and histone modification: Patterns and paradigms. Nat Rev Genet 10: 295–304 [DOI] [PubMed] [Google Scholar]
- Chahrour M, Jung SY, Shaw C, Zhou X, Wong ST, Qin J, Zoghbi HY 2008. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science 320: 1224–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y, Sun L, Kokura K, Horton JR, Fukuda M, Espejo A, Izumi V, Koomen JM, Bedford MT, Zhang X, et al. 2011. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat Commun 2: 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME 2003. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science 302: 885–889 [DOI] [PubMed] [Google Scholar]
- *.Cheng X 2014. Structures of epigenetic reading modules and synthetic chromatin. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018747 [DOI] [Google Scholar]
- Cooper DN, Youssoufian H 1988. The CpG dinucleotide and human genetic disease. Hum Genet 78: 151–155 [DOI] [PubMed] [Google Scholar]
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. 2011. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470: 419–423 [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese R, Lewin J, Backdahl L, Krispin M, Wasserkort R, Eckhardt F, Beck S 2011. Genome-wide screen for differential DNA methylation associated with neural cell differentiation in mouse. PLoS One 6: e26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton AM, Bird A 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis K, Fan T, Geiman T, Yan Q, Muegge K 2001. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev 15: 2940–2944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge JE, Okano M, Dick F, Tsujimoto N, Chen T, Wang S, Ueda Y, Dyson N, Li E 2005. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem 280: 17986–17991 [DOI] [PubMed] [Google Scholar]
- Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schubeler D, et al. 2008. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 27: 2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M 2003. The ICF syndrome, a DNA methyltransferase 3B deficiency and immunodeficiency disease. Clin Immunol 109: 17–28 [DOI] [PubMed] [Google Scholar]
- *.Elgin S, Reuter G 2013. Positive effect variegation, heterochromatin formation and gene silencing in Drosophila. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a017780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S 2006. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 20: 3089–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Y 2001. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev 15: 827–832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL 1992. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci 89: 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser SM, Li E 2011. Epigenetics and disease: Pharmaceutical opportunities. Preface. Prog Drug Res 67: v–viii [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R 2003. Induction of tumors in mice by genomic hypomethylation. Science 300: 489–492 [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, McDowell TL, Raman S, O’Rourke DM, Garrick D, Ayyub H, Higgs DR 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat Genet 24: 368–371 [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477: 606–610 [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A 2001. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet 27: 322–326 [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A 2007. Reversal of neurological defects in a mouse model of Rett syndrome. Science 315: 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, Haaf T, El-Maarri O, Reik W, Walter J, Surani MA 2002. Epigenetic reprogramming in mouse primordial germ cells. Mech Dev 117: 15–23 [DOI] [PubMed] [Google Scholar]
- Harbers K, Schnieke H, Stuhlmann H, Jahner D, Jaenisch B 1981. DNA methylation and gene expression; Endogenous retroviral genome becomes infectious after molecular cloning. Proc Natl Acad Sci 78: 7609–7613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129: 1983–1993 [DOI] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. 2011. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Hardeland U, Ng H-H, Jiricny J, Bird A 1999. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature 401: 301–304 [DOI] [PubMed] [Google Scholar]
- Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A 2001. Closely related proteins Mbd2 and Mbd3 play distinctive but interacting roles in mouse development. Genes Dev 15: 710–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB 1996. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci 93: 9821–9826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R, Pugh JE 1975. DNA modification mechanisms and gene activity during development. Science 187: 226–232 [PubMed] [Google Scholar]
- Hon GC, Hawkins RD, Caballero OL, Lo C, Lister R, Pelizzola M, Valsesia A, Ye Z, Kuan S, Edsall LE, et al. 2012. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res 22: 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins A, Mullen A, Lee H, Barner K, High F, Hendrich B, Bird A, Reiner S 2002. Gene silencing quantitatively controls the function of a developmental trans-activator. Mol Cell 10: 81–91 [DOI] [PubMed] [Google Scholar]
- Inoue A, Zhang Y 2011. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Shen L, Dai Q, He C, Zhang Y 2011. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res 21: 1670–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE 2011. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci 108: 3642–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. 2009. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y 2010. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466: 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y 2011. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahner D, Stuhlmann H, Stewart CL, Harbers K, Lohler J, Simon I, Jaenisch R 1982. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature 298: 623–628 [DOI] [PubMed] [Google Scholar]
- Jeddeloh JA, Stokes TL, Richards EJ 1999. Maintenance of genomic methylation requires a SW12/SNF2-like protein. Nat Genet 22: 94–97 [DOI] [PubMed] [Google Scholar]
- *.Jones P, Baylin S 2014. Epigenetics and cancer. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Taylor SM 1980. Cellular differentiation, cytidine analogues and DNA methylation. Cell 20: 85–93 [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19: 187–191 [DOI] [PubMed] [Google Scholar]
- Kaneda M, Sado T, Hata K, Okano M, Tsujimoto N, Li E, Sasaki H 2004. Role of de novo DNA methyltransferases in initiation of genomic imprinting and X-chromosome inactivation. Cold Spring Harb Symp Quant Biol 69: 125–129 [DOI] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G 2008. Transient cyclical methylation of promoter DNA. Nature 452: 112–115 [DOI] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. 2008. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet 40: 741–750 [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N 2009. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science 324: 929–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, Peters AH 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13: 1192–1200 [DOI] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Gos KA, Jaenisch R, Li E 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122: 3195–3205 [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69: 915–926 [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R 1993. Role for DNA methylation in genomic imprinting. Nature 366: 362–365 [DOI] [PubMed] [Google Scholar]
- Li H, Rauch T, Chen ZX, Szabo PE, Riggs AD, Pfeifer GP 2006. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem 281: 19489–19500 [DOI] [PubMed] [Google Scholar]
- Lioy DT, Garg SK, Monaghan CE, Raber J, Foust KD, Kaspar BK, Hirrlinger PG, Kirchhoff F, Bissonnette JM, Ballas N, et al. 2011. A role for glia in the progression of Rett’s syndrome. Nature 475: 497–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubs H, Abidi F, Bier JA, Abuelo D, Ouzts L, Voeller K, Fennell E, Stevenson RE, Schwartz CE, Arena F 1999. XLMR syndrome characterized by multiple respiratory infections, hypertelorism, severe CNS deterioration and early death localizes to distal Xq28. Am J Med Genet 85: 243–248 [DOI] [PubMed] [Google Scholar]
- Maiti A, Drohat AC 2011. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem 286: 35334–35338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE 2003. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302: 890–893 [DOI] [PubMed] [Google Scholar]
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T 2000. Demethylation of the zygotic paternal genome. Nature 403: 501–502 [DOI] [PubMed] [Google Scholar]
- McKeon C, Ohkubo H, Pastan I, de Crombrugghe B 1982. Unusual methylation pattern of the α 2 (I) collagen gene. Cell 29: 203–210 [DOI] [PubMed] [Google Scholar]
- Meehan RR, Lewis JD, McKay S, Kleiner EL, Bird AP 1989. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell 58: 499–507 [DOI] [PubMed] [Google Scholar]
- Meins M, Lehmann J, Gerresheim F, Herchenbach J, Hagedorn M, Hameister K, Epplen JT 2005. Submicroscopic duplication in Xq28 causes increased expression of the MECP2 gene in a boy with severe mental retardation and features of Rett syndrome. J Med Genet 42: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. 2008. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452: 45–50 [DOI] [PubMed] [Google Scholar]
- Millar CB, Guy J, Sansom OJ, Selfridge J, MacDougall E, Hendrich B, Keightley PD, Bishop SM, Clarke AR, Bird A 2002. Enhanced CpG mutability and tumorigenesis in MBD4-deficient mice. Science 297: 403–405 [DOI] [PubMed] [Google Scholar]
- Mohandas T, Sparkes RS, Shapiro LJ 1981. Reactivation of an inactive human X-chromosome: Evidence for X-inactivation by DNA methylation. Science 211: 393–396 [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schubeler D 2008. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766 [DOI] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435: 903–910 [DOI] [PubMed] [Google Scholar]
- Nan X, Ng H-H, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386–389 [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E 1998a. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 19: 219–220 [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E 1998b. Dnmt2 is not required for de novo and maintenance methylation of viral DNA in embryonic stem cells. Nucleic Acids Res 26: 2536–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99: 247–257 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH 2008. The colorful history of active DNA demethylation. Cell 133: 1145–1148 [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. 2007. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448: 714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J 2000. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 10: 475–478 [DOI] [PubMed] [Google Scholar]
- Peprah E 2012. Fragile X syndrome: The FMR1 CGG repeat distribution among world populations. Ann Hum Genet 76: 178–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffeneder T, Hackner B, Truss M, Munzel M, Muller M, Deiml CA, Hagemeier C, Carell T 2011. The discovery of 5-formylcytosine in embryonic stem cell DNA. Angew Chem Int Ed Engl 50: 7008–7012 [DOI] [PubMed] [Google Scholar]
- *.Pikaard C, Mittelsten Scheid O 2014. Epigenetic regulation in plants. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai J, Bhagwat AS, Posfai G, Roberts RJ 1989. Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res 17: 2421–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G, Bird A, Prokhortchouk E 2001. The p120 catenin partner Kaiso is a DNA methylation-dependent transcriptional repressor. Genes Dev 15: 1613–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs AD 1975. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 14: 9–25 [DOI] [PubMed] [Google Scholar]
- Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al. 2012. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 19: 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougier N, Bourc’his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E 1998. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev 12: 2108–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I 2004. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell 15: 595–605 [DOI] [PubMed] [Google Scholar]
- Sasaki H, Matsui Y 2008. Epigenetic events in mammalian germ-cell development: Reprogramming and beyond. Nat Rev Genet 9: 129–140 [DOI] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr AR, James KD, Turner DJ, Andrews R, Bird AP 2010. Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell 37: 457–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. 2011. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495 [DOI] [PubMed] [Google Scholar]
- Stewart CL, Stuhlmann H, Jahner D, Jaenisch R 1982. De novo methylation and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci 79: 4098–4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Surani A, Reik W 2014. Germline and pluripotent stem cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. 2009. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414: 277–283 [DOI] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost J, Gut IG 2007. DNA methylation analysis by pyrosequencing. Nat Protoc 2: 2265–2275 [DOI] [PubMed] [Google Scholar]
- Valinluck V, Sowers LC 2007. Endogenous cytosine damage products alter the site selectivity of human DNA maintenance methyltransferase DNMT1. Cancer Res 67: 946–950 [DOI] [PubMed] [Google Scholar]
- Villa R, Morey L, Raker VA, Buschbeck M, Gutierrez A, De Santis F, Corsaro M, Varas F, Bossi D, Minucci S, et al. 2006. The methyl-CpG binding protein MBD1 is required for PML-RARα function. Proc Natl Acad Sci 103: 1400–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439: 871–874 [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP 1999. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet 23: 62–66 [DOI] [PubMed] [Google Scholar]
- Watt F, Molloy PL 1988. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus late promoter. Genes Dev 2: 1136–1143 [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, Dymov S, Szyf M, Meaney MJ 2004. Epigenetic programming by maternal behavior. Nat Neurosci 7: 847–854 [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet 37: 853–862 [DOI] [PubMed] [Google Scholar]
- Williams K, Christensen J, Pedersen MT, Johansen JV, Cloos PA, Rappsilber J, Helin K 2011. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 473: 343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf SF, Jolly DJ, Lunnen KD, Friedman T, Migeon BR 1984. Methylation of the hypoxanthine phosphoribosyltransferase locus on the human X-chromosome: Implications for X-chromosome inactivation. Proc Natl Acad Sci 81: 2806–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J 2011. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun 2: 241. [DOI] [PubMed] [Google Scholar]
- Wu SC, Zhang Y 2010. Active DNA demethylation: Many roads lead to Rome. Nat Rev Mol Cell Biol 11: 607–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Zhang Y 2011. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes Dev 25: 2436–2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329: 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, D’Alessio AC, Ito S, Xia K, Wang Z, Cui K, Zhao K, Sun YE, Zhang Y 2011. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473: 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y, Mann MR, Lee SS, Marh J, McCarrey JR, Yanagimachi R, Bartolomei MS 2003. Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc Natl Acad Sci 100: 12207–12212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, Weng Z, Rando OJ, Fazzio TG 2011. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 147: 1498–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HG, Chan DW, Reynolds AB, Qin J, Wong J 2003. N-CoR mediates DNA methylation-dependent repression through a methyl CpG binding protein Kaiso. Mol Cell 12: 723–734 [DOI] [PubMed] [Google Scholar]
- You JS, Jones PA 2012. Cancer genetics and epigenetics: Two sides of the same coin? Cancer Cell 22: 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. 2012. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell 149: 1368–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D 1998. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289 [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, et al. 2006. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52: 255–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK 2009. Active DNA demethylation mediated by DNA glycosylases. Annu Rev Genet 43: 143–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
WWW RESOURCES
- http://www.illumina.com/products/methylation_450_beadchipkits.ilmn. Illumina methylation chip.
- http://www.sequenom.com/Sites/Genetic-Analysis/Applications/DNA-Methylation. Sequenom EpiTYPER.