Abstract
All morphologically complex life on Earth, beyond the level of cyanobacteria, is eukaryotic. All eukaryotes share a common ancestor that was already a complex cell. Despite their biochemical virtuosity, prokaryotes show little tendency to evolve eukaryotic traits or large genomes. Here I argue that prokaryotes are constrained by their membrane bioenergetics, for fundamental reasons relating to the origin of life. Eukaryotes arose in a rare endosymbiosis between two prokaryotes, which broke the energetic constraints on prokaryotes and gave rise to mitochondria. Loss of almost all mitochondrial genes produced an extreme genomic asymmetry, in which tiny mitochondrial genomes support, energetically, a massive nuclear genome, giving eukaryotes three to five orders of magnitude more energy per gene than prokaryotes. The requirement for endosymbiosis radically altered selection on eukaryotes, potentially explaining the evolution of unique traits, including the nucleus, sex, two sexes, speciation, and aging.
Bacteria respire across their cell membranes and show little tendency to evolve complex traits. In eukaryotes, mitochondria energetically support the nuclear genome; this may have enabled the evolution of complex traits.
Evolutionary theory has enormous explanatory power and is understood in detail at the molecular genetic level, yet it cannot easily predict even the past. The history of life on Earth is troubling. Life apparently arose very early, perhaps 4 billion years ago, but then remained essentially bacterial for probably some 2–3 billion years. Bacteria and archaea explored almost every conceivable metabolic niche and still dominate in terms of biomass. Yet, in morphological diversity and genomic complexity, bacteria barely begin to compare with eukaryotes, even at the level of cells, let alone multicellular plants and animals. Eukaryotes are monophyletic and share a common ancestor that by definition arose only once, probably between 1.5 and 2 billion years ago, although the dates are poorly constrained (Knoll et al. 2006; Parfrey et al. 2011). The eukaryotic common ancestor already had a nucleus, nuclear pore complexes, introns and exons, straight chromosomes, mitosis and meiotic sex, a dynamic cytoskeleton, an endoplasmic reticulum, and mitochondria, making it difficult to trace the evolution of these traits from a prokaryotic state (Koonin 2010). The “eukaryotic niche”—limited metabolic diversity but enormous morphological complexity—was never invaded by prokaryotes. In short, life arose early, stagnated in morphological complexity for several billion years, and then rather abruptly gave rise to a single group—the eukaryotes—which explored the morphological realm of life in ways never seen in bacteria or archaea.
Consider the possibility of life evolving on other planets. Would it follow a similar trajectory? If not, why not? Evolutionary theory gives little insight. The perplexing history of life on Earth conceals a paradox relating to natural selection. If basal eukaryotic traits such as the nucleus, meiotic sex, and phagocytosis arose by selection, starting with a prokaryotic ancestor, and each step offered some small advantage over the last, then why don’t the same traits arise repeatedly in prokaryotes too? Prokaryotes made many a start. There are examples of bacteria or archaea with nucleus-like structures (Lindsay et al. 2001), recombination (Smith et al. 1993), linear chromosomes (Bentley et al. 2002), internal membranes (Pinevich 1997), multiple replicons (Robinson and Bell 2007), giant size (Schulz and Jorgensen 2001), extreme polyploidy (Mendell et al. 2008), a dynamic cytoskeleton (Vats and Rothfield 2009), predation (Davidov and Jurkevitch 2009), parasitism (Moran 2007), introns and exons (Simon and Zimmerly 2008), intercellular signaling (Waters and Bassler 2005), endocytosis-like processes (Lonhienne et al. 2010), and even endosymbionts (Wujek 1979; von Dohlen et al. 2001). Yet, for each of these traits, bacteria and archaea stopped well short of the baroque complexity of eukaryotes. Compare this with the evolution of eyes. From a simple, light-sensitive spot in an early metazoan, morphologically disparate eyes arose on scores of occasions (Vopalensky and Kozmic 2009). This is exactly what evolutionary theory predicts. Each step offers an advantage in its own ecological setting, so morphologically different eyes arise on multiple occasions. Why is this not the case for traits such as the nucleus, meiotic sex, and phagocytosis? To suggest that lateral gene transfer (LGT) or bacterial conjugation is equivalent to meiotic sex will not do: Neither involves a systematic and reciprocal exchange of alleles across the entire genome.
The simplest explanation is a bottleneck. The “big bang” radiation of major eukaryotic supergroups, combined with the apparent absence of surviving evolutionary intermediates between prokaryotes and the last eukaryotic common ancestor, does indeed hint at a bottleneck at the origin of eukaryotes. There is no shortage of environmental possibilities, from snowball glaciations to rising atmospheric oxygen. The most widely held explanation contends that when oxygen levels rose after the great oxidation event, some proto-eukaryotic cells acquired mitochondria, which protected them against oxygen toxicity (Andersson and Kurland 1999) and enabled them to exploit oxygen as a terminal electron acceptor in respiration (Sagan 1967), giving the first eukaryotes an enormous competitive advantage. They swiftly occupied new niches made available by oxygen, outcompeting to extinction any other prokaryotes that tried subsequently to invade this niche (de Duve 2007; Gross and Bhattacharya 2010). But this is an evolutionary “just-so story” and has no evidence to support it. The idea that mitochondria might protect against oxygen toxicity is nonsense: The single-electron donors of respiratory chains are among the most potent free-radical generators known. And what was to stop facultatively aerobic bacteria—from which the mitochondria evolved, hence already present—from occupying the aerobic niche first?
In fact, the limited evidence available suggests that oxygen had little to do with it (Müller et al. 2012; van der Giezen and Lenton 2012). A large, diverse group of morphologically simple protists dubbed archezoa are the key here. The archezoa appear to lack mitochondria; and three decades ago, looked to branch deeply in the eukaryotic tree. Cavalier-Smith postulated that some archezoa might be primitively amitochondriate: surviving evolutionary intermediates between prokaryotes and eukaryotes (Cavalier-Smith 1987, 1989). But 20 years of careful molecular biology and phylogenetics have shown that all known archezoa possess specialized organelles that derive from mitochondria, namely hydrogenosomes or mitosomes (Keeling 1998; Embley and Martin 2006; van der Giezen 2009; Archibald 2011). The archezoa are obviously not real evolutionary intermediates, and radical developments in phylogenomics have transformed the eukaryotic tree to a “big-bang” radiation with no early branching archezoa (Koonin 2010). The archezoa remain significant not because they are genuine evolutionary intermediates, but because they are true ecological intermediates. Critically, they were not outcompeted to extinction by more sophisticated aerobic eukaryotes. On the contrary, they lost their capacity for aerobic respiration and depend instead on anaerobic fermentations, yet remain, morphologically, more complex than bacteria or archaea.
The fact that the archezoa are a phylogenetically disparate group that arose on multiple occasions is equally significant. The “intermediate” niche is viable and was invaded many times, without the new arrivals being outcompeted to extinction by existing cells, or vice versa. Yet each time the invader was an anaerobic eukaryote, which adapted by reductive evolution to the niche—not bacteria or archaea evolving slightly greater complexity. What is the likelihood of this bias? Given at least 20 independent origins of archezoa (van der Giezen 2009; Müller et al. 2012), the probability of these ecological intermediates arising each time from the eukaryotes rather than prokaryotes is less than one in a million. It is far more parsimonious to assume that there was something about the structure of eukaryotes that facilitated their invasion of this intermediate niche; and, conversely, something about the structure of prokaryotes that tended to preclude their evolution of greater morphological complexity. But this quite reasonable statement is loaded because it implies that prokaryotes existed for nearly 4 billion years, and throughout that time showed no tendency to evolve greater morphological complexity. In stark contrast, eukaryotes arose just once, a seemingly improbable event.
Here I argue that the constraint on prokaryotes was bioenergetic. There was, indeed, a bottleneck at the origin of eukaryotes, but it was biological (restrictive), not environmental (selective). It related to the physical structure of prokaryotic cells: Both bacteria and archaea respire across their plasma membrane. I make three key points, which arguably apply to life elsewhere in the universe, and are therefore proposed as biological principles that could guide our understanding of life generally: (1) chemiosmotic coupling is as universal as the genetic code, for fundamental reasons relating to the origin of life; (2) prokaryotes are constrained by chemiosmotic coupling across their plasma membrane, but eukaryotes escaped this constraint through a rare and stochastic endosymbiosis between two prokaryotes, giving them orders of magnitude more energy per gene; and (3) this endosymbiosis, in turn, produced a unique genomic asymmetry, transforming the selection pressures acting on eukaryotes and driving the evolution of unique eukaryotic traits.
ENERGETIC CONSTRAINTS ON THE ORIGIN OF LIFE
Why are all cells powered by chemiosmotic coupling? In place of straightforward chemical coupling between “high-energy” moieties generated by substrate-level phosphorylation, chemiosmosis uses a gradient of protons (or sodium ions) across a membrane. First proposed by Peter Mitchell (1961), the mechanism remained controversial for two decades. Part of the reason for this was surely that the interpolation of an electrochemical ion gradient across a membrane seemed little short of crazy. Why would nature come up with something so odd? Since then, research on chemiosmotic coupling has focused mainly on the structure and function of respiratory complexes involved in generating and harnessing proton gradients. This work has been extremely successful, culminating in the recent atomic-level resolution of the structure of complex I (Baradaran et al. 2013). From the beginning, however, Mitchell’s interests went beyond the precise mechanisms of coupling into questions about how cells interact with their environment. As early as 1957, Mitchell published a paper on the origin of life, in which he wrote:
I cannot consider the organism without its environment … from a formal point of view the two may be regarded as equivalent phases between which dynamic contact is maintained by the membranes that separate and link them (Mitchell 1957).
This line of Mitchell’s thinking has been less successful, but I would argue is equally prescient. There has been remarkably little interest in why such a counterintuitive mechanism of energy transduction evolved. As we know it, chemiosmotic coupling requires a combination of sophisticated molecular machines, such as the ATP synthase (a nanoscale rotary motor) and an ion-tight membrane. Given that the earliest membranes—probably fatty acid vesicles or something similar—were extremely permeable to protons, the idea that chemiosmotic coupling could have been the earliest form of energy transduction has not enjoyed much popularity (Pohorille and Deamer 2009; Mulkidjanian et al. 2012). But if it arose subsequently, why did it displace all other forms of energy transduction to become universal? In fact, the underexplored possibility that chemiosmotic coupling was the primordial energy source is compelling—but only in a very specific setting.
Natural Proton Gradients in Alkaline Hydrothermal Vents
Michael Russell and colleagues have argued for two decades that natural proton and redox gradients in alkaline hydrothermal vents could have driven the origin of life (Russell et al. 1993, 2013; Russell and Hall 1997; Martin and Russell 2003, 2007; Martin et al. 2008). Organic chemists have been very successful at the batch synthesis of organics, including amino acids and even activated nucleotides (Powner et al. 2009), but there are intractable problems with the conception of a primordial soup, not least how to drive and focus the continuous exergonic reactions needed to generate RNA, DNA, proteins, and lipids at high enough rates and concentrations for them to form growing, dividing, cell-like structures, capable of evolution by natural selection (Lane et al. 2010). Many of these difficulties are solved by alkaline hydrothermal vents, at least as they should have existed 4 billion years ago, when high CO2 levels (0.1–1 bar) and anoxia meant that the oceans should have been mildly acidic (pH 5–6) and rich in dissolved transition metals, notably ferrous iron (Sleep et al. 2011; Arndt and Nisbet 2012). These vents are natural far-from-equilibrium electrochemical reactors, which provide a continuous flow of H2 (in millimolar quantities), CO2, and NH3 through a labyrinth of thin-walled micropores containing catalytic Fe(Ni)S minerals. Thermophoresis through these labyrinths concentrates organics to high levels, potentially driving the beginnings of biochemistry (Baaske et al. 2007). The single most appealing aspect of such vents is that their compartments are analogous to autotrophic cells, right down to the magnitude (150–300 mV) and polarity (acid outside) of proton gradients across thin inorganic walls (Russell and Hall 1997; Lane et al. 2010).
But there are two big problems with alkaline hydrothermal vents. The first is the starting point: H2 and CO2. In terms of thermodynamics, H2 and CO2 should react to form organics: The synthesis of total cell biomass is exergonic under alkaline hydrothermal conditions between 25°C and 125°C (Amend and McCollom 2009; Amend et al. 2013). In practice, there is an energetic barrier to the reduction of CO2, with the step from formate (CHOO−) to formaldehyde (CH2O) being strongly endergonic (Maden 2000). This has proven difficult to break down under plausibly abiotic conditions. Methanogens and acetogens live from exactly this reaction, via the ancient acetyl CoA pathway drawing on numerous enzymes with catalytic Fe(Ni)S centers that resemble the minerals found in vents (Russell and Martin 2004). However, attempts to reduce CO2 with H2 using Fe(Ni)S minerals in the laboratory have so far proved disappointing. This difficulty is hardly trivial and prompted Wächtershäuser (1988) to surmise that it was impossible for life to have started from CO2 and H2 for that very reason.
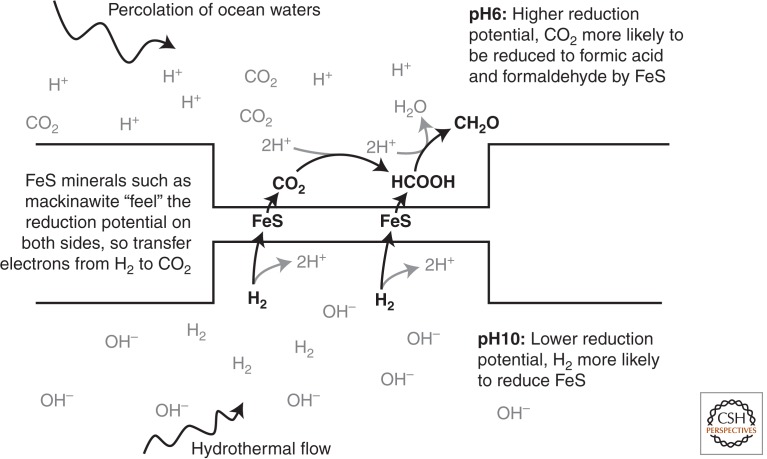
The second problem relates to the proton gradients themselves. Specifically, how could proton flux be harnessed to do useful work in an abiotic vent system in the absence of proteins? One possible answer could solve both problems. In redox reactions that involve protons, the reduction potential depends on pH (proton concentration) and falls by ∼60 mV for every unit rise in pH. In the case of H2, the standard reduction potential of the H+/H2 couple at pH 7, Eo′, is –414 mV. In contrast, the Eo′ of the CO2/HCOOH couple is –430 mV, and of the HCOOH/CH2O couple is –580 mV. Thus, at pH 7, it is, indeed, impossible for H2 to reduce CO2 to CH2O. But neither the vent fluids nor the oceans are at neutral pH. The actual reduction potential (Eh) of the H+/H2 couple falls to –590 mV at pH 10; that is, H2 is far more reducing. In contrast, the Eh of the CO2/HCOOH couple at pH 6 rises to –370 mV, and the Eh of the HCOOH/CH2O couple rises to –520 mV; that is, CO2 is more easily reduced. Thus, H2 at pH 10 could theoretically reduce CO2 at pH 6 to make formaldehyde. That might sound improbable but is, in fact, exactly what alkaline hydrothermal vents can offer. Electrons could be transferred from H2 at pH 10 to CO2 at pH 6 across a thin, semiconducting barrier composed of FeS minerals. FeS minerals such as mackinawite are themselves protonated, with a pKa at ∼pH 7. If the inorganic barrier is thin enough for FeS to “feel” the distinct reduction potentials in two adjacent compartments, one containing CO2 in ocean water at pH 6 and the other containing H2 in alkaline hydrothermal fluids at pH 10, there should be a transfer of electrons from H2 to CO2, to produce simple organics such as formaldehyde, breaking the energetic barrier (Fig. 1).
Figure 1.
Proposed vectorial reduction of CO2 by H2 across a thin FeS barrier. The reduction potential (Eh) of the H+/H2 couple is –590 mV at pH 10, whereas the Eh of the CO2/HCOOH couple at pH 6 is –370 mV, and the Eh of the HCOOH/CH2O is –520 mV. A semiconducting FeS barrier should “feel” the distinct reduction potentials in both compartments and transfer electrons from H2 to CO2, to produce simple organics such as formaldehyde (CH2O).
From Harvesting to Generating Proton Gradients
Hydrothermal flow and percolation of ocean waters should drive the continuous reduction of CO2 to organics, as suggested by thermodynamics, while simultaneously concentrating them by thermophoresis. Not only organic syntheses are favored under these conditions, but some higher-order assemblies too, such as fatty-acid vesicles (Budin et al. 2009) and polymerization (Amend et al. 2013; Mast et al. 2013). This is not the place to discuss the origin of protocells. Suffice it to say that natural proton gradients can provide the energy needed not only at the earliest stages, but also subsequently in the world of protocells, ribosomes, genes, and proteins, still within the vent (Lane and Martin 2012). For example, methanogens use proton gradients to reduce ferredoxin via the energy-converting hydrogenase Ech, driving CO2 reduction within cells (Buckel and Thauer 2013). Ultimately, natural proton gradients could drive ATP synthesis directly via an ATP synthase embedded in a lipid membrane. Such a system works well in the absence of active pumping, but importantly, only if the membrane is leaky to protons (Lane and Martin 2012). Although leaky membranes rapidly dissipate proton gradients, in a vent setting they are replenished by hydrothermal flow. In the absence of active pumping, it is imperative that the membrane be leaky to protons and hydroxide ions, or else protons enter the cell faster than they can leave, equilibrating the concentration gradient. That cuts off cells from their energy supply and should kill any that evolved “modern” ion-tight membranes before they had active ion pumps. The universality of the ATP synthase in the absence of a common membrane or active pumping machinery in bacteria and archaea is consistent with this scenario (Lane and Martin 2012).
Thus, natural proton gradients could, in principle, drive both carbon and energy metabolism in vents, leading to the evolution of protocells. But this is only half a chemiosmotic circuit. If cells that depend on natural ion gradients need to be leaky, how did they tighten off their membranes to protons and other ions and begin pumping themselves? The origin of active pumping is costly and difficult and might have forced the deep divergence between archaea and bacteria, as described elsewhere (Lane and Martin 2012). Yet it was strictly necessary on thermodynamic grounds. In methanogens (archaea) and acetogens (bacteria), H2 and CO2 provide the carbon and energy needed for growth, what Everett Shock called “a free lunch that you’re paid to eat” (Shock et al. 1998). It is no accident that both groups are obligately chemiosmotic. The problem is the energetic barrier to reducing CO2 with H2. In vents, the barrier is broken by natural proton gradients, but to escape (or survive in regions that lack gradients) means finding another way to break the energy barrier. Methanogens and acetogens do so by an ingenious process called flavin-based electron bifurcation (Li et al. 2008; Kaster et al. 2011), which uses the exergonic steps of the reaction between H2 and CO2 to generate a proton (or sodium) gradient, reestablishing what vents provide for free. This process seems to be necessary. The energy released by the reaction of H2 and CO2 is less than double the energy input needed to overcome the barrier to their reaction. In terms of stoichiometric chemistry, 1 ATP must be consumed to gain ∼1.5 ATPs (Martin and Russell 2007; Thauer et al. 2008). Obviously 0.5 ATP is not possible, so, in fact, 1 ATP must be spent to gain 1 ATP. This simple fact precludes growth by stoichiometric chemistry, such as substrate-level phosphorylation (Martin and Russell 2007). The beauty of chemiosmotic coupling is that it transcends chemistry. It allows cells to “save up loose change”—substoichiometric energy conservation (Lane 2010). A single reaction can be repeated indefinitely to generate an ion electrochemical potential, which can then be used to drive ATP synthesis and growth.
What does all this mean? The origin of life is a matter of probabilities. Alkaline hydrothermal vents are produced by a geological process called “serpentinization,” which is no more than the reaction of minerals such as olivine with water at low temperatures (Martin et al. 2008; Sleep et al. 2011). Both olivine and water are ubiquitous in interstellar space and are likely to be common on exoplanets. Serpentinization is therefore likely to be widespread. It is an exergonic reaction that produces warm alkaline fluids and hydrogen gas in large quantities. Alkaline vents are therefore likely to be abundant on any wet, rocky planet. Given a CO2 atmosphere (again commonplace), such vents can be relied on to produce natural microporous systems replete with proton gradients, Fe(Ni)S minerals, and the H2/CO2 redox couple. That means that the “shopping list” for life is just rock, water, and CO2. But for thermodynamic reasons, these conditions, in turn, are likely to force the emergence of cells that depend on proton gradients and ultimately chemiosmotic coupling. Perhaps life can arise some other way; but as a matter of probability, life elsewhere is most likely to resemble bacteria on Earth, right down to chemiosmotic coupling across a membrane. If so, then the evolution of complex life will face the same problems anywhere in the universe.
ENERGETICS OF THE TRANSITION FROM PROKARYOTES TO EUKARYOTES
With the evolution of photosynthesis and the accumulation of higher potential electron acceptors—ultimately oxygen—membrane bioenergetics could have been supplanted by other forms of energy transduction. This did not happen, probably for two reasons. First, chemiosmotic coupling is extremely versatile, because it allows almost any combination of electron donor and acceptor to be plugged into a basic electrical circuit. The proteins required form a limited “redox protein construction kit” (Baymann et al. 2003; Schoepp-Cothenet et al. 2013), which is easily passed around by lateral gene transfer, enabling swift adaptation to new niches. Second, by decoupling an exergonic reaction from ATP synthesis, growth can be sustained using many redox couples, which in terms of stoichiometric chemistry should not support life. Substoichiometric energy conservation allows the last drops of energy to be squeezed out of any reaction. Although membrane bioenergetics was probably forced into existence by a highly constrained set of circumstances, nothing supplants chemiosmotic coupling because nothing else is as versatile or efficient.
Yet, for all their metabolic virtuosity, neither bacteria nor archaea ever evolved the morphological complexity that typifies eukaryotes. Despite their deep genetic and biochemical divergence, both these groups are constrained by their energetics: Both respire across their plasma membrane. This does not constrain real prokaryotes in any way, it merely sets limits on what is possible. We do not see large prokaryotic cells crammed with internal compartments encoded by outsized genomes because these are not energetically sustainable in prokaryotes. The problem is energy per gene (Lane and Martin 2010).
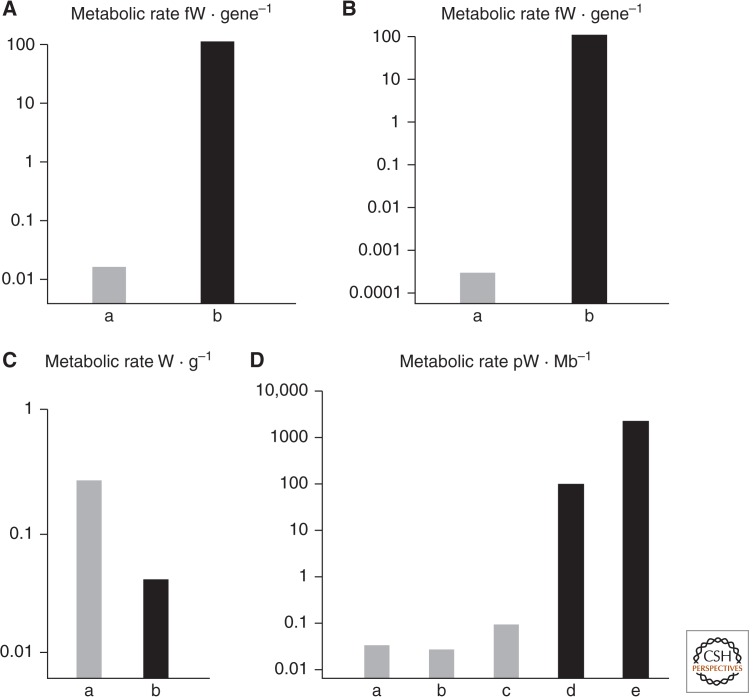
In terms of mean metabolic rate per gram, bacteria respire about threefold faster than eukaryotic protists (Fig. 2). But in terms of mean metabolic rate per cell, protists respire around 5000 times faster than bacteria. The difference just reflects mean cell volume; protists are around 15,000 times larger than bacteria. This difference nearly vanishes again if the metabolic rate of protists is considered in relation to genome size. Despite their enormous mean difference in size, bacteria and protists have nearly equal energy per megabase of DNA. Therefore, eukaryotes become larger, scale up respiration accordingly, and use their energy to support a much larger genome. In microbes, ∼75%–80% of the ATP budget of the cell is spent on protein synthesis (Harold 1986). To a first approximation, then, energy per gene equates to energy per protein. Standardizing to energy per gene, protists have ∼5000-fold more energy to spend on protein synthesis than bacteria (Lane and Martin 2010).
Figure 2.
Energetics of genome size in eukaryotes and prokaryotes. (A) Mean energy per gene in prokaryotes versus eukaryotes equalized for genome size. (Gray) Prokaryotes; (black) eukaryotes. Note the log scale. (B) Mean energy per gene in prokaryotes versus eukaryotes equalized for genome size and cell volume (see text). (Gray) Prokaryotes; (black) eukaryotes. Note the log scale. (C) Mean energy per gram in prokaryotes versus eukaryotes. (D) Power per haploid genome (energy per gene × number of genes in one haploid genome) in (lane a) Escherichia coli; (lane b) Thiomargarita; (lane c) Epulopiscium; (lane d) Chlamydomonas; and (lane e) Amoeba proteus. Note the log scale and broad agreement with derived mean values in A and B. (Figure based on data from Lane 2011a.)
Scaling of Cell Volume and Genome Copy Number
But this difference really reflects cell volume, which should be standardized too. That requires scaling up a bacterium to eukaryote size, 15,000-fold in volume. This demands some assumptions about respiration and protein synthesis. Bacteria and archaea respire over their plasma membrane and thus are subject to surface-area-to-volume constraints. Increasing their linear dimension 25-fold increases their surface area 625-fold, and volume 15,000-fold. ATP synthesis could therefore increase 625-fold, but such an increase would require a 625-fold increase in the number of respiratory proteins, ATP synthase enzymes, and all other molecular machinery needed to transcribe and translate the genes. Transcription from a single bacterial genome could hardly be increased 625-fold. We must then scale up the number of genomes accordingly. To a superficial first approximation, scaling up ATP synthesis 625-fold would require haploid genome number to increase by 625-fold. Energy per gene would remain unchanged. If we take internal volume into consideration, the same principles apply. Protein synthesis could not increase 15,000-fold from a single genome, but if the number of haploid genomes were increased 15,000-fold, energy per gene would fall by 25-fold. So scaling up a bacterium to mean eukaryotic volume would cut energy per gene by 5000 × 25 = 125,000-fold (Lane 2011a). All else being equal, a eukaryotic-sized bacterium should have about five orders of magnitude less energy per gene than the eukaryote (Lane and Martin 2010; Lane 2011a).
These numbers may seem absurd but are supported by the few examples of giant bacteria known, notably Epulopiscium and Thiomargarita. These cells are larger than most protists and show extreme polyploidy. Epulopiscium has tens of thousands of copies of its complete genome (Mendell et al. 2008), whereas Thiomargarita has around 15,000 copies (Lane and Martin 2010; Angert 2012). In both cases, the genomes are placed next to the plasma membrane, and the internal volume of the cell is metabolically inactive (being a giant vacuole in the case of Thiomargarita). Thus, despite their giant size, their energy per gene is exactly equivalent to that of E. coli (Fig. 2), as predicted by the scaling argument above. In comparison, eukaryotic protists such as Euglena and Amoeba proteus have orders of magnitude more energy per gene (Fig. 2).
How do eukaryotes escape the scaling problem? Their chimeric ancestry is the key. Phylogenomic studies point to an endosymbiosis between an archaeon host cell and a bacterial endosymbiont, the ancestor of the mitochondria, at the origin of eukaryotes (Rivera and Lake 2004; Pisani et al. 2007; Cox et al. 2008; Williams et al. 2012). In an endosymbiosis, a population of bacteria is internalized. Assuming metabolic syntrophy, the endosymbionts provide an immediate advantage to the host cell. In the case of the hydrogen hypothesis, for example, the endosymbionts provide the methanogen host cell with the H2 it needs (Martin and Müller 1998). Having more endosymbionts helps because they produce more H2. Critically, endosymbionts are autonomous cells that replicate themselves and compete with each other. There is a general evolutionary tendency for endosymbionts to lose genes and become genomically more streamlined because the internal environment is relatively constant compared with the outside world (Lane and Archibald 2008). Gene loss is favored because it speeds up replication; hence, the more streamlined endosymbionts prevail. In the case of mitochondria, which began as proteobacteria with several thousand genes, the process of gene loss ultimately reduced them to organelles. The energetic benefits of this process are astonishing (see Table 1). If 5% of unnecessary genes (e.g., those required for cell wall synthesis) are lost from each of 100 endosymbionts, the total rate of ATP synthesis is not affected at all, but the overheads of protein synthesis are reduced by 50 billion ATPs. Over a 24-h life cycle, this is a saving of 580,000 ATPs per second. In terms of respiratory balance (ATP/ADP ratio, rate of free-radical leak, and membrane potential), it is better for the host cell to consume these ATPs in its own growth, or on better supply networks (such as a dynamic cytoskeleton) that provide endosymbionts with the substrates they need.
Table 1.
Potential ATP savings as a result of loss of endosymbiont proteins
| Energy savings | ATP cost | Actin costs | ATP cost |
|---|---|---|---|
| Number of proteins (5% of 4-Mb genome) | 200 | Length of monomer—29 nm | - |
| Endosymbiont genomes | 100 | Monomers per micrometer | 35 |
| Copies of each protein | 2000 | Residues per monomer | 374 |
| Amino acids per protein | 250 | Dimers in actin filament | 2 |
| ATPs per peptide bond | 5 | ATPs per peptide bond | 5 |
| Total per 24-h life cycle | 50 × 109 | Total per micrometer of actin | 131,000 |
| Total per second | 580,000 | Micrometers per second | 4.5 |
Assuming a loss of 5% of genes from a standard bacterial genome in 100 endosymbionts, the total ATP savings amount to 50 billion ATPs over a 24-h life cycle, or 580,000 ATP/sec. If these “savings” were diverted into the assembly of a dynamic actin cytoskeleton, they could, in principle, support the de novo synthesis and assembly of 4.5 µm of actin cytoskeleton per second (an absurd 0.4 m over 24 h).
The Requirement for Core Bioenergetic Genomes
Why could bacteria not internalize their bioenergetic membranes in the same way as mitochondria, overcoming their surface-area-to-volume constraints? Some bacteria, including cyanobacteria, do have extensively invaginated internal membranes, but never scale them up over several orders of magnitude to eukaryotic sizes (Fig. 3). The difference lies in mitochondrial genes. Mitochondria never lost their genomes altogether, except when they also lost the capacity for respiration (van der Giezen 2009). Their core genomes invariably encode the same group of membrane-integral respiratory proteins, plus the ribosomal and transfer RNAs needed to express them locally (Gray et al. 2004). The most compelling explanation for the retention of these genes is that because local transcription and translation enable a swift, proportionate response to shifts in substrate availability, oxygen tension, and free-radical leak, maintaining effective coupling of electron flow to ATP synthesis, as argued by the CoRR (co-location for redox regulation) hypothesis (Allen 1993, 2003). Local transcription is indispensable in the mitochondria, but not in other membrane systems such as the endoplasmic reticulum (ER), because the costs and benefits are much greater, while the time window is far shorter. Mitochondrial membrane potential is 150–200 mV, and the membrane is ∼5 nm thick, giving a field strength of 30 MV/m, equal to a bolt of lightning. The penalty for losing control over this colossal electrical potential is collapsing ATP availability, high rates of free-radical leak, and cell death. Mitochondrial genes are expressed in response to local (matrix) changes in free-radical leak, which modulate redox-sensitive transcription factors such as mt topoisomerase-1 (Lane 2011b), via oxidation of protein thiols (F Boege, pers. comm.). This can correct for imbalances in electron flow in a matter of minutes, where and when needed, staving off cell death for a little longer. Suffocation at the organism level takes minutes; failure to control respiration in the mitochondria carries an equally swift and lethal penalty.
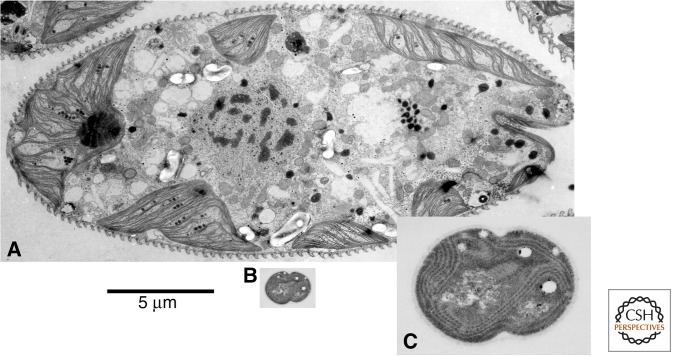
Figure 3.
Size and morphological complexity of eukaryotic alga versus cyanobacterium. Approximately scaled comparison of (A) the eukaryotic alga Euglena with (B) the relatively large complex cyanobacterium Synechocystis, here approximately to scale. Despite its extensive internal thylakoid membranes (magnified in C) and moderate polyploidy (100–200 copies of nucleoid), Synechocystis is approximately 1500 times smaller by volume. (Courtesy of Mark Farmer, University of Georgia.)
If genes are required to control respiration, then large cyanobacteria with thylakoid membranes such as Synechocystis should face similar problems to Epulopiscium and Thiomargarita; and, indeed, despite being much smaller than those behemoths (a few micrometers in diameter), they carry several hundred copies of their full genome, undermining their energy per gene in exactly the same way (Griese et al. 2011). Thus, to generalize: The problem with substantially expanding the surface area of bioenergetic membranes, regardless of whether they are convoluted internal membranes or the plasma membrane itself, is that multiple copies of local bioenergetic genomes are needed to control chemiosmotic coupling. If these genomes are complete copies of the full bacterial genome, then no energetic advantage is gained.
The resemblance of remnant mitochondrial genomes to bacterial plasmids might suggest the use of “respiratory” plasmids positioned next to bioenergetic membranes as another possibility. Plasmids containing the genes for respiration (probably a couple of hundred genes in total) are not known. Apart from the difficulties involved in placing exactly the right genes on a plasmid and positioning multiple giant plasmids next to the requisite membranes, the immediate problem is intracellular transport. There is no advantage to being larger without the intracellular infrastructure to support greater size, but this would take many generations to evolve, during which time the larger bacteria should be selected against. Normal bacteria are not deficient in ATP, so simply producing more ATP has no benefit. This is a very different situation from endosymbiosis, where there is a syntrophic benefit from the beginning. The host cell benefits from endosymbionts, not because it gains more ATP, but because it gains the substrate it needs to grow at all (e.g., H2). The more endosymbionts, the more substrate, and hence the advantage to greater size. Thus, there is an advantage to endosymbionts from the start, but no obvious advantage to being larger purely for the sake of making more ATP.
Endosymbiosis in Prokaryotes Gives Rise to Extreme Genomic Asymmetry
Thus, only endosymbiotic gene loss supports a larger nuclear genome. The total amount of DNA in a cell may not change, but its distribution does. Eukaryotes display an extreme genomic asymmetry, in which hundreds or thousands of tiny mitochondrial genomes support, energetically, a huge nuclear genome, typically three to four orders of magnitude larger than the largest known prokaryotic genomes (Gregory 2005). In contrast, in giant bacteria with extreme polyploidy, such gene loss is not possible. Bacterial nucleoids are not autonomous selective units and not capable of competing among themselves for succession. They remain stable as standard “bacterial” genomes over many generations, each one several megabases in size, unlike mitochondrial DNA, which in vertebrates is reduced to a mere ∼20 kb. Thus, endosymbiosis is necessary for the extreme genomic asymmetry that supports nuclear genome expansion. Reams of DNA is not the same thing as complexity, but it forms the genetic raw material for the proliferation of gene families—3000 new gene families at the origin of eukaryotes (Koonin et al. 2004; Fritz-Laylin et al. 2010)—and the exploration of protein sequence space that underpins eukaryotic genomic and morphological complexity. The evolution of traits like phagocytosis, which require hundreds if not thousands of new genes, must have incurred a massive energetic cost for experimentation with new protein families, a cost that is readily borne by eukaryotes but was never met by bacteria or archaea (Lane and Martin 2010; Lane 2011a). In this regard, it is revealing that phagocytosis might have arisen on three separate occasions in eukaryotes (Yutin et al. 2009).
Endosymbiosis is common in eukaryotes capable of phagocytosis, but extremely rare in prokaryotes, although a single free-living example is known (Wujek 1979), so it is possible. The rare occurrence of endosymbiosis in the absence of phagocytosis is also attested by endosymbionts in fungi, which are no more phagocytic than bacteria (Minerdi et al. 2002). It is therefore unlikely that natural selection, even acting over geological epochs on infinite prokaryotic populations, will give rise to eukaryotic complexity, except by way of a rare endosymbiosis. This could, in part, explain the long delay before eukaryotes arose. Resolving the intimate conflicts between prokaryotic host cell and endosymbiont must have been equally difficult (Blackstone 2013) and could explain the evolution of unique eukaryotic traits.
MITOCHONDRIA AND THE EVOLUTION OF BASAL EUKARYOTIC TRAITS
The retention of mitochondrial genes is significant because it means that all complex eukaryotic cells must balance the requirements of two genomes in every cell. From the beginning, the two genomes were necessary for the chimeric cell to function because each depended on the other for growth—a true symbiosis. The loss of endosymbiont genes and transfer of others to the nucleus helped integrate the two cells into one, but the fact that this process was never—can never be—completed means that eukaryotes remain fundamentally chimeric. The precise selection pressures that mitochondria placed on their host cells must have changed over time, but the dominant leitmotif of eukaryotic evolution, differing radically from anything in bacteria and archaea, is the need for intergenomic coadaptation, generation after generation. I suggest that this requirement offers productive and testable insights into why eukaryotes evolved many unique traits that are never seen in prokaryotes.
The earliest problem produced by endosymbiosis was likely to be a high mutation rate, especially aberrant recombinations deriving from group II mobile self-splicing introns. These retroelements probably derived from the endosymbiont and proliferated through the host cell genome, possibly fragmenting circular chromosomes into linear pieces, before decaying into fixed introns. Introns require splicing out of transcripts, and the spliceosome that does so derives from bacterial group II introns, hinting at a common origin for both spliceosomes and eukaryotic introns (Lambowitz and Zimmerly 2004; Roy and Gilbert 2006). As pointed out by Martin and Koonin (2006), because splicing is much slower than ribosomal translation, unless introns are spliced out before reaching the ribosome, aberrant proteins are translated with intron sequences intact, potentially causing an error catastrophe. Physically separating transcription from translation, by inserting a barrier between the two—the nuclear membrane—solves the problem, giving a strong selection pressure for the evolution of the nucleus in eukaryotes that does not exist in prokaryotes (Martin and Koonin 2006).
The high mutation rate might also have driven the origin of meiosis and sex (Lane 2011a). The maintenance of sex was the “queen” of biological questions in the 20th century, but most work compared sex with clonal reproduction. Without recombination, selection must operate on whole chromosomes. This means that mildly deleterious mutations accumulate, and selective sweeps for particular variants wipe out variation in linked genes throughout a population—selective interference (Keightley and Otto 2006). Recombination breaks up chromosomes and forms new combinations of alleles that can be subject to natural selection. Thus, sex increases variance, which facilitates selection, improving fitness over time as unfit alleles are purged. Sex works best if selection is strong: if there is a high mutation rate, continuously generating unfit alleles, or if there is strong selection for fit alleles, such as those that confer resistance to parasites (Keightley and Otto 2006). The problem is that bacteria are not clonal but maintain fluid chromosomes by LGT. Thus, the origin of syngamy, meiosis, and reciprocal recombination across the entire genome from LGT in bacteria is a more difficult question. Two factors stand out here, which can be modeled explicitly. First, intron invasion should have produced a high mutation rate in early eukaryotes (as well as breaking up chromosomes into linear fragments, causing problems with the normal cell cycle) (Koonin 2009). Second, the acquisition of mitochondria enabled the expansion of nuclear genome size over several orders of magnitude (Lane and Martin 2010). Even if the mutation rate remained constant (e.g., one detrimental mutation in 5000 genes per generation), expanding genome size to 20,000 genes causes problems that might not be soluble by LGT, which is piecemeal and nonreciprocal. Another intriguing possibility is that systematic recombination of nuclear genes could aid their coadaptation to rapidly mutating mitochondrial genes by minimizing selective interference.
The Requirement for Mitonuclear Coadaptation
The requirement for mitonuclear coadaptation is central to eukaryotic evolution. Rapidly mutating mitochondrial genes cannot be purged by sex because each selective unit—each host cell—contains a mixed population of mitochondria. But similar principles apply. Just as sex increases the variance in a population, so does the uniparental inheritance of mitochondria. Zygotes that receive mitochondria from only one of the two fusing gametes are more likely to have a clonal population of mitochondria (especially if there is a sampling of the mitochondrial population during gamete formation). Different zygotes then have different clones of mitochondria, increasing the variance across the population. This facilitates selection for the best mitochondria, at the level of the host cell, in much the same way as sex (Hadjivasiliou et al. 2012, 2013). In contrast, biparental inheritance of mitochondria decreases variance between zygotes and interferes with selection (Hadjivasiliou et al. 2012, 2013). This is one of the deepest distinctions between the two sexes, even in isogametic algae and fungi: One sex passes on mitochondria, and the other does not (Hurst and Hamilton 1992). The subsequent evolution of anisogamy, in which oocytes pass on a large clonal population of mitochondria while sperm have their tiny populations of mitochondria destroyed, is the gateway to true sexes—again, unknown in bacteria or archaea.
The existence of two sexes facilitates selection for mitochondrial integrity, but also exaggerates the disparity in the tempo and mode of evolution of mitochondrial and nuclear genomes. Nuclear genes are recombined by sex every generation (at least in most multicellular eukaryotes). Recombination, in turn, facilitates selection against mildly deleterious mutations, which can slow the evolution rate. In contrast, mitochondrial genes are inherited asexually down the maternal line. The evolution rate of mitochondrial genes is 20- to 40-fold faster than the nuclear mean in animals and other groups such as fungi (Pesole et al. 1999; Nabholz et al. 2009). Although the nucleotide substitution rate is substantially lower in plant mitochondria, their genomes are more prone to recombinatorial errors; hence, the overall mutation rate can still be high (Galtier 2011). Both mitochondrial and nuclear genomes encode key subunits of the respiratory chain, which must interact with each other with nanoscopic precision for effective electron transfer and ATP synthesis (Lane 2011b,c). All biochemistry and physiology depend on ATP; hence, a failure to interact correctly affects growth and development, fertility, and survival. Nothing could be more central to fitness. Yet these critical respiratory proteins are encoded by two genomes that are inherited in contrasting ways and that diverge from each other sharply. This bizarre situation is little short of outrageous but is dictated by the requirement for local mitochondrial genomes, combined with a tendency to transfer any genes to the nucleus that can be physically moved. Thus, the mosaic respiratory chain is no freak accident but a necessary feature of all complex eukaryotic cells—and if the thesis in this article is correct, all complex life anywhere in the universe.
What happens if the two genomes do not match properly and electrons fail to flow freely down the mosaic respiratory chains? An impedance of electron flow means that the respiratory complexes become more highly reduced, as electrons cannot flow onward. Complexes I and III are replete with reduced low-potential FeS centers that react readily with oxygen. This produces oxygen free radicals, notably the superoxide radical, O2•−, and other reactive oxygen species. These oxidize membrane lipids such as cardiolipin, which, in turn, release cytochrome c into the intermembrane space and cytosol. At the same time, ATP synthesis dwindles with electron flow. This triad of factors—a fall in ATP levels, free-radical leak, and cytochrome c release—is the classic trigger of programmed cell death in animals and plants and was greeted with “general stupefaction” (Hengartner 1998) when discovered in the mid 1990s. Why would a respectable respiratory protein lead a double life as the harbinger of death? From the perspective of eukaryotic evolution, however, it makes perfect sense (Lane 2011b,c). Given the evolutionary divergence of the mitochondrial and nuclear genomes, combined with the necessity for them to work together closely, selection for mitonuclear coadaptation is necessary. Apoptosis is a form of functional selection against cells, and by extension whole organisms, with incompatible mitochondrial and nuclear genomes (Lane 2011c).
The strength of selection for mitonuclear coadaptation in each generation is unknown, but there is abundant evidence of selection for coadaptation over evolutionary time in many eukaryotic groups including animals, plants, and fungi. This evidence includes a concordance between the evolutionary rates of mitochondrial and nuclear genes encoding respiratory-chain subunits (Blier et al 2001; Mishmar et al. 2006; Gershoni et al. 2010); a decline in respiratory function in nuclear–cytoplasmic hybrids (cybrids) (Barrientos et al. 1998; Dey et al. 2000; Yamaoka et al. 2000); and hybrid breakdown in introgressed populations caused by mitonuclear incompatibilities (Burton et al. 2006; Ellison and Burton 2008; Burton and Barreto 2012). The rapid rates of mitochondrial divergence in some species, notably copepods—some 50-fold faster than the nuclear mean—means that mitonuclear incompatibility is the dominant factor in hybrid breakdown (Burton and Barreto 2012). How commonly this contributes to the origin of species is unknown, but given the generality of fast mitochondrial evolution rates in animals, mitonuclear incompatibilities could play an important role (Gershoni et al. 2009; Lane 2009; Chou and Leu 2010; Burton and Barreto 2012).
Evolutionary Trade-Offs Relating to Respiratory Function
Theoretically, the quality of mitonuclear coadaptation ought to depend on aerobic requirements. Organisms with high aerobic demands, such as powered flight, need fast electron flow and efficient respiration, and hence excellent mitonuclear coadaptation (Lane 2011c). Mitochondrial and nuclear genomes that are even slightly incompatible should therefore trigger apoptosis, eliminating defective embryos early in development. Strong purifying selection on mitonuclear incompatibility is predicted to constrain the evolution of mitochondrial DNA, which is certainly the case in birds (Nabholz et al. 2009). Little is known about how such selection operates, but it is likely to involve mitochondrial free-radical leak.
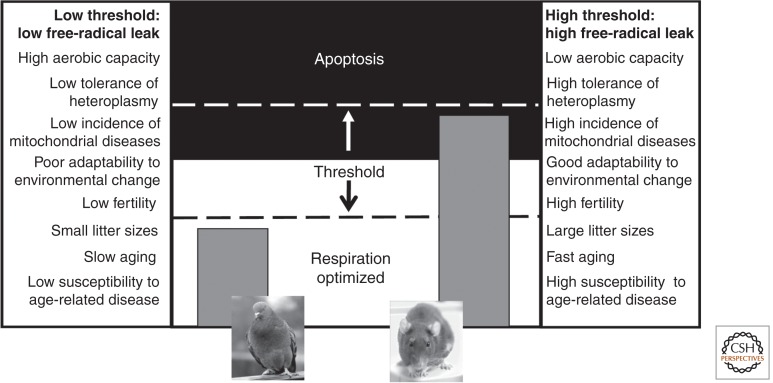
Species with high aerobic requirements would benefit from a lower apoptotic threshold; that is, a moderate free-radical leak signals a mild incompatibility, triggering apoptosis (Lane 2011c). High rates of apoptosis lower fertility because embryos with mildly incompatible genomes are terminated before term. Conversely, species with low aerobic requirements (such as rats) should tolerate much higher rates of free-radical leak before triggering apoptosis, giving them a high apoptotic threshold. The benefit in this case is much greater fertility because fewer embryos need to be terminated. More variation in mitochondrial DNA could also be tolerated, potentially facilitating adaptation to different climates and diets (Wallace 2013), albeit at greater risk of mitochondrial diseases. These simple considerations imply that there is a trade-off between fertility, adaptability, and aerobic capacity (Fig. 4). Insofar as a low free-radical leak is linked with a long life span and a lower risk of age-related diseases, the stringency of selection for mitonuclear coadaptation could influence rates of aging and susceptibility to age-related diseases (Lane 2011b).
Figure 4.
The apoptotic threshold is variable and determines fitness. The threshold for apoptosis can be raised or lowered in principle (center panel). If the threshold is low (left) even a small increase in free-radical leak triggers apoptosis. A low tolerance for free-radical leaks selects for good mitonuclear coadaptation, with predicted costs and benefits listed in the left-hand panel, correspondingly to a K reproductive strategy. High tolerance for free-radical leaks raises the threshold for apoptosis. This relaxes selection for mitonuclear coadaptation, corresponding to an r reproductive strategy, with the costs and benefits listed in the right-hand panel.
CONCLUDING REMARKS
Much is known about the mechanisms of evolution and natural selection, but this knowledge gives little insight into the actual trajectory of life on Earth. A consideration of membrane bioenergetics offers striking insights into why life arose so early here, why all life on Earth is chemiosmotic, why prokaryotes stalled in their morphological complexity, why eukaryotes arose only once in 4 billion years and then evolved a large number of traits that have puzzled biologists for a century, and why these traits are never seen in prokaryotes. This perspective is as yet a tentative outline, but makes several predictions about the traits of both prokaryotic and eukaryotic life that can be tested by mathematical modeling and experiment. If true, energetic constraints dictated the evolution of life as much as genetics. These principles should apply equally to life elsewhere in the universe, making the bold if unsettling prediction that any complex alien life will share many traits with life on Earth, from mitochondria to sex, for the very same reasons.
ACKNOWLEDGMENTS
I thank Bill Martin, University of Düsseldorf, for numerous insights, discussions, shared enthusiasms, and joint authorship of several papers on which this review is based; and Andrew Pomiankowski at UCL for many stimulating discussions, and common purpose in developing population genetic models of eukaryotic evolution. I thank Don Braben and David Price at UCL for their support in the UCL Provost’s Venture Research Fellowship, which made this work possible. I am grateful to the Leverhulme Trust for supporting my research on the origin of life.
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
- Allen JF 1993. Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J Theor Biol 165: 609–631 [DOI] [PubMed] [Google Scholar]
- Allen JF 2003. The function of genomes in bioenergetic organelles. Phil Trans R Soc Lond B 358: 19–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend JP, McCollom TM 2009. Energetics of biomolecule synthesis on early Earth. In Chemical evolution II: From the origins of life to modern society (ed. Zaikowski L, et al. ), pp. 63–94 American Chemical Society, Washington, DC [Google Scholar]
- Amend JP, LaRowe DE, McCollom TM, Shock EL 2013. The energetics of organic synthesis inside and outside the cell. Phil Trans R Soc Lond B 368: 20120255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Kurland CG 1999. Origins of mitochondria and hydrogenosomes. Curr Opin Microbiol 2: 535–541 [DOI] [PubMed] [Google Scholar]
- Angert ER 2012. DNA replication and genomic architecture of very large bacteria. Annu Rev Microbiol 66: 197–212 [DOI] [PubMed] [Google Scholar]
- Archibald JM 2011. Origin of eukaryotic cells: 40 years on. Symbiosis 54: 69–86 [Google Scholar]
- Arndt N, Nisbet E 2012. Processes on the young Earth and the habitats of early life. Annu Rev Earth Planet Sci 40: 521–549 [Google Scholar]
- Baaske P, Weinert FM, Duhr S, Lemke KH, Russell MJ, Braun D 2007. Extreme accumulation of nucleotides in simulated hydrothermal pore systems. Proc Nat Acad Sci 104: 9346–9351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradaran R, Berrisford JM, Minhas GS, Sazanov LA 2013. Crystal structure of the entire respiratory complex. Nature 494: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Kenyon L, Moraes CT 1998. Human xenomitochondrial cybrids. Cellular models of mitochondrial complex I deficiency. J Biol Chem 273: 14210–14217 [DOI] [PubMed] [Google Scholar]
- Baymann F, Lebrun E, Brugna M, Schoepp-Cothenet B, Giudici-Orticoni M-T, Nitschke W 2003. The redox protein construction kit: Pre-LUCA evolution of energy conserving enzymes. Phil Trans R Soc Lond B 358: 267–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeño-Tárraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417: 141–147 [DOI] [PubMed] [Google Scholar]
- Blackstone NW 2013. Why did eukaryotes evolve only once? Genetic and energetic aspects of conflict and conflict mediation. Phil Trans R Soc Lond B 368: 20120266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blier P, Dufresne F, Burton RS 2001. Natural selection and the evolution of mtDNA-encoded peptides: Evidence for intergenomic co-adaptation. Trends Genet 17: 400–406 [DOI] [PubMed] [Google Scholar]
- Buckel W, Thauer RK 2013. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochem Biophys Acta Bioenergetics 1827: 94–113 [DOI] [PubMed] [Google Scholar]
- Budin I, Bruckner RJ, Szostak JW 2009. Formation of protocell-like vesicles in a thermal diffusion column. J Am Chem Soc 131: 9628–9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RS, Barreto FS 2012. A disproportionate role for mtDNA in Dobzhansky–Muller incompatibilities? Mol Ecol 21: 4942–4957 [DOI] [PubMed] [Google Scholar]
- Burton RS, Ellison CK, Harrison JS 2006. The sorry state of F2 hybrids: Consequences of rapid mitochondrial DNA evolution in allopatric populations. Am Nat 168: S14–S24 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 1987. Eukaryotes with no mitochondria. Nature 326: 332–333 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T 1989. Archaebacteria and Archezoa. Nature 339: 100–101 [DOI] [PubMed] [Google Scholar]
- Chou J-Y, Leu J-Y 2010. Speciation through cytonuclear incompatibility: Insights from yeast and implications for higher eukaryotes. BioEssays 32: 401–411 [DOI] [PubMed] [Google Scholar]
- Cox CJ, Foster PG, Hirt RP, Harris SR, Embley TM 2008. The archaebacterial origin of eukaryotes. Proc Natl Acad Sci 105: 20356–20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidov Y, Jurkevitch E 2009. Predation between prokaryotes and the origin of eukaryotes. BioEssays 31: 748–757 [DOI] [PubMed] [Google Scholar]
- de Duve C 2007. The origin of eukaryotes: A reappraisal. Nat Rev Genet 8: 395–403 [DOI] [PubMed] [Google Scholar]
- Dey R, Barrientos A, Moraes CT 2000. Functional constraints of nuclear–mitochondrial DNA interactions in xenomitochondrial rodent cell lines. J Biol Chem 275: 31520–31527 [DOI] [PubMed] [Google Scholar]
- Ellison CK, Burton RS 2008. Interpopulation hybrid breakdown maps to the mitochondrial genome. Evolution 62: 631–638 [DOI] [PubMed] [Google Scholar]
- Embley TM, Martin W 2006. Eukaryotic evolution, changes and challenges. Nature 440: 623–630 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, et al. 2010. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell 140: 631–642 [DOI] [PubMed] [Google Scholar]
- Galtier N 2011. The intriguing evolutionary dynamics of plant mitochondrial DNA. BMC Biol 9: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershoni M, Templeton AR, Mishmar D 2009. Mitochondrial biogenesis as a major motive force of speciation. BioEssays 31: 642–650 [DOI] [PubMed] [Google Scholar]
- Gershoni M, Fuchs A, Shani N, Fridman Y, Corral-Debrinski M, Aharoni A, Frishman D, Mishmar D 2010. Coevolution predicts direct interactions between mtDNA-encoded and nDNA-encoded subunits of oxidative phosphorylation complex I. J Mol Biol 404: 158–171 [DOI] [PubMed] [Google Scholar]
- Gray MW, Lang BF, Burger G 2004. Mitochondria of protists. Annu Rev Genet 38: 477–524 [DOI] [PubMed] [Google Scholar]
- Gregory TR 2005. Synergy between sequence and size in large-scale genomics. Nat Rev Genet 6: 699–708 [DOI] [PubMed] [Google Scholar]
- Griese M, Lange C, Soppa J 2011. Ploidy in cyanobacteria. FEMS Microbiol Lett 323: 124–31 [DOI] [PubMed] [Google Scholar]
- Gross J, Bhattacharya D 2010. Uniting sex and eukaryote origins in an emerging oxygenic world. Biol Direct 5: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N 2012. Selection for mitonuclear coadaptation could favour the evolution of two sexes. Proc R Soc B 279: 1865–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjivasiliou Z, Lane N, Seymour R, Pomiankowski A 2013. Dynamics of mitochondrial inheritance in the evolution of binary mating types and two sexes. Proc R Soc B 280: 20131920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold FM 1986. The vital force: A study of bioenergetics. Freeman, New York [Google Scholar]
- Hengartner MO 1998. Death cycle and Swiss army knives. Nature 391: 441–442 [DOI] [PubMed] [Google Scholar]
- Hurst LD, Hamilton WD 1992. Cytoplasmic fusion and the nature of sexes. Proc R Soc Lond B 247: 189–194 [Google Scholar]
- Kaster A-K, Moll J, Parey K, Thauer RK 2011. Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic Archaea. Proc Natl Acad Sci 108: 2981–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ 1998. A kingdom’s progress: Archezoa and the origin of eukaryotes. BioEssays 20: 87–95 [Google Scholar]
- Keightley PD, Otto SP 2006. Selective interference among deleterious mutations favours sex and recombination in finite populations regardless of the nature of epistasis. Nature 443: 89–92 [DOI] [PubMed] [Google Scholar]
- Knoll AH, Javaux EJ, Hewitt D, Cohen P 2006. Eukaryotic organisms in Proterozoic oceans. Phil Trans Roy Soc B 361: 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E 2009. Intron-dominated genomes of early ancestors of eukaryotes. J Heredity 100: 618–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV 2010. The origin and early evolution of eukaryotes in the light of phylogenomics. Genome Biol 11: 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makrova KS, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, et al. 2004. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol 5: R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2004. Mobile group II introns. Ann Rev Genet 38: 1–35 [DOI] [PubMed] [Google Scholar]
- Lane N 2009. On the origin of barcodes. Nature 462: 272–274 [DOI] [PubMed] [Google Scholar]
- Lane N 2010. Why are cells powered by proton gradients? Nature Edu 3: 18 [Google Scholar]
- Lane N 2011a. Energetics and genetics across the prokaryote–eukaryote divide. Biol Direct 6: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane N 2011b. Mitonuclear match: Optimizing fitness and fertility over generations drives ageing within generations. BioEssays 33: 860–869 [DOI] [PubMed] [Google Scholar]
- Lane N 2011c. The costs of breathing. Science 334: 184–185 [DOI] [PubMed] [Google Scholar]
- Lane CE, Archibald JM 2008. The eukaryotic tree of life: Endosymbiosis takes its TOL. Trends Ecol Evol 23: 268–275 [DOI] [PubMed] [Google Scholar]
- Lane N, Martin W 2010. The energetics of genome complexity. Nature 467: 929–934 [DOI] [PubMed] [Google Scholar]
- Lane N, Martin W 2012. The origin of membrane bioenergetics. Cell 151: 1406–1416 [DOI] [PubMed] [Google Scholar]
- Lane N, Allen JF, Martin W 2010. How did LUCA make a living? Chemiosmosis in the origin of life. BioEssays 32: 271–280 [DOI] [PubMed] [Google Scholar]
- Li F, Hinderberger J, Seedorf H, Zhang J, Buckel W, Thauer RK 2008. Coupled ferredoxin and crotonyl coenzyme A (CoA) reduction with NADH catalyzed by the butyryl-CoA dehydrogenase/Etf complex from Clostridium kluyveri. J Bacteriol 190: 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay MR, Webb RI, Strous M, Jetten MS, Butler MK, Forde RJ, Fuerst JA 2001. Cell compartmentalisation in planctomycetes: Novel types of structural organisation for the bacterial cell. Arch Microbiol 175: 413–429 [DOI] [PubMed] [Google Scholar]
- Lonhienne TG, Sagulenko E, Webb RI, Lee KC, Franke J, Devos DP, Nouwens A, Carroll BJ, Fuerst JA 2010. Endocytosis-like protein uptake in the bacterium Gemmata obscuriglobus. Proc Natl Acad Sci 107: 12883–12888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden BEH 2000. Tetrahydrofolate and tetrahydromethanopterin compared: Functionally distinct carriers in C1 metabolism. Biochem J 350: 609–629 [PMC free article] [PubMed] [Google Scholar]
- Martin W, Koonin EV 2006. Introns and the origin of nucleus–cytosol compartmentalization. Nature 440: 41–45 [DOI] [PubMed] [Google Scholar]
- Martin W, Müller M 1998. The hydrogen hypothesis for the first eukaryote. Nature 392: 37–41 [DOI] [PubMed] [Google Scholar]
- Martin W, Russell M 2003. On the origins of cells: A hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Phil Trans R Soc Lond B 358: 59–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Russell MJ 2007. On the origin of biochemistry at an alkaline hydrothermal vent. Phil Trans R Soc Lond B 367: 1887–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Baross J, Kelley D, Russell MJ 2008. Hydrothermal vents and the origin of life. Nat Rev Microbiol 6: 805–814 [DOI] [PubMed] [Google Scholar]
- Mast CB, Schink S, Gerland U, Braun D 2013. Escalation of polymerization in a thermal gradient. Proc Natl Acad Sci 110: 8030–8035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JE, Clements KD, Choat JH, Angert ER 2008. Extreme polyploidy in a large bacterium. Proc Natl Acad Sci 105: 6730–6734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minerdi D, Bianciotto V, Bonfante P 2002. Endosymbiotic bacteria in mycorrhizal fungi: From their morphology to genomic sequences. Plant Soil 244: 211–219 [Google Scholar]
- Mishmar D, Ruiz-Pesini E, Mondragon-Palomino M, Procaccio V, Gaut B, Wallace DC 2006. Adaptive selection of mitochondrial complex I subunits during primate radiation. Gene 378: 11–18 [DOI] [PubMed] [Google Scholar]
- Mitchell P 1957. The origin of life and the formation and organising functions of natural membranes. In Proceedings of the First International Symposium on the Origin of Life on the Earth (ed. Oparin AI, et al. ), pp. 229–234 Academy of Sciences, Moscow [Google Scholar]
- Mitchell P 1961. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191: 144–148 [DOI] [PubMed] [Google Scholar]
- Moran NA 2007. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci 104: 8627–8633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkidjanian AY, Bychkov AY, Dibrova DV, Galperin MY, Koonin EV 2012. Origin of first cells at terrestrial, anoxic geothermal fields. Proc Natl Acad Sci 109: E821–E830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Mentel M, van Hellemond JJ, Henze K, Woehle C, Gould SB, Yu RY, van der Giezen M, Tielens AG, Martin W 2012. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev 76: 444–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabholz B, Glemin S, Galtier N 2009. The erratic mitochondrial clock: Variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol Biol 9: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahra DJG, Knoll AH, Katz LA 2011. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci 108: 13624–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G, Gissi C, De Chirico A, Saccone C 1999. Nucleotide substitution rate of mammalian mitochondrial genomes. J Mol Evol 48: 427–434 [DOI] [PubMed] [Google Scholar]
- Pinevich AV 1997. Intracytoplasmic membrane structures in bacteria. Endocyt Cell Res 12: 9–40 [Google Scholar]
- Pisani D, Cotton JA, McInerney JO 2007. Supertrees disentangle the chimeric origin of eukaryotic genomes. Mol Biol Evol 24: 1752–1760 [DOI] [PubMed] [Google Scholar]
- Pohorille A, Deamer DW 2009. Self-assembly and function of primitive cell membranes. Res Microbiol 160: 449–456 [DOI] [PubMed] [Google Scholar]
- Powner MW, Gerland B, Sutherland JD 2009. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature 459: 239–242 [DOI] [PubMed] [Google Scholar]
- Rivera MC, Lake JA 2004. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature 431: 152–155 [DOI] [PubMed] [Google Scholar]
- Robinson NP, Bell SD 2007. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc Natl Acad Sci 104: 5806–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W 2006. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat Rev Genet 7: 211–221 [DOI] [PubMed] [Google Scholar]
- Russell MJ, Hall AJ 1997. The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J Geol Soc Lond 154: 377–402 [DOI] [PubMed] [Google Scholar]
- Russell MJ, Martin W 2004. The rocky roots of the acetyl-CoA pathway. Trends Biochem Sci 29: 358–363 [DOI] [PubMed] [Google Scholar]
- Russell MJ, Daniel RM, Hall A 1993. On the emergence of life via catalytic iron-sulphide membranes. Terra Nova 5: 343–347 [Google Scholar]
- Russell MJ, Nitschke W, Branscomb E 2013. The inevitable journey to being. Phil Trans R Soc Lond B 368: 20120254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagan L 1967. On the origin of mitosing cells. J Theoret Biol 14: 225–274 [DOI] [PubMed] [Google Scholar]
- Schoepp-Cothenet B, van Lis R, Atteia A, Baymann F, Capowiez L, Ducluzeau A-L, Duval S, ten Brink F, Russell MJ, Nitschke W 2013. On the universal core of bioenergetics. BBA Bioenergetics 1827: 79–93 [DOI] [PubMed] [Google Scholar]
- Schulz HN, Jorgensen BB 2001. Big bacteria. Annu Rev Microbiol 55: 105–137 [DOI] [PubMed] [Google Scholar]
- Shock EL, McCollom TM, Schulte MD 1998. The emergence of metabolism from within hydrothermal systems. In Thermophiles: The keys to molecular evolution and the origin of life (ed. Wiegel J, Adams MWW), pp. 59–76 Taylor & Francis, London [Google Scholar]
- Simon DM, Zimmerly S 2008. A diversity of uncharacterized retroelements in bacteria. Nucleic Acids Res 36: 7219–7229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleep NH, Bird DK, Pope EC 2011. Serpentinite and the dawn of life. Phil Trans R Soc Lond B 366: 2857–2869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JM, Smith NH, O’Rourke M, Spratt BG 1993. How clonal are bacteria? Proc Natl Acad Sci 90: 4384–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R 2008. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat Rev Microbiol 6: 579–591 [DOI] [PubMed] [Google Scholar]
- van der Giezen M 2009. Hydrogenosomes and mitosomes: Conservation and evolution of functions. J Eukaryot Microbiol 56: 221–231 [DOI] [PubMed] [Google Scholar]
- van der Giezen M, Lenton TM 2012. The rise of oxygen and complex life. Eukaryot Microbiol 59: 111–113 [DOI] [PubMed] [Google Scholar]
- Vats P, Yu J, Rothfield L 2009. The dynamic nature of the bacterial cytoskeleton. Cell Mol Life Sci 66: 3353–3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Dohlen CD, Kohler S, Alsop ST, McManus WR 2001. Mealybug β-proteobacterial symbionts contain γ-proteobacterial symbionts. Nature 412: 433–436 [DOI] [PubMed] [Google Scholar]
- Vopalensky P, Kozmic Z 2009. Eye evolution: Common use and independent recruitment of genetic components. Phil Trans R Soc Lond B 364: 2819–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtershäuser G 1988. Pyrite formation, the first energy source for life: A hypothesis. Syst Appl Microbiol 10: 207–210 [Google Scholar]
- Wallace DC 2013. Bioenergetics in human evolution and disease: Implications for the origins of biological complexity and the missing genetic variation of common diseases. Phil Trans R Soc Lond B 368: 20120267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters CM, Bassler BL 2005. Quorum sensing: Cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol 21: 319–346 [DOI] [PubMed] [Google Scholar]
- Williams TA, Foster PG, Nye TMW, Cox CJ, Embley TM 2012. A congruent phylogenomic signal places eukaryotes within the archaea. Proc R Soc Lond B 279: 4870–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wujek DE 1979. Intracellular bacteria in the blue-green-alga Pleurocapsa minor. Trans Am Microsc Soc 98: 143–145 [Google Scholar]
- Yamaoka M, Isobe K, Shitara H, Yonekawa H, Miyabayashi S, Hayashi JI 2000. Complete repopulation of mouse mitochondrial DNA-less cells with rat mitochondrial DNA restores mitochondrial translation but not mitochondrial respiratory function. Genetics 155: 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Wolf MY, Wolf YI, Koonin EV 2009. The origins of phagocytosis and eukaryogenesis. Biol Direct 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]