Abstract
Hepatitis C virus (HCV) infection affects more than 170 million people. The high genetic variability of HCV and the rapid development of drug-resistant strains are driving the urgent search for new direct-acting antiviral agents. A new class of agents has recently been developed that are believed to target the HCV protein NS5A although precisely where they interact and how they affect function is unknown. Here we describe an in vitro assay based on microscale thermophoresis and demonstrate that two clinically relevant inhibitors bind tightly to NS5A domain 1 and inhibit RNA binding. Conversely, RNA binding inhibits compound binding. The compounds bind more weakly to known resistance mutants L31V and Y93H. The compounds do not affect NS5A dimerisation. We propose that current NS5A inhibitors act by favouring a dimeric structure of NS5A that does not bind RNA.
Hepatitis C virus (HCV) infection, a major cause of acute hepatitis and chronic liver disease, is a widespread problem (http://www.who.int). Although the current standard of care, a combination of pegylated interferon, ribavirin and a protease inhibitor1, has improved efficacy over earlier treatments2,3 it continues to suffer from limitations including long length of treatment and additional side effects1,2,3. An entirely new class of agents exemplified by BMS-790052 (daclatasvir, Bristol-Myers Squibb, Fig. 1a)4 and AZD7295 (AstraZeneca, Fig. 1b)5 are amongst the most potent antivirals ever discovered with broad activity against HCV genotypes and subtypes. Identified from cell-based HCV genotype 1b replicon assays they have been described as NS5A inhibitors on the basis of resistance mutants4,6, pull-down assays4,7 and chemical cross-linking7, although there is no confirmed mechanism of action. It is possible that the inhibitors disrupt the interaction of NS5A with other proteins; current estimates put the NS5A interactome at 132 human proteins, demonstrating the importance and variety of protein interactions in NS5A functions8.
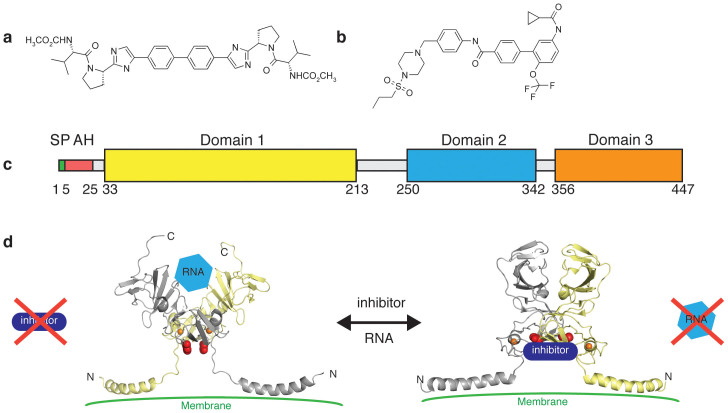
Figure 1. Hepatitis C NS5A and inhibitor structures.
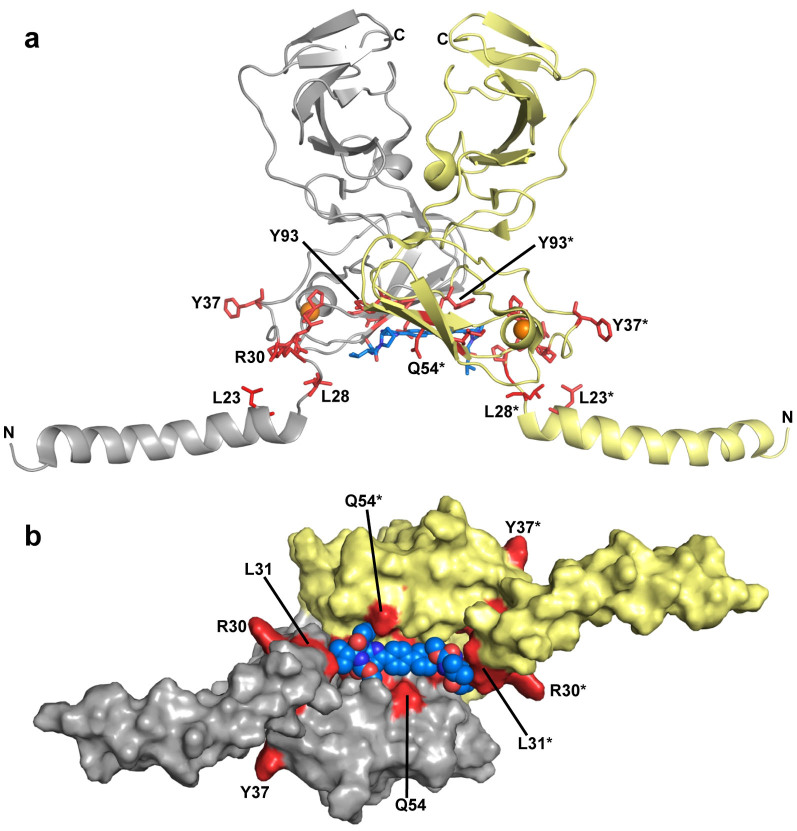
Chemical structures of (a) BMS-790052 and (b) AZD7295. (c) NS5A domain structure. SP denotes the signal peptide and AH is an N-terminal amphipathic helix. (d) Proposed mechanism of action of NS5A inhibition. Compound binding to NS5A domain 1 prevents RNA binding. Models of observed NS5A1–191 dimer conformations (based on crystal structures PDB ID: 1ZH110 and 3FQQ11 and the NMR structure PDB ID: 1R7G19) displayed as cartoon and coloured by monomer. The location of Y93 in each monomer is indicated by red cpk and the Zn2+ ions are orange spheres.
In HCV infected cells, NS5A is produced as part of the viral polyprotein. Once cleaved from the polyprotein, NS5A localises to membranes where it binds to the 3′-end of newly synthesised viral RNA and participates in genome replication, in part through interactions with the viral RNA-dependent RNA polymerase NS5B2,8,9,10. NS5A is a 447 residue RNA binding phosphoprotein consisting of three cytoplasmic domains (Fig. 1c). Across the HCV genotypes, domain 1 is well conserved with two crystal structures of domain 1 revealing the same zinc-binding monomer conformation but different dimer organisations (Fig. 1d)10,11. Whether either homodimer assembly is physiologically relevant has been the subject of debate7,11,12,13,14,15. Domains 2 and 3, however, are more varied in sequence16 and, in isolation, are disordered in solution16,17,18. Domain 1 is preceded by an N-terminal amphipathic α-helix that is thought to anchor NS5A to the endoplasmic reticulum-derived membrane10,19.
Here we describe the first in vitro NS5A binding assay using a recently developed technique called microscale thermophoresis (MST)20, a fluorescence-based method, which can measure the binding of small molecules to proteins in solution. We show that BMS-790052 and AZD7295 bind tightly and specifically to domain 1 of NS5A. Furthermore, we show that both compounds inhibit RNA binding to NS5A, believed to be necessary for HCV replication, and bind significantly more weakly to the common resistance mutants Y93H and L31V. We demonstrate that the compounds bind to NS5A dimers and describe computational models that explain their mode of inhibition. The in vitro protein assay described here will be a valuable tool in the development of next generation NS5A inhibitors for the treatment of HCV infection.
Results
In vitro NS5A binding assay
We have used MST20 to develop an in vitro NS5A binding assay. MST data arise from the spontaneous movement of fluorescently labelled protein in a glass capillary tube along induced microscopic temperature gradients generated by an infra-red laser. The thermophoretic movement can be monitored by measuring the local changes in fluorescence intensity emitted from the labelled protein. The rate of this thermophoresis is dependent on a number of factors including the solvent structure around a molecule, and crucially is altered by the binding of a ligand. Measuring the thermophoretic behaviour of a protein in the presence of differing ligand concentrations by MST allows quantitative analysis of molecular interactions in solution on the microlitre scale. The MST technique has previously been used to investigate protein-protein21,22, small organic molecule-protein23,24,25 and antibody-protein interactions26.
MST was initially used to confirm the interaction of short, single stranded RNA [consisting of four, five or ten uridines (4rU, 5rU or 10rU respectively) or five adenines (5rA)] or double stranded 5rU (ds(5rU)) with NS5A domain 1. In this study we explored binding to two different NS5A constructs: HCV genotype 1b (con1) residues 33–202 (NS5A33–202) and residues 26–202 (NS5A26–202). The majority of studies in this work were performed with the former construct, which expresses at relatively high levels and results in a well behaved protein that has been used previously in crystallographic studies11. Extended constructs have been reported by others to be very difficult to handle due to the stickiness of the amphipathic N-terminal helix and proteolytic susceptibility of domains 2 and 3. The NS5A26–202 construct did not express well and produced a protein that was prone to aggregation and thus difficult to handle. This construct was used to explore one of the documented resistance mutants (L31V) to BMS-790052.
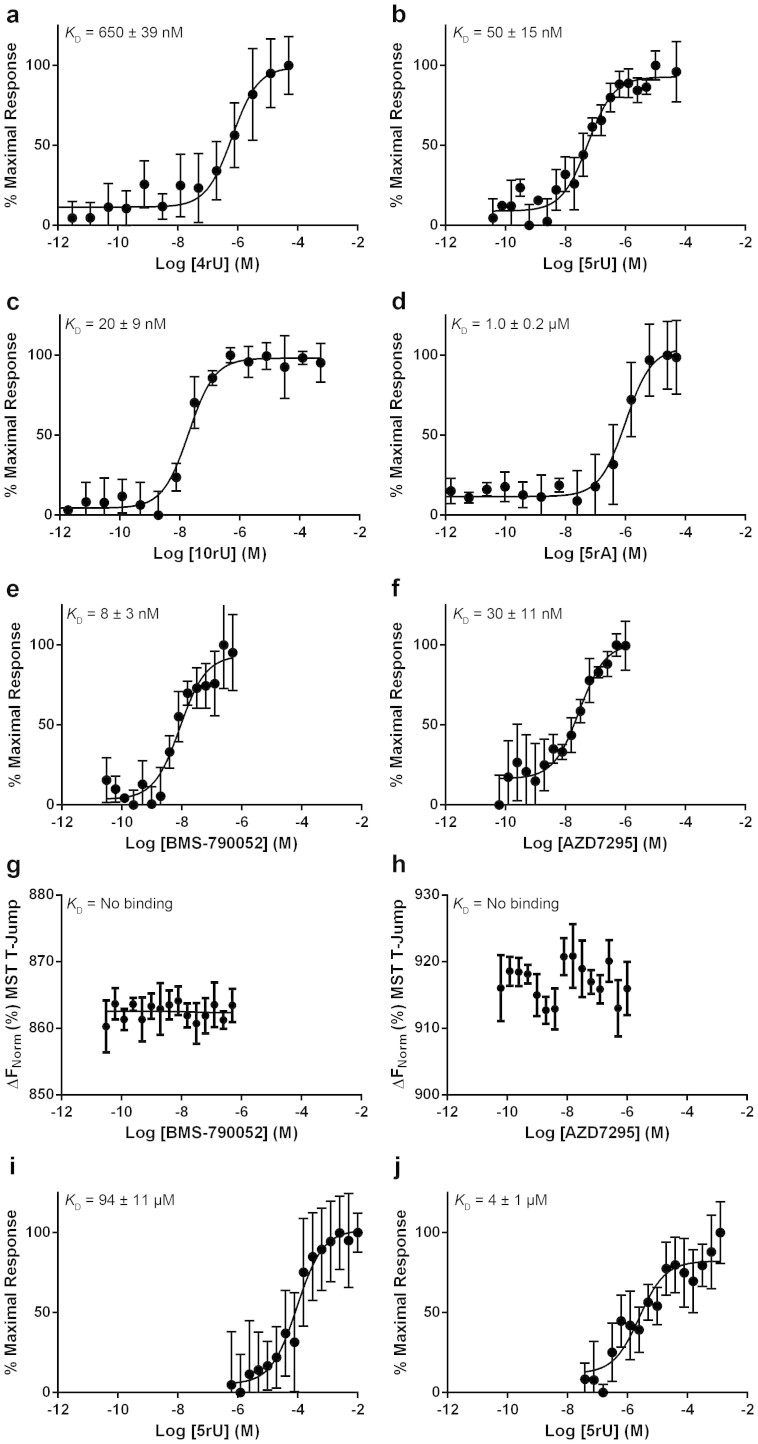
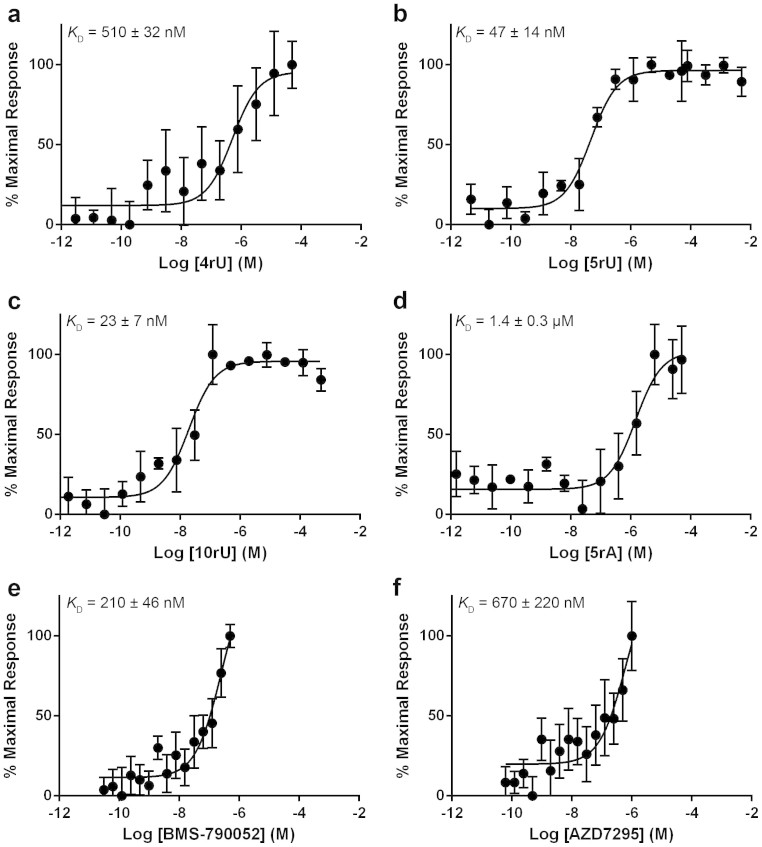
The MST data show that NS5A33–202 binds 4rU with a KD of 650 ± 39, 5rU with a KD of 50 ± 15, 10rU with a KD of 20 ± 9 nM and 5rA with a KD of 1.0 ± 0.2 μM (Fig. 2a–d). There was no statistically significant difference between the MST data obtained for RNA binding to the NS5A26–202 (Fig. 3a–d, p values of 0.6, 0.9, 0.9 and 0.3 for 4rU, 5rU, 10rU and 5rA, respectively) and NS5A33–202 constructs. The MST data are consistent with published filter-binding and fluorescence polarisation assays for genotype 1b (con1) NS5A (KD of 600 ± 200 nM for 4rU, 100 ± 20 nM for 5rU, 100 ± 10 nM for 10rU and 1000 ± 200 nM for 5rA)14. There was no detectable binding to double stranded 5rU by any of the NS5A constructs.
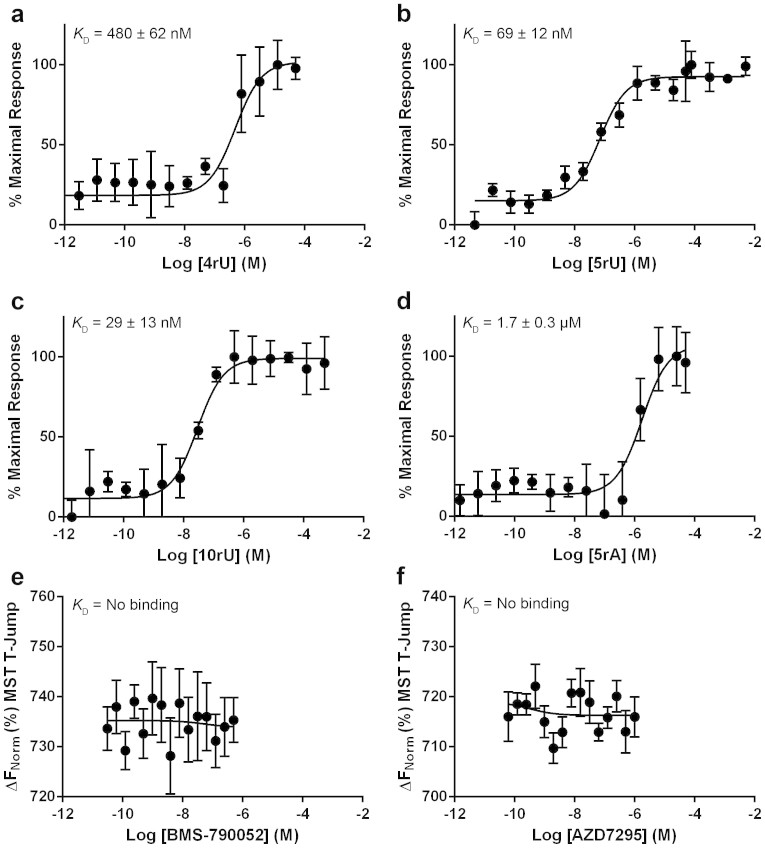
Figure 2. Binding to domain 1 NS5A33–202 genotype 1b (con1).
Binding of (a) 4rU, (b) 5rU, (c) 10rU, (d) 5rA, (e) BMS-790052, (f) AZD7295, (g) BMS-790052 in the presence of 5-fold excess 5rU, (h) AZD7295 in the presence of 5-fold excess 5rU, (i) 5rU in the presence of 0.25 μM BMS-790052, or (j) 5rU in the presence of 0.5 μM AZD7295 to NS5A33–202. Values on the Y-axis represent the percentage of the maximal thermophoretic response observed, or in the case of no detectable binding values on the Y-axis represent the thermophoretic shift of the labelled protein. All binding curves were determined in triplicate by MST and are represented as the mean ± SD.
Figure 3. Binding to domain 1 NS5A26–202 genotype 1b (con1).
Binding of (a) 4rU, (b) 5rU, (c) 10rU, (d) 5rA, (e) BMS-790052 or (f) AZD7295 to NS5A26–202. Values on the Y-axis represent the percentage of the maximal thermophoretic response observed. All binding curves were determined in triplicate by MST and are represented as the mean ± SD.
Overall, this part of the study showed both NS5A constructs bound various RNA fragments in accord with published values and thus validated the MST assay.
NS5A inhibitors bind to domain 1 of NS5A and compete with RNA binding
We then investigated the binding of BMS-790052 and AZD7295: they both bound tightly to NS5A33–202 with affinities in the low nM range (Fig. 2e, KD = 8 ± 3 nM and Fig. 2f, KD = 30 ± 11 nM, respectively). Surprisingly, the NS5A26–202 construct bound more weakly to BMS-790052 (Fig. 3e, KD of 210 ± 46 nM) and AZD7295 (Fig. 3f, KD of 670 ± 220 nM) than NS5A33–202. We hypothesise that the NS5A N-terminal tail (residues 26 to 32) in the absence of the N-terminal amphipathic helix (residues 5–25) may compete for the same binding site as the inhibitor.
Incubation of NS5A33–202 with 100-fold excess 5rU prior to adding compound eliminated binding of both BMS-790052 and AZD7295 (Fig. 2g and 2h respectively). When NS5A33–202 was pre-incubated with either 0.25 μM BMS-790052 or 0.5 μM AZD7295, the affinity of NS5A33–202 for 5rU was dramatically shifted from the low nM (KD = 50 ± 15 nM, Fig. 2b) to μM (KD = 94 ± 11 μM, Fig. 2i and KD = 4 ± 1 μM, Fig. 2j, respectively).
Overall, this part of the study showed unambiguously that both BMS-790052 and AZD7295 bind directly to a site located in NS5A domain 1 with nanomolar affinity. Previously, it was suspected that BMS-790052 bound to domain 1 based on resistance mutations although it couldn't be ruled out that the compound was binding to an interacting protein2,4,6. The MST data demonstrate a direct, tight interaction between the compounds and NS5A domain 1 and, most importantly, show that RNA binding inhibits compound binding and, conversely, compound binding inhibits RNA binding. Thus a possible mechanism of action for BMS-790052 and AZD7295 is that they directly reduce the affinity of NS5A for viral RNA.
NS5A inhibitors bind more weakly to common resistance mutants
Mechanism of action studies have revealed that a number of mutants conferring resistance against both BMS-790052 and AZD7295 mapped to the NS5A region of the viral genome6,27,28,29. Two of the most common mutation sites are Y93H/N/C and L31V/M/F which confer 19- to 47,000-fold and 23- to 3,350-fold resistance against BMS-790052, respectively4,6. We thus predicted that compound binding to these common resistance mutants would be compromised, but that the mutants would be unlikely to affect RNA binding significantly since that would compromise NS5A function.
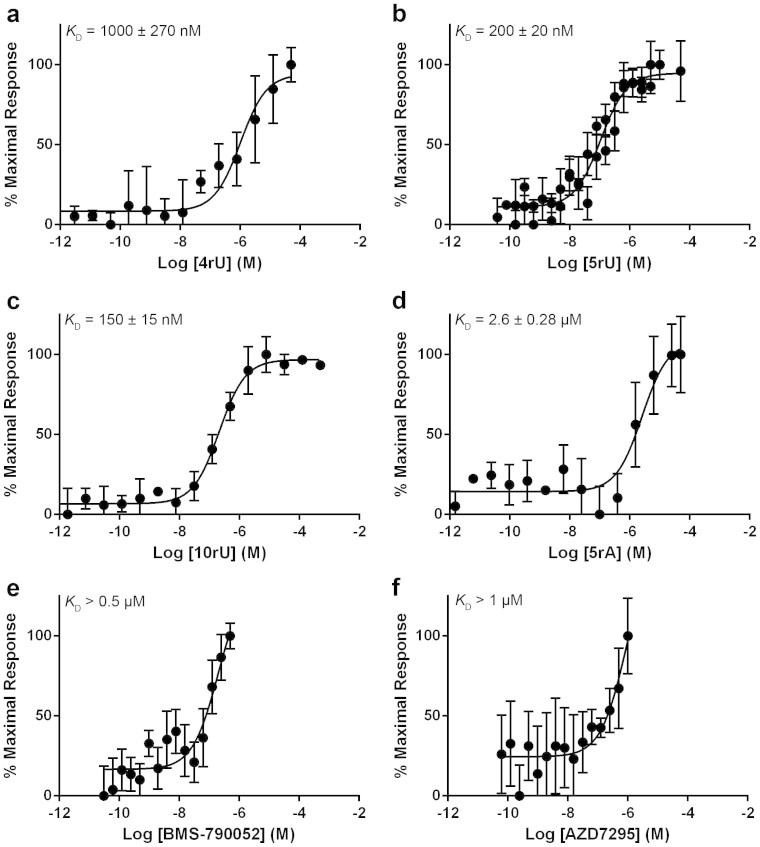
Both BMS-790052 (KD > 0.5 μM; Fig. 4e) and AZD7295 (KD > 1 μM; Fig. 4f) showed significantly weaker binding to the resistance mutant Y93H NS5A33–202, although we were unable to determine a precise KD due to their limited solubility (<1.6 μg/ml at pH 7.0). The L31V NS5A26–202 resistance mutation did not show any detectable binding to either inhibitor (Fig. 5e and f).
Figure 4. Binding to domain 1 NS5A33–202 Y93H genotype 1b (con1).
Binding of (a) 4rU, (b) 5rU, (c) 10rU, (d) 5rA, (e) BMS-790052 or (f) AZD7295 to NS5A33–202 Y93H. Values on the Y-axis represent the percentage of the maximal thermophoretic response observed. All binding curves were determined in triplicate by MST and are represented as the mean ± SD.
Figure 5. Binding to domain 1 NS5A26–202 L31V genotype 1b (con1).
Binding of (a) 4rU, (b) 5rU, (c) 10rU, (d) 5rA, (e) BMS-790052 or (f) AZD7295 to NS5A26–202 L31V. Values on the Y-axis represent the percentage of the maximal thermophoretic response observed, or in the case of no detectable binding values on the Y-axis represent the thermophoretic shift of the labelled protein. All binding curves were determined in triplicate by MST and are represented as the mean ± SD.
In contrast, RNA binding was unaffected for the L31V NS5A26–202 construct compared to wild type NS5A26–202 (p values of 0.9, 0.3, 0.7 and 0.5 for 4rU, 5rU, 10rU and 5rA, respectively, Fig. 5a–d). The Y93H NS5A33–202 resistance mutation had little effect upon the binding of 4rU or 5rU compared to wild type (KD of 1000 ± 270 nM and 200 ± 20 nM respectively, Fig. 4 a and b); however, there was a statistically significant difference in the binding of 10rU compared to wild type NS5A33–202 (Fig. 4c, KD of 150 ± 15 nM, p < 0.01) and 5rA (Fig. 4d, KD of 2.6 ± 0.28 μM, p < 0.01).
Overall, this part of the study showed that common resistance mutants to clinically relevant NS5A inhibitors act by directly reducing affinity towards the compounds with little affect on RNA binding.
NS5A inhibitors do not affect NS5A dimerisation
The observation that many NS5A inhibitors are roughly symmetrical has led to suggestions they might bind across the NS5A dimer interface4,7,11,13,15. As the presence of NS5A monomers in solution has been reported in the literature14, we wanted to explore whether the inhibitors directly bind to NS5A dimers, monomers or both species.
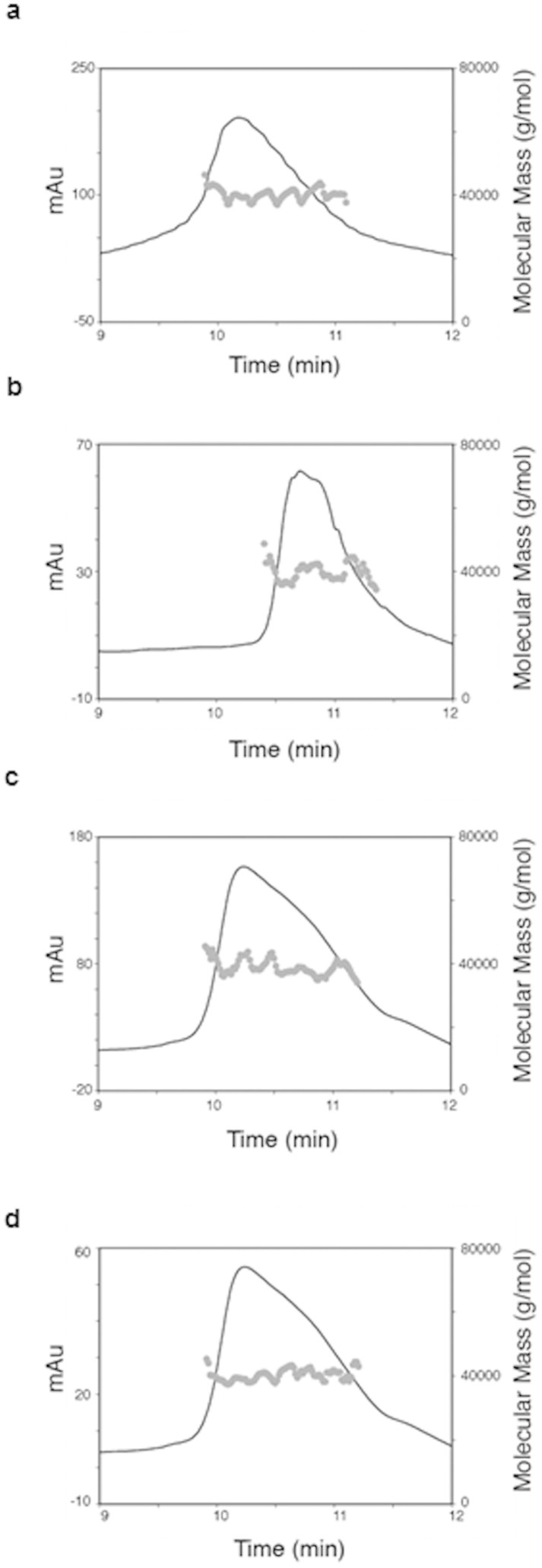
Multi-Angle Laser Light Scattering (MALLS) analysis of the NS5A33–202 construct shows that it assembles almost exclusively as dimers in solution (Fig. 6a, average molecular weight 39 ± 5 kDa). The addition of 5rU, BMS-790052 or AZD7295 did not affect the dimerisation state (Fig. 6b–d, average molecular weights of 38 ± 6, 41 ± 7 and 40 ± 5 kDa respectively). These results agree with previous findings that RNA or compounds do not affect dimerisation14,15.
Figure 6. SEC-MALLS analysis of domain 1 NS5A33–202 genotype 1b (con1) oligomerisation state.
(a) Solution molecular weight analysis of NS5A33–202 domain 1. (b) Molecular weight analysis in the presence of 5rU, (c) BMS-790052, (d) AZD7295. All molecular weights were determined by MALLS and are shown as grey points, with the black trace representing the absorbance at 280 nm recorded by an AKTApurifier.
In silico docking of inhibitors to NS5A domain 1 models
Having shown that the inhibitors bind to NS5A domain 1 and that common resistance mutants affect their binding, we wished to define the details of the binding site. Extensive crystallographic studies by us, and others, have failed to observe the direct binding of inhibitors to the protein. Hence as a first step we computationally docked the inhibitors into the two different published dimer structures. Since some resistance mutations are located in part of the N-terminal region of domain 1 missing in the crystal structures, we produced models of NS5A1–191 based on the 1ZH110 and 3FQQ11 crystal structures together with the 1R7G19 NMR model of the NS5A amphipathic helix (Fig. 1d).
In silico docking of inhibitors to the 3FQQ dimer
In the 3FQQ dimer model, the compounds bind in a well-defined cleft at the dimer interface containing most of the reported BMS-790052 resistance mutation sites, including Y93 and L31. Flexible loops from each monomer, comprising residues 26–32, form part of a hydrophobic pocket into which the prolyl substituents of BMS-790052 bind (Figs. 7, 8e,f). The proposed compound binding site is formed by the following residues: I52, I52*, Q54, Q54*, T56, T56*, P58, P58*, Q62 and Q62* form the “walls” of the site; Y93, Y93*, T95 and T95*are located in the base of the site; K26, L28, P29, R30, L31, P32, V34, S38, I52*, T56, P58, G60, Q62, G96* and P97* form the largely hydrophobic pockets located at either end of the monomer-monomer interface (* denotes a residue from the second monomer).
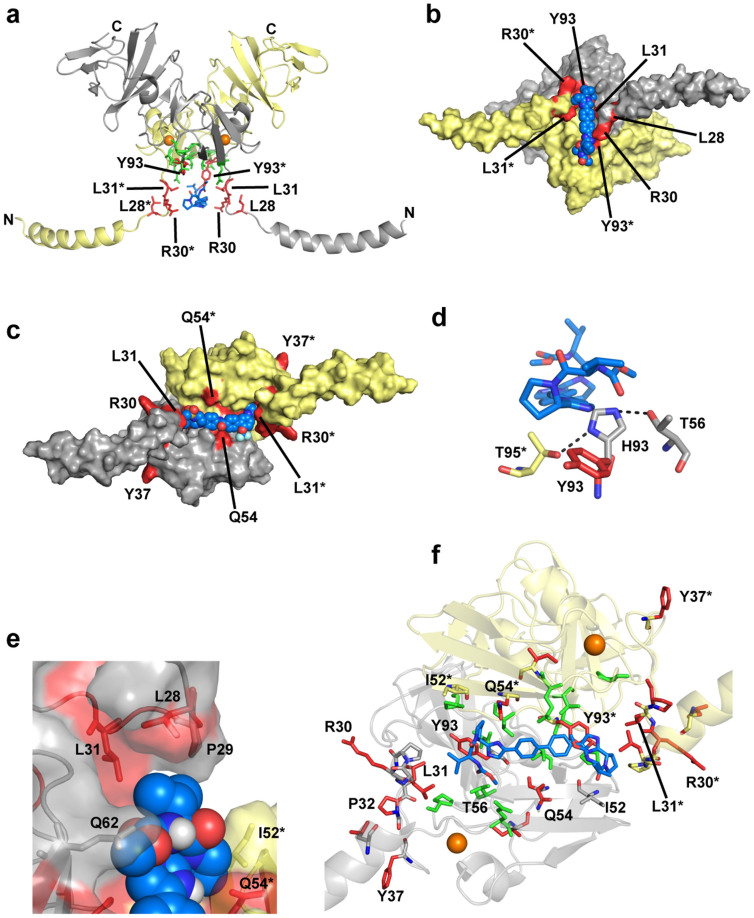
Figure 7. Computational docking of BMS-790052 to NS5A1–191.
(a) The 3FQQ dimer model, coloured as in Fig. 1d. Known BMS-790052 resistance mutation residues for genotype 1b (L23, L31, P32, F37, Q54, P58, I63, Y93)6 and genotype 1a (M28, Q30, L31, P32, H54, Y93)6 are in red. BMS-790052 (blue sticks) docks into the channel formed by the dimer and interacts with many of the resistance mutation residues. (b) The 3FQQ dimer model shown as a molecular surface. View is a rotation of −90° about the X-axis from that shown in (a), looking directly down onto the cleft at the dimer interface. BMS-790052 depicted as blue and red cpk. * denotes residues from the yellow monomer.
Figure 8. Docking of inhibitors and mapping of drug resistance and genotype variation onto the alternate NS5A1–191 dimer models.
The same monomer colour scheme is used as in Figs. 1 and 7. (a) The 1ZH1 dimer model. BMS-790052 (blue sticks) has been docked into a pocket at the dimer interface formed by the concave protein surface (where Y93 is located) and the two flexible loops consisting of residues 26–32. The locations of resistance mutation residues (red sticks) and genotype difference residues (green sticks) around the putative inhibitor binding site are indicated. (b) The 1ZH1 dimer model shown as a molecular surface, known resistance mutation residues are coloured red. View is a rotation of −90° about the X-axis from that shown in (a) looking directly down onto the pocket at the dimer interface. BMS-790052 depicted as blue and red cpk. (c) AZD7295 (blue, red and cyan cpk) docked into the 3FQQ dimer model (shown as a molecular surface; same view of NS5A1–191 as shown in Fig. 7b). Known resistance mutation residues are in red. (d) Model of the 3FQQ dimer Y93H resistance mutation with BMS-790052 docked (blue and red sticks). Showing a close up view of Y93 (red sticks) in wild type and the mutated residue Y93H (grey and blue sticks). Dashed lines indicate possible hydrogen bonds. (e) Close up view of the 3FQQ dimer model hydrophobic pocket. There are two of these hydrophobic pockets in the 3FQQ dimer model, located at each end of the cleft formed by the monomer-monomer interface. BMS-790052 shown as blue, red and white cpk. NS5A1–191 shown as cartoon and transparent molecular surface. Side-chains of residues depicted in stick format. (f) Same orientation of BMS-790052 (blue sticks) docked into the 3FQQ dimer model as shown in Fig. 7b; NS5A depicted as a transparent cartoon. The side-chains of resistance mutation residues (red sticks) and genotype difference residues (green sticks) around the putative inhibitor binding site are shown. Other residues interacting with the compound are shown as grey sticks. * denotes residues from the yellow monomer.
The docked pose of BMS-790052 shown in Figs. 7 and 8d,e,f is one example of how this compound can bind into the site and the interactions that it can potentially make are: the biphenyl of BMS-790052 sits above and interacts with T95 and T95*; the biphenyl-imidazole rings can interact with I52, I52*, Q54, Q54*, T56, T56*, P58, P58*, Q62 and Q62* (i.e. the “walls”); the imidazole-prolyl rings interact with Y93 and Y93*; each prolyl isopropyl substituent can make hydrophobic interactions with L28, P29 and L31; depending on the prolyl ester group orientation, hydrogen bonds with T56, T56*, Q62 and Q62* are possible.
In silico docking of inhibitors to the 1ZH1 dimer
In the 1ZH1 dimer model a deep cleft at the interior dimer interface has a positive electrostatic potential and provides a convincing RNA binding site (Fig. 1d). However, in this model, none of the resistance mutations are located in the deep cleft, rather the majority are located on the opposite side of the protein to the interior dimer interface (Fig. 8a). Hence it seems unlikely that the putative RNA binding site is also the inhibitor binding site.
The inclusion of residues 1–35 into the 1ZH1 dimer model creates another potential compound binding site, located between the NS5A dimer convex exterior surface and the membrane, which contains most of the known BMS-790052 resistance mutation sites (Fig. 8a,b). The flexible loops, residues 26–32, form the walls of the hydrophobic pocket into which the biphenyl section of BMS-790052 may bind. The possible compound binding site is formed by the following residues: L28, L28*, P29, P29*, R30, R30*, L31, L31*, P32 and P32* form the “walls” of the site; Q54, Q54*, T56, T56*, G60, G60*, Q62, Q62*, Y93, Y93*, T95 and T95* are located on the concave base of the site.
An example of how BMS-790052 can bind into this site is shown in Fig. 8a,b. In contrast to the 3FQQ dimer model, the biphenyl moiety is not in direct contact with NS5A surface residues but interacts with L31 and L31* in the flexible loops. The interaction with NS5A surface residues is confined to the imidazole-prolyl rings and the prolyl substituents via Q54, Q54*, Q62, Q62*, Y93 and Y93*; as a consequence the compound makes fewer contacts with residues located on the convex surface compared to the 3FQQ dimer model. Depending on the prolyl ester group orientation, hydrogen bonds with R30, R30*, Q54, Q54*, Q62, Q62*, Y93 and Y93* are possible. Each Y93 lies on the NS5A convex surface at either end of the pocket. Given the flexibility of the loops (residues 26–32) and the concave nature of the protein surface, this site is less convincing than that in the 3FQQ dimer model.
Like BMS-790052, AZD7295 can also adopt a range of conformations in the proposed compound binding sites of the 3FQQ and 1ZH1 dimer models. One example is shown in Fig. 8c. Although both compounds can bind into the same site, the specific interactions made by each inhibitor differ due to their different chemical structures.
Thus the 1ZH1 dimer model provides a suitable RNA binding site without a convincing compound binding site, while the 3FQQ model does the reverse, showing a compound binding site that explains published data without revealing a probable RNA binding site. This leads to the hypothesis that NS5A inhibitors act by favouring a dimeric structure that does not bind RNA.
Mapping genotype differences and resistance mutations onto the NS5A1–191 dimer models
Any proposed mechanism of action should be consistent with the extensive literature on inhibitor activity against NS5A genotypes and subtypes. Replicon data for the activity of BMS-790052 towards a wide range of NS5A genotypes and subtypes is available. For most genotypes and subtypes, the EC50 is in the range 2–150 pM, however notable exceptions are genotype 1a (J1), 2a (J6) and 3a (S52) which have EC50's of 1,100, 14,000 and 900 pM respectively4,6,28,30,31,32,33. We mapped the differences between the genotype 1b (con1) and genotype 1a (J1), 2a (J6) and 3a (S52) NS5A sequences for residues 26–191 onto the 3FQQ and 1ZH1 dimer models of genotype 1b (con1). The residue differences around the proposed compound binding sites for both dimer models (Fig. 8a,f) are as follows: L27 in genotype 1b (con1) is replaced by I27 in genotype 3a; L28 is replaced by M28 in genotype 1a and 3a and in genotype 2a it is F28; R30 is replaced by H30 in genotype 1a, K30 in genotype 2a and A30 in genotype 3a; L31 is replaced by M31 in genotype 2a; I52 is replaced by V52 in genotype 3a; Q54 is replaced by H54 in genotype 1a, T54 in genotype 2a and S54 in genotype 3a; T56 is replaced by R56 in genotype 1a, 2a and 3a; P58 is replaced by H58 in genotype 1a; Q62 is replaced by the polar E62 in genotype 1a, N62 in genotype 2a and S62 in genotype 3a; A92 is replaced by C92 in genotype 2a and in genotype 3a the analogous residue is E92; T95 and P97 are replaced by E95 and Q97, respectively, in genotype 2a. Many of these genotype 1a and 1b residues (residues 28, 30, 31, 54 and 58) are also mutated in response to replicon treatment with BMS-790052, i.e. they are resistance mutation residues6,34.
In genotype 1b (con1) the environment around Y93 may play a significant role in preserving the activity of BMS-790052 (depicted in Fig. 8d for the 3FQQ NS5A dimer) compared to genotypes 1a, 2a and 3a. In the 3FQQ NS5A dimer, Y93 is flanked on either side by T56 (from the same NS5A monomer as Y93) and T95* (from the opposing NS5A monomer). As described above, T56 is replaced by the larger charged R56 in genotype 1a (H77), 2a (J6) and 3a (S52) and T95 is replaced by the negatively charged E95 in genotype 2a (J6). Replacing T56 and/or T95* with larger charged residues would prevent BMS-790052 from binding as deeply into the 3FQQ NS5A dimer interface cleft compared to wild type NS5A. In contrast, in the 1ZH1 NS5A dimer model Y93 is flanked by T56 and G33 (all from the same NS5A monomer). Larger charged side-chains at residue 56 are readily accommodated in the 1ZH1 NS5A dimer model without impacting on BMS-790052 binding (G33 is conserved across genotypes).
It has also been observed that the effect of the Y93H/C resistance mutation on BMS-790052 replicon activity for genotype 1b (con1) is minimal (~5-fold decrease in EC504) compared to other genotypes; for example, genotype 1a (H77), 2a (J6) and 3a (S52) exhibit 220-fold4, 143-fold31 and 2,111-fold31 decreases respectively. In contrast to the genotype 1b (con1) replicon data, a much larger decrease in BMS-790052 binding was observed in the MST in vitro assay (>50-fold decrease in KD). In the 3FQQ NS5A dimer, replacing the common resistance site residue Y93 with histidine (i.e. Y93H) enables the side-chain to interact with both T56 (from the same NS5A monomer as Y93H) and T95* (from the other NS5A monomer). The Y93H resistance mutation was modelled in the 3FQQ dimer and, while some of the possible histidine side-chain conformations did not impact on the binding of BMS-790052, other conformations of the histidine side-chain prevented BMS-790052 from binding as deeply into the cleft compared to wild type NS5A (as illustrated in Fig. 8d). In the 1ZH1 NS5A dimer model Y93H is flanked on either side by T56 and G33 (both from the same NS5A monomer as Y93H), and an interaction between Y93H and T56 should not impact on the binding of BMS-790052. For genotype 1b (con1), the common L31V resistance mutation confers 23-fold resistance against BMS-790052 in replicon assays6, whereas in the MST in vitro assay there was no detectable binding of the inhibitor to the L31V NS5A26–20 mutant. In both NS5A dimer models, the L31V mutation may reduce inhibitor activity by destabilising the conformation of the residue 26–32 loop. In the 3FQQ dimer model the L31V mutation would also enlarge the hydrophobic pocket located at either end of the dimer interface, thereby reducing the potential for the compound to interact with both pockets simultaneously (Fig. 8e).
Overall, the putative inhibitor binding site in the 3FQQ dimer model (Fig. 7) is consistent with the behaviour of the common resistance mutants L31V and Y93H and also with known BMS-790052 activity towards different genotypes. In contrast, the less plausible inhibitor binding site in the 1ZH1 dimer (Fig. 8a,b) is not well supported by the published resistance and genotype data.
Discussion
In summary, we have shown that MST can be used to demonstrate and measure the binding of RNA or inhibitors to NS5A in the absence of other proteins, an issue which has complicated the interpretation of cell-based approaches. We have shown that both BMS-790052 and AZD7295 bind directly to a site located in NS5A domain 1 with nanomolar affinity. Most importantly, we show that RNA binding inhibits compound binding and, conversely, compound binding inhibits RNA binding. Furthermore, we show that common resistance mutants act by directly reducing affinity towards the compounds with little affect on RNA binding. We show that neither RNA nor the compounds affect dimerisation, consistent with previously published observations using different approaches. Finally, computational docking of the inhibitors reveals a compelling compound binding site that is consistent with known resistance mutations and their activity towards different HCV genotypes.
During the preparation of our manuscript O'Boyle and coworkers published7 a cell-based study which strongly supports many of our own conclusions. They used chemically labelled (biotinylated and azide cross-linked) stilbene BMS-790052-like compounds in HCV replicon cells to demonstrate tight and specific binding to the NS5A protein. Furthermore, mass spectrometry of labelled NS5A suggested the compounds bound to domain 1, close to residue L31, which is consistent with our docking results. In addition, cross-linking results suggested the compounds bind close to the dimer interface as we predict from our docking studies.
Our hypothesis for a mechanism of action of NS5A inhibition is that there are distinct RNA binding and inhibitor binding forms of the NS5A domain 1 dimer, represented by the 1ZH1 and 3FQQ dimers, respectively, which are dynamically interconverting. The process is summarised in Fig. 1d. RNA binding traps the NS5A domain 1 dimer in the form that the inhibitor does not bind to, and vice versa. The model provides a molecular basis as to why the common Y93H resistance mutant does not significantly affect RNA binding (it is located far from the RNA-binding cleft in the 1ZH1 dimer, Fig. 8a,b) but greatly affects inhibitor binding (it is located in the compound binding site in the 3FQQ dimer) (Figs. 7, 8c,d,f). This model also explains why previous attempts to show direct binding using surface plasmon resonance failed12, because the technique does not allow free interconversion between dimer states. The advantage of the MST technique is that it is solution-based and allows conversion between NS5A dimer states. Attempts to demonstrate direct binding between NS5A and BMS-790052 using isothermal calorimetry have been unsuccesful12, perhaps due to the propensity of NS5A to aggregate readily which can mask titration curves. The computational docking work also explains why compound binding has not been seen by X-ray crystallography: the putative compound binding site in the 3FQQ dimer is buried due to crystal contacts. The model presented here is consistent with the recent FRET studies conducted by Bhattacharya and co-workers12 who demonstrated that binding of an NS5A inhibitor in cell culture caused conformational changes in domain 1 resulting in the formation of large NS5A aggregates.
It has been suggested by others that the two types of NS5A dimers may co-assemble into large oligomeric arrays that channel RNA substrates within HCV-infected cells11. Furthermore, recent data suggests that BMS-790052 prevents correct NS5A assembly into functional replication complexes35. Our evidence for the direct interaction, in solution, of inhibitors with NS5A domain 1 and the potential impact of this interaction on dimer conformation supports this model for the mechanism of action of NS5A protein. BMS-790052 inhibits HCV replicon cells at picomolar concentrations that correspond to one inhibitor molecule for every thousand NS5A molecules. It has been suggested by others that disruption of multimer function by a single inhibitor molecule, via transmission of conformational changes in the same and adjacent polymers, may explain the extraordinary potency of NS5A inhibitors in cells2,4,7. The ability of one inhibitor molecule to disrupt the NS5A functional network explains why we see low nanomolar binding for the interaction of BMS-790052 with NS5A in vitro when others measure picomolar inhibition in cell-based replicon assays.
Many NS5A inhibitors in clinical trials appear similar to the chemical structure of the BMS-790052 compound series2,13. Our work in identifying the likely binding site for current inhibitors and the development of a direct binding assay provides a rational framework for refining the molecular mechanism of action, the discovery of entirely novel NS5A inhibitor scaffolds and possible application to related viruses that have NS5A homologues.
Methods
Materials
BMS-790052 and AZD7295 were gifts from Biota Pharmaceuticals and were synthesised according to published methods36,37,38. In line with the published work on the domain 1 structure10,11, we expressed and purified recombinant HCV genotype 1b (con1) NS5A33–202. Briefly, the constructs containing the NS5A33–202 protein fused to an N-terminal 6-His tag and TEV protease cleavage site were synthesised by Genscript and cloned into pET15b using NdeI and HindIII restriction sites. Protein production was performed in 5 L baffled flasks containing 2 L of TB supplemented with 100 μg/mL ampicillin. After inoculation with 20 mL of an overnight culture, the culture was grown at 37°C and 130 rpm in an Inforce shaker for 6 hours, prior to cooling to 16°C and induction with 100 μM IPTG. The culture was grown overnight at 16°C prior to being harvested at 5000 × g for 20 min. The cell pellets were washed in PBS and stored at −80°C until ready for purification.
Cell pellets were resuspended in 25 mM Tris, 500 mM NaCl, 20 mM imidazole, 10% glycerol pH 8.0 with Roche Complete EDTA free protease inhibitor cocktail and DNase. Cells were lysed by French Press, and clarified by centrifugation at 20000 × g for 30 min. The supernatant was then passed over a Ni HiTrap column (GE Healthcare) using AKTApurifier, washed in 25 mM Tris, 500 mM NaCl, 20 mM imidazole, 10% glycerol pH 8.0, and the protein eluted using a gradient up to 25 mM Tris, 500 mM NaCl, 400 mM imidazole, 10% glycerol pH 8.0. The purified protein was dialysed into 25 mM Tris, 500 mM NaCl, 20 mM imidazole, 10% glycerol pH 8.0 and the tag cleaved using an N-terminal 6-His tagged TEV protease. This was then repassed over a Ni HiTrap column to remove the TEV protease and cleaved tag. The protein was then concentrated using a 10 kDa MWCO Amicon concentrator (Millipore) and further purified by size exclusion chromatography using a Superdex 75 26/60 column (GE Healthcare) equilibrated in 25 mM Tris, 250 mM NaCl, 10% glycerol pH 8.0 (Supplementary Fig. S1).
A longer length NS5A construct was explored to characterise the role of other resistance mutation sites outside our original construct: residues 26 to 202 (Supplementary Fig. S1). The Y93H NS5A33–202 mutant, NS5A26–202 and the L31V NS5A26–202 mutant were expressed and purified in a similar manner to NS5A33–202. Filter-binding assays have previously shown that NS5A binds short stretches of uridine-rich oligonucleotides14,39 with the minimum RNA length determined to be five bases. In our study we examined binding to single stranded RNA consisting of four, five or ten uridines (4rU, 5rU and 10rU respectively), five adenines (5rA); as well as double stranded RNA formed through the annealing of 5rU and 5rA (ds(5rU)) (Integrated DNA Technologies).
MST
Solution MST binding studies were performed using standard protocols on a Monolith NT.115 (Nanotemper Technologies). Briefly, recombinant NS5A26–202, NS5A33–202, L31V NS5A26–202, Y93H NS5A33–202 or control protein was labelled using the RED-NHS (Amine Reactive) Protein Labelling Kit (Nanotemper Technologies). NS5A was mixed with either RNA, BMS-790052 or AZD7295 with a final buffer condition of 25 mM Tris-HCl, pH 8.0, containing 250 mM NaCl, 10% glycerol, 0.05% Tween-20 and 5% DMSO. Each replicate contained a 16 step 2- to 4-fold serial dilution series. The protein concentration (12 nM) was chosen such that the observed fluorescence was approximately 1000 units at 40% LED power. The samples were loaded into hydrophobic capillaries and heated at 40% laser power for 30 sec, followed by 5 sec cooling. The data were normalised against the baseline obtained in the absence of any inhibitor, and the maximal response obtained at the highest concentration of inhibitor. The dissociation constant KD was obtained by plotting the normalised fluorescence Fnorm against the logarithm of the different concentrations of the dilution series resulting in a sigmoidal binding curve that could be directly fitted with a non-linear solution of the law of mass action. All experiments were performed with a minimum of 3 replicates and the normalised fluorescence temperature jump curves were analysed using GraphPad Prism (Version 5, GraphPad, San Diego, CA, USA). KD's were compared by one-way Anova with Tukey posttest and p < 0.01 was considered to be statistically significant. A Hill slope analysis suggested 1 inhibitor molecule binds per NS5A dimer. No measurable interactions with several unrelated proteins including insulin-regulate membrane aminopeptidase, nicastrin and carbonic anhydrase were observed by MST for either compound. In another control we added excess EDTA to NS5A33–202 to remove metal from the Zn2+ binding site (leading to destabilisation of the protein fold) and no binding to RNA or either compound was observed.
MALLS
MALLS was carried out both in batch mode using a syringe pump set to 0.1 ml/min and through size-exclusion chromatography (SEC) using a BioSep-SEC S3000 column (Phenomenex) equilibrated in a buffer containing 25 mM Tris-HCl, pH 8.0, 250 mM NaCl, 10% glycerol, 0.05% Tween-20, 5% DMSO and in the presence and absence of 5 mM β-mercaptoethanol. The column was run at a flow rate of 0.35 ml/min on an AKTApurifier (GE Healthcare) coupled to a miniDawn MALLS detector and an Optilab T-rEX refractive index detector (Wyatt Technology). The molecular masses of bovine serum albumin (used as a control) and NS5A33–202 were calculated using a dn/dc value of 0.185. NS5A33–202 was run at a range of concentrations from 0.3–4 mg/ml. The overall weight-average molar mass for each sample was determined from the concentrations of each molar mass at all points by use of ASTRA 5 software (Wyatt Technologies).
Modelling
The inhibitors BMS-790052 and AZD7295 were constructed and geometrically optimised within SYBYL-X 2.0 (Certara, L. P., http://www.tripos.com) using the MMFF94s forcefield and partial atomic charges, conjugate gradient convergence method. Termination of the optimisation was achieved when the gradient difference of successive steps was < 0.05 kcals/mol•Å. Since residues 26–32, located on a flexible loop connecting the Zn2+ binding domain (residues 33–213) to the amphipathic helix (residues 1–25), is a site of many resistance mutations, we decided to include this region and the amphipathic helix in our models. To construct the 3FQQ dimer model for NS5A1–191, residues 1–27 of the NMR structure 1R7G19 were combined with the 3FQQ A and B chains (residues 33–191)11. The loops containing residues 26–32 were built manually in a conformation that would allow the amphipathic helices (residues 1–25) to interact with the cell membrane and for L28 and L31 to point towards the putative compound binding pocket (Figs. 1d, 7, 8e,f). The side-chains of the NS5A1–191 3FQQ dimer model were geometry optimised for 1,000 iterations (or until the gradient of successive iterations was < 0.05 kcal/mol•Å) using the same protocol as above. The 1ZH1 dimer model (Figs. 1d, 8a,b) was generated in an analogous fashion. The inhibitors were then docked into each of the NS5A1–191 dimer models using the docking algorithm Surflex (within SYBYL-X 2.0, Certara L. P., http://tripos.com). For both the 3FQQ and 1ZH1 dimer models, the protomol was generated by manually selecting the residues lining the pocket at the monomer-monomer interface, a threshold of 0.50 and a bloat value of 2. The docking mode used for both NS5A1–191 dimer models was GeomX and protein flexibility was allowed, all other parameters were at default values. The C-Score scoring function was used and the top 100 ranked poses of each inhibitor were retained for examination. Both BMS-790052 and AZD7295 are conformationally flexible and can adopt a range of orientations within the docking sites of both dimers. The orientations shown in Figs. 7 and 8 are selected examples. Figs. 1, 7 and 8 were produced using PyMOL (http://pymol.org).
Author Contributions
D.B.A. designed and performed the MST binding experiments in Figs. 2–5 and the MALLS experiments in Fig. 6. L.D. also performed some of the MST experiments. T.L.N. did the modelling and computational docking in Figs. 1, 7 and 8. J.W., C.J.M. and M.W.P. designed and supervised various aspects of the study. All authors wrote and revised the paper.
Supplementary Material
Supplementary Information
Acknowledgments
We thank Biota Pharmaceuticals for gifts of the inhibitors. This work was supported by a capital grant from the Australian Cancer Research Foundation and infrastructure funding from the Victorian Government Operational Infrastructure Support Scheme to St Vincent's Institute. D.B.A. was supported by a Victoria Fellowship from the Victorian Government and the Leslie (Les) J. Fleming Churchill Fellowship from The Winston Churchill Memorial Trust. M.W.P. is a National Health and Medical Research Council of Australia Research Fellow.
Footnotes
The laboratory where the study took place received funding from anti-infectives company Biota Pharmaceuticals on hepatitis C projects although not on the studies described in this work. C.J.M. is a Principal Research Associate at St Vincent's Institute and was an employee of Biota Pharmaceuticals. J.W., T.L.N. and M.W.P. were consultants to Biota Pharmaceuticals.
References
- Ghany M. G., Nelson D. R., Strader D. B., Thomas D. L. & Seeff L. B. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology 54, 1433–1444 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda O. & Targett-Adams P. Small molecule inhibitors of the hepatitis C virus-encoded NS5A protein. Virus Res. 170, 1–14 (2012). [DOI] [PubMed] [Google Scholar]
- Suk-Fong Lok A. HCV NS5A Inhibitors in Development. Clin. Liver Dis. 17, 111–121 (2013). [DOI] [PubMed] [Google Scholar]
- Gao M. et al. Chemical genetics strategy identifies an HCV NS5A inhibitor with a potent clinical effect. Nature 465, 96–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gane E. et al. Antiviral activity, pharmacokinetics and tolerability of AZD7295, a novel NS5A inhibitor, in a placebo-controlled multiple ascending dose study in HCV genotype 1 and 3 patients. J. Hepatol. 52, S464 (2010). [Google Scholar]
- Fridell R. A., Qiu D., Wang C., Valera L. & Gao M. Resistance analysis of the hepatitis C virus NS5A inhibitor BMS-790052 in an in vitro replicon system. Antimicrob. Agents Chemother. 54, 3641–3650 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Boyle I. D. R. et al. Characterizations of HCV NS5A replication complex inhibitors. Virology 10.1016/j.virol.2013.06.032 (2013). [DOI] [PubMed] [Google Scholar]
- Tripathi L. P. et al. Understanding the biological context of NS5A-host interactions in HCV infection: a network-based approach. J. Proteome Res. 12, 2537–2551 (2013). [DOI] [PubMed] [Google Scholar]
- Targett-Adams P. et al. Small molecules targeting hepatitis C virus-encoded NS5A cause subcellular redistribution of their target: insights into compound modes of action. J. Virol. 85, 6353–6368 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellinghuisen T. L., Marcotrigiano J. & Rice C. M. Structure of the zinc-binding domain of an essential component of the hepatitis C virus replicase. Nature 435, 374–379 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love R. A., Brodsky O., Hickey M. J., Wells P. A. & Cronin C. N. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 83, 4395–4403 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D. et al. Pharmacologic disruption of hepatitis C non-structural 5A (NS5A) intra and intermolecular conformations. J. Gen. Virol. 10.1099/vir.0.054569-0 (2013). [DOI] [PubMed] [Google Scholar]
- Hamatake R., Maynard A. & Kazmierski W. M. in Annu. Rep. Med. Chem. Vol. 47 (ed Desai Manoj, C.) Ch. 22, 331–345 (Academic Press, 2012). [Google Scholar]
- Hwang J. et al. Hepatitis C virus nonstructural protein 5A: biochemical characterization of a novel structural class of RNA-binding proteins. J. Virol. 84, 12480–12491 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P. J. et al. Correlation between NS5A dimerization and HCV replication. J. Biol. Chem. 287, 30861–30873 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanoulle X., Badillo A., Verdegem D., Penin F. & Lippens G. The domain 2 of the HCV NS5A protein is intrinsically unstructured. Protein Pept. Lett. 17, 1012–1018 (2010). [DOI] [PubMed] [Google Scholar]
- Liang Y., Ye H., Kang C. B. & Yoon H. S. Domain 2 of nonstructural protein 5A (NS5A) of hepatitis C virus is natively unfolded. Biochemistry 46, 11550–11558 (2007). [DOI] [PubMed] [Google Scholar]
- Verdegem D. et al. Domain 3 of NS5A protein from the hepatitis C virus has intrinsic alpha-helical propensity and is a substrate of cyclophilin A. J. Biol. Chem. 286, 20441–20454 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penin F. et al. Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 279, 40835–40843 (2004). [DOI] [PubMed] [Google Scholar]
- Zillner K. et al. Microscale thermophoresis as a sensitive method to quantify protein: nucleic acid interactions in solution. Methods Mol. Biol. 815, 241–252 (2012). [DOI] [PubMed] [Google Scholar]
- Lin C. C. et al. Inhibition of basal FGF receptor signaling by dimeric Grb2. Cell 149, 1514–1524 (2012). [DOI] [PubMed] [Google Scholar]
- Arbel N., Ben-Hail D. & Shoshan-Barmatz V. Mediation of the antiapoptotic activity of Bcl-xL protein upon interaction with VDAC1 protein. J. Biol. Chem. 287, 23152–23161 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patnaik S. et al. Discovery, structure-activity relationship, and biological evaluation of noninhibitory small molecule chaperones of glucocerebrosidase. J. Med. Chem. 55, 5734–5748 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brvar M. et al. Structure-based discovery of substituted 4,5′-bithiazoles as novel DNA gyrase inhibitors. J. Med. Chem. 55, 6413–6426 (2012). [DOI] [PubMed] [Google Scholar]
- Bosco E. E. et al. Rational design of small molecule inhibitors targeting the Rac GTPase-p67(phox) signaling axis in inflammation. Chem. Biol. 19, 228–242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippok S. et al. Direct detection of antibody concentration and affinity in human serum using microscale thermophoresis. Anal. Chem. 84, 3523–3530 (2012). [DOI] [PubMed] [Google Scholar]
- Carter M. C. et al. Diphenylcarboxamides as inhibitors of HCV non-structural protein NS5a. Antiviral Res. 86, A19 (2010). [Google Scholar]
- Fridell R. A. et al. Distinct functions of NS5A in hepatitis C virus RNA replication uncovered by studies with the NS5A inhibitor BMS-790052. J. Virol. 85, 7312–7320 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridell R. A. et al. Genotypic and phenotypic analysis of variants resistant to hepatitis C virus nonstructural protein 5A replication complex inhibitor BMS-790052 in humans: in vitro and in vivo correlations. Hepatology 54, 1924–1935 (2011). [DOI] [PubMed] [Google Scholar]
- Qiu D. et al. The effects of NS5A inhibitors on NS5A phosphorylation, polyprotein processing and localization. J. Gen. Virol. 92, 2502–2511 (2011). [DOI] [PubMed] [Google Scholar]
- Scheel T. K., Gottwein J. M., Mikkelsen L. S., Jensen T. B. & Bukh J. Recombinant HCV variants with NS5A from genotypes 1-7 have different sensitivities to an NS5A inhibitor but not interferon-alpha. Gastroenterology 140, 1032–1042 (2011). [DOI] [PubMed] [Google Scholar]
- Wang C. et al. In vitro activity of BMS-790052 on hepatitis C virus genotype 4 NS5A. Antimicrob. Agents Chemother. 56, 1588–1590 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. et al. In Vitro Activity of Daclatasvir on Hepatitis C Virus Genotype 3 NS5A. Antimicrob. Agents Chemother. 57, 611–613 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm J. A. et al. Identification of hepatitis C virus NS5A inhibitors. J. Virol. 84, 482–491 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. et al. The hepatitis C virus NS5A inhibitor (BMS-790052) alters the subcellular localization of the NS5A non-structural viral protein. Virology 414, 10–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M. C., Cockerill S., Flack S. S. & Wheelhouse C. J. Heterocyclic derivatives and their use in treating Hepatits C. Great Britain patent WO/2009/034390 (2009).
- Pack S. K., Geng P., Smith M. J. & Hamm J. Process for synthesizing compounds useful for treating Hepatitis C. United States patent WO/2009/020825 (2009).
- Wheelhouse C. J. et al. Biphenyl derivatives and their using in treating Hepatitis C. Great Britain patent WO/2007/031791 (2007).
- Foster T. L., Belyaeva T., Stonehouse N. J., Pearson A. R. & Harris M. All three domains of the hepatitis C virus nonstructural NS5A protein contribute to RNA binding. J. Virol. 84, 9267–9277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information