Abstract
Using the hyperthermophile Pyrococcus furiosus, we have delineated several key steps in CRISPR (clustered regularly interspaced short palindromic repeats)–Cas (CRISPR-associated) invader defence pathways. P. furiosus has seven transcriptionally active CRISPR loci that together encode a total of 200 crRNAs (CRISPR RNAs). The 27 Cas proteins in this organism represent three distinct pathways and are primarily encoded in two large gene clusters. The Cas6 protein dices CRISPR locus transcripts to generate individual invader-targeting crRNAs. The mature crRNAs include a signature sequence element (the 5′ tag) derived from the CRISPR locus repeat sequence that is important for function. crRNAs are tailored into distinct species and integrated into three distinct crRNA–Cas protein complexes that are all candidate effector complexes. The complex formed by the Cmr [Cas module RAMP (repeat-associated mysterious proteins)] (subtype III-B) proteins cleaves complementary target RNAs and can be programmed to cleave novel target RNAs in a prokaryotic RNAi-like manner. Evidence suggests that the other two CRISPR–Cas systems in P. furiosus, Csa (Cas subtype Apern) (subtype I-A) and Cst (Cas subtype Tneap) (subtype I-B), target invaders at the DNA level. Studies of the CRISPR–Cas systems from P. furiosus are yielding fundamental knowledge of mechanisms of crRNA biogenesis and silencing for three of the diverse CRISPR–Cas pathways, and reveal that organisms such as P. furiosus possess an arsenal of multiple RNA-guided mechanisms to resist diverse invaders. Our knowledge of the fascinating CRISPR–Cas pathways is leading in turn to our ability to co-opt these systems for exciting new biomedical and biotechnological applications.
Keywords: clustered regularly interspaced short palindromic repeats (CRISPR), CRISPR-associated (Cas), non-coding RNA, prokaryotic immunity, Pyrococcus furiosus, virus
Small RNA-guided prokaryotic immunity
To defend themselves from potentially hostile viruses and other invading mobile genetic elements, most archaea and many bacteria rely on small RNA and protein-based CRISPR (clustered regularly interspaced short palindromic repeats)–Cas (CRISPR-associated) adaptive immune systems [1–5]. CRISPR loci consist of tandem arrays of identical repeat sequences interspersed with variable ‘spacer’ sequences acquired from the invaders [6–9]. CRISPR loci are transcribed, and the primary CRISPR RNA transcripts undergo specific processing to generate functionally active mature crRNAs (CRISPR RNAs) [10–13]. Thus CRISPR loci capture virus and plasmid DNA sequences and produce RNAs containing the captured sequences that identify and eliminate the corresponding invaders. The protein components of the immune systems are encoded by specific modules of cas genes, which are typically found near the CRISPR locus [14,15]. Together, the crRNAs and Cas proteins provide RNA-directed immunity against diverse genomic invaders in a wide range of prokaryotic organisms.
Multiple distinct CRISPR–Cas systems have been identified, which differ with regard to cas gene content, CRISPR repeat sequences and mechanistic processes. Ten subtypes of CRISPR–Cas systems were initially identified on the basis of modules of specific cas genes [14,16] and named after an organism that contains only this subtype-specific cas gene cluster (e.g. Cse, Cas-subtype Escherichia coli) [14]. The CRISPR–Cas systems were recently refined and renamed to recognize categorization into three major types (I–III) (i.e. subtypes I-A to I-F, II-A and II-B, and III-A and III-B, wherein, for example, subtype I-E corresponds to the Cse system) [17]. The distribution of the diverse CRISPR–Cas modules between prokaryotic genomes is highly variable as a result of apparent lateral gene transfer [17,18].Many bacteria and a few archaea lack CRISPR–Cas systems completely. Moreover, some organisms contain a single CRISPR–Cas system, whereas other organisms, including Pyrococcus furiosus, harbour multiple distinct CRISPR–Cas systems.
The CRISPR–Cas systems of P. furiosus
Pyrococcus furiosus (roughly translated as ‘mad fireball’) is a hyperthermophilic euryarchaeaon of the order Thermococcales. P. furiosus was initially isolated from thermal marine sediments off the coast of Vulcano island, Italy, and exhibits a remarkable optimal growth temperature of ~100°C [19]. Sequenced genomes from six distinct pyrococcal species are available and reveal interesting species-specific diversity in CRISPR–Cas genes and gene organization [20]. We chose P. furiosus as a model system to unlock the mysteries of CRISPR–Cas immune systems for several compelling reasons. The inherent thermal stability of the proteins and complexes greatly facilitates detailed mechanistic and structural studies with both recombinant and native complexes. The newly established genetic manipulability of P. furiosus [21–23] has further increased the power of P. furiosus as a model system and expanded the types of analysis that can be performed. Moreover, as detailed next, P. furiosus offers the opportunity to investigate three distinct CRISPR–Cas defence pathways.
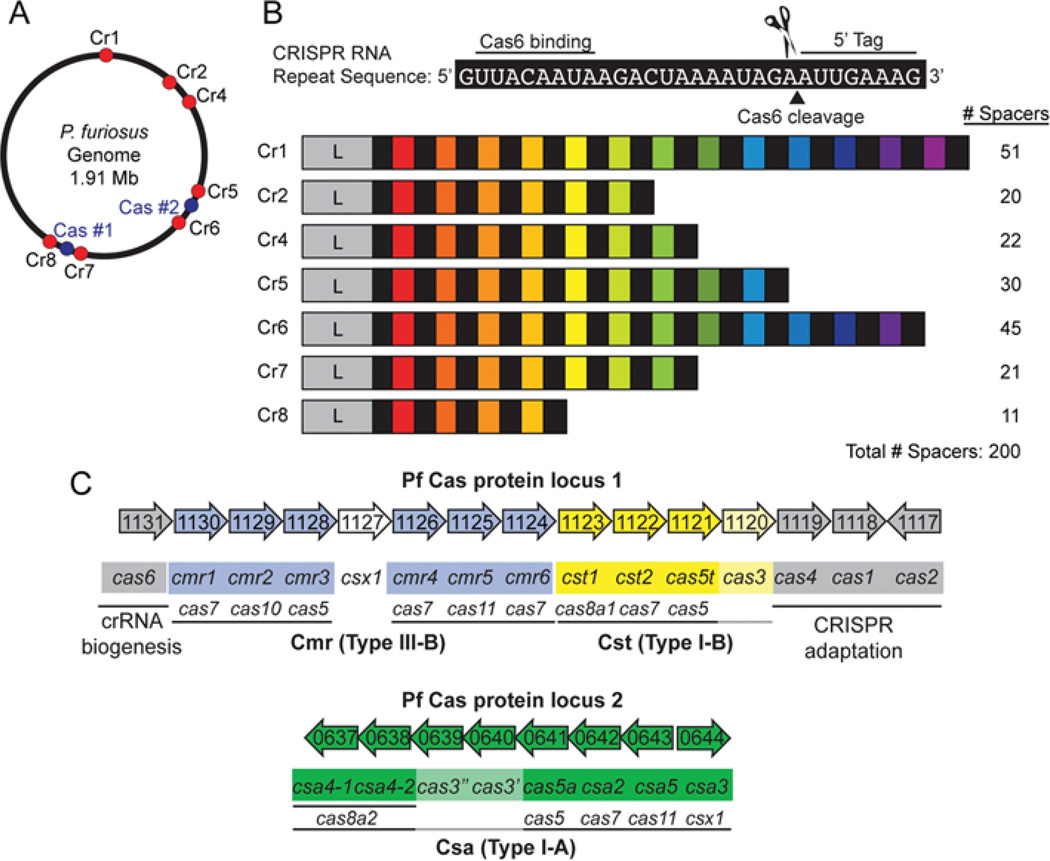
The genome of P. furiosus strains DSM 3638 and COM1 include seven separate CRISPR loci (distributed throughout the genome) that together encode a total of 200 potential crRNAs (Figures 1A and 1B). All seven CRISPR loci share a common 30 bp repeat sequence interspaced by variable numbers of unique spacers (Figure 1B). The majority of spacer sequences are 37 bp in length, but range in size from 34 to 59 bp. The origins of the spacers are unknown as none map to the few currently known Thermococcales viruses or plasmids [24–27]. The P. furiosus repeat RNA is unstructured in solution [28], consistent with the classification of the P. furiosus repeat sequence in the ‘unfolded archaeal cluster 6’ family [29]. Adjacent to each CRISPR array is a ~500 bp leader region; these are highly conserved among the seven CRISPR loci and contain transcriptional start sites [30] and probably also regulatory elements needed for novel spacer acquisition.
Figure 1. P. furiosus CRISPR–Cas systems.
(A) Distribution of the CRISPR loci (Cr1, Cr2, Cr4–Cr8; red) and two major cas gene clusters (Cas; blue) throughout the P. furiosus genome. (B) CRISPRs comprise leader regions (L) preceding series of repeat (black) and spacer (coloured) elements. The actual numbers of spacers are indicated. The consensus crRNA repeat sequence found in the seven CRISPR loci is shown above with the Cas6-binding and cleavage sites, and the 8-nt 5′ tag crRNA sequence indicated. (C) Genome organization of predicted P. furiosus (Pf) cas genes. Subtype III-B Cmr (blue), subtype I-B Cst (yellow), subtype I-A Csa (green) and core cas genes (grey) are indicated, and recent cas gene superfamily designations [15] are indicated below [14]. The known or predicted general function of the core cas genes is indicated.
The genes encoding Cas proteins are primarily organized in two large gene clusters in P. furiosus (Figures 1A and 1C). The first cas gene cluster encodes the core Cas proteins (grey), the Cmr [Cas module RAMP (repeat-associated mysterious proteins)] proteins (blue) and the Cst (Cas subtype Tneap) proteins (yellow). Csa (Cas subtype Apern) proteins (green) are encoded in the second cas gene cluster. The core cas genes are found in many organisms with different CRISPR– Cas systems and include proteins implicated in new spacer acquisition (Cas1, Cas2 and Cas4 proteins) as well as crRNA biogenesis (Cas6 protein). Together, the P. furiosus genome encodes proteins of three predicted effector immune complexes: Cmr (subtype III-B), Csa (subtype I-A) and Cst (subtype I-B) [14,15]. Additional copies of genes encoding predicted Cmr1, Cas6, Cas4 and three other possible Cas proteins are unlinked and scattered elsewhere in the genome [13] for a total of 27 known or predicted Cas proteins in this organism.
Expression and processing of crRNAs
By a combination of deep RNA sequencing and Northern blot analyses, we have determined that all seven P. furiosus CRISPR loci are transcribed and give rise to processed mature crRNA species [13,31]. We have mapped the transcription start sites and promoter elements within the CRISPR leader regions [30]. All crRNAs and Cas proteins appear to be expressed in the absence of infection [13,28,30,31], consistent with an immune system operating in ‘surveillance mode’. We also found that crRNAs containing the most recently acquired invader sequences (encoded proximal to the leader) are generally more abundant [13,30,31].
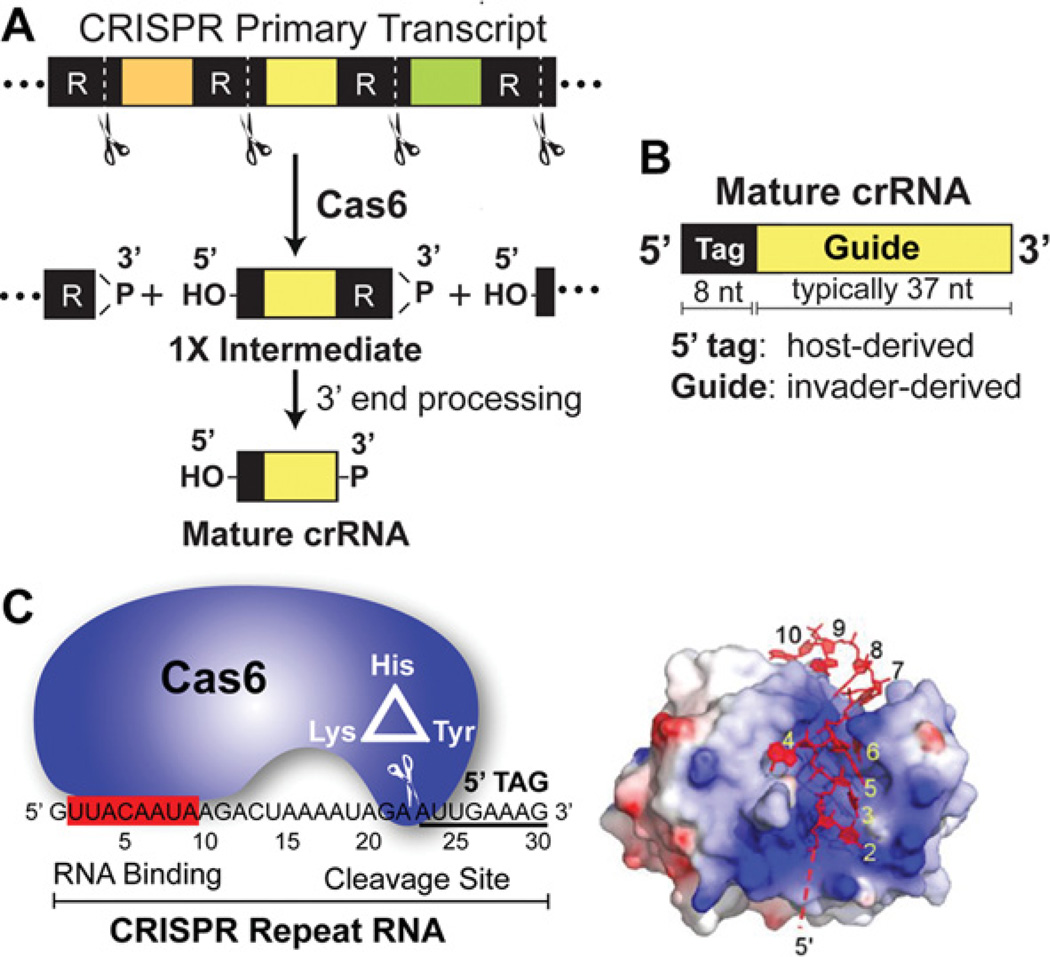
Our studies revealed that P. furiosus Cas6 is responsible for primary processing of precursor CRISPR RNA transcripts and plays a critical role in producing individual mature crRNAs [11,28,32] (Figure 2A). We found that Cas6 is a riboendonuclease that makes a single cut within each repeat sequence of the CRISPR transcript, reducing it step-by-step to intermediate RNAs that we call 1× RNAs (Figures 1B and 2A). The 1× RNA intermediates each contain an 8-nt segment of the repeat at the 5′ end (5′-AUUGAAAG-3′) that is retained in the mature crRNAs (the 5′ tag), an invader-targeting or guide sequence, and 22 nt of repeat sequence at the 3′ end (Figure 2A). Cleavage by P. furiosus Cas6 produces RNAs with 5′ hydroxy and 2′–3′ cyclic phosphate chemical end groups, consistent with its divalent metal-independent cleavage activity [11,28]. Further 3′ end trimming of 1× crRNA intermediates (by unknown mechanisms) produces the various mature crRNA species (Figures 2A and 2B). Cas6 and Cas6-like enzymes also play a critical role in producing mature crRNAs (and generates 8-nt 5′ tag sequences) in other Type I and III CRISPR–Cas systems [1,10,33–35].
Figure 2. Cas6 and crRNA biogenesis.
(A) Cas6 cleaves within the repeat regions of the CRISPR transcript to liberate the individual embedded crRNAs. The products of Cas6 cleavage (1× crRNAs) undergo 3′ end trimming. (B) The architecture of a mature crRNA. This abundant crRNA form includes the 5′ repeat-derived tag and complete guide sequence. Other mature crRNAs in P. furiosus contain some repeat sequences at their 3′ ends or slightly less than full-length guide sequences (not depicted). (C) Cas6 is an endoribonuclease critical for crRNA biogenesis. Cas6 interacts with the 5′ region of the CRISPR repeat RNA (red box) and cleaves at a distal site using an active site composed of three key amino acids (indicated). The Cas6-binding region in the crRNA interacts with the positively charged cleft of Cas6.
In a series of biochemical and structural studies, we determined how Cas6 specifically recognizes and cleaves precursor crRNAs [11,28,32]. Cas6 recognizes the upstream region of the repeat sequence (nucleotides 2–9) and cleaves at a distal downstream site (between nucleotides 22 and 23; 8 nt upstream of the 5′ end of the guide sequence) using an active site composed of a triad of amino acids (tyrosine, histidine and lysine) that resemble the catalytic triad found in tRNA intron-splicing endonuclease [11,28,36]. Our data also indicate that the distance between the binding site and cleavage site, as well as the identity of the nucleotide at the cleavage site, is critical forCas6 action [11,28,32]. A co-crystal structure of P. furiosus Cas6 bound with a non-hydrolysable CRISPR repeat RNA revealed that nucleotides 2–9 contact the protein along the positively charged central cleft on the opposite face of the protein to where cleavage occurs, suggesting that the repeat RNA wraps around through the central cleft to the putative active site [32]. In vivo, Cas6 stably associates with its cleavage product (1× crRNA intermediates), but is not associated with mature crRNAs (that have undergone 3′ trimming, resulting in loss of the identified Cas6-binding site) [28].
The Cmr RNA-targeting defence system
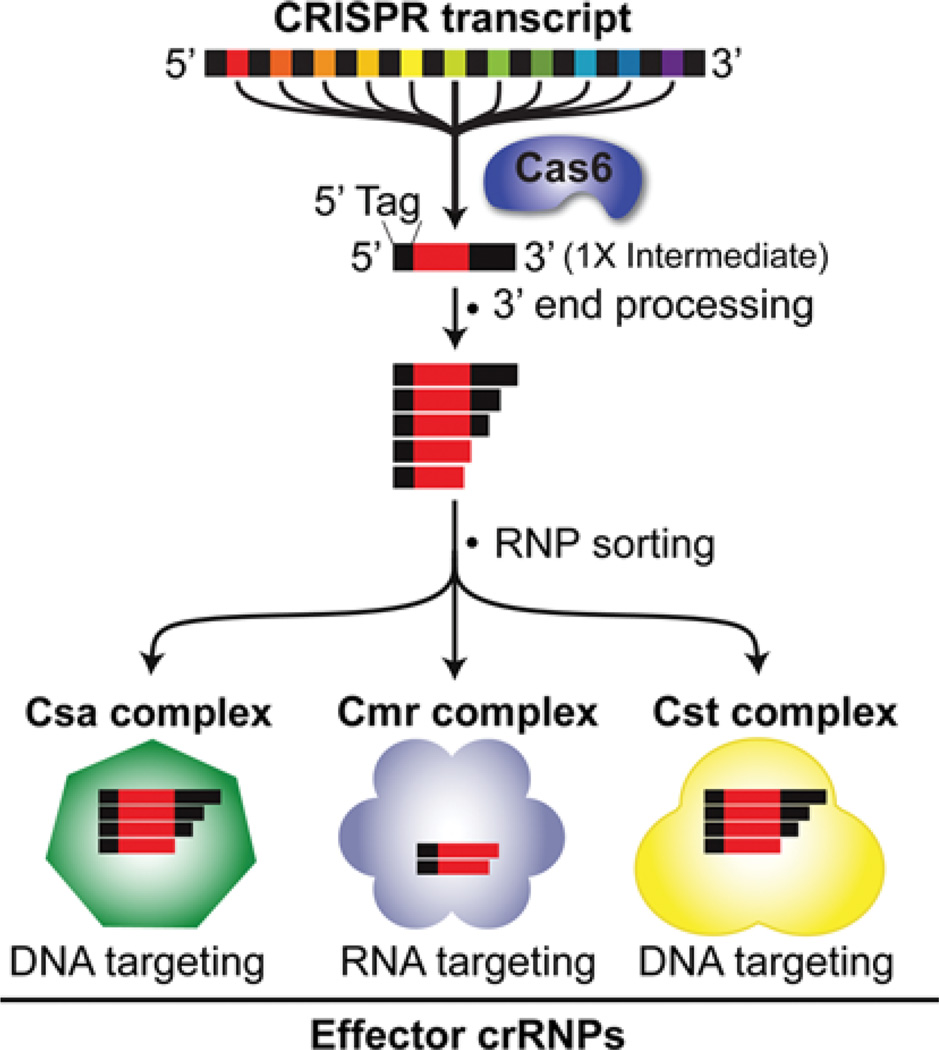
We have isolated and characterized the Cmr effector complex from P. furiosus strains DSM 3638 and COM1 (Figure 3). We identified two crRNA species and six Cmr protein subunits associated with native complexes by RNA deep sequencing and MS [13]. The complexes specifically cleave complementary RNAs in vitro. The two crRNAs associated with the Cmr complex (45 and 39 nt in length) share the common 8-nt 5′ tag (produced by Cas6 cleavage), but have guide sequences (complementary to the target RNA) of 37 and 31 nt respectively, regardless of the length of the guide sequence encoded by the CRISPR spacer [13,30,31]. We reconstituted functional complexes using the recombinant Cmr proteins and synthetic crRNAs. Mutational analysis showed that the 5′ tag is critical for the function of the Cmr crRNP (CRISPR ribonucleoprotein), probably as a Cmr protein recognition site [30]. All six Cmr proteins are important for efficient target RNA cleavage (C.R. Hale, R.M. Terns and M.P. Terns, unpublished work).
Figure 3. crRNAs are loaded into three distinct crRNP immune effector complexes in P. furiosus.
Following release by Cas6 and further 3′ end trimming, crRNAs of specific size formed from all seven CRISPR loci become incorporated into the three distinct crRNP complexes containing Cmr, Csa or Cst Cas proteins. There is evidence that Cmr crRNPs cleave target RNAs, whereas Csa and Cst complexes cleave invader DNA.
The structural organization of the P. furiosus Cmr complex and roles of the individual Cmr proteins and crRNA subunits in the target RNA cleavage reaction are currently under investigation. X-ray structures have been obtained for P. furiosus Cmr2 [37,38], the Cmr2–Cmr3 heterodimer [39,40] and Cmr5 [41]. Cmr2 belongs to the Cas10 family of Type III CRISPR–Cas system signature proteins [17]. Cmr2 is the largest of the Cmr proteins and is composed of several domains including an N-terminal permutated histidine–aspartate (HD) superfamily hydrolase domain, zinc-finger domain, ferredoxin-like fold (cyclase/polymerase domains) and a highly conserved nucleotide/metal-binding domain [37,38]. Cmr2 was a lead candidate for the catalytic component in the Cmr complex cleavage, but mutational analysis ruled out a role for both the HD domain and the nucleotide/metal-binding domain of Cmr2 in target RNA cleavage [37]. Cmr5 is the smallest subunit of the complex, is mostly α-helical in nature [41] and associates physically with Cmr4 ([41], and C.R. Hale, R.M. Terns and M.P. Terns, unpublished work). Cmr1, Cmr3, Cmr4 and Cmr6 are predicted RNA-binding proteins as they contain one or two predicted ferredoxin-fold-like domains that are common to many Cas proteins and resemble RRMs (RNA-recognition motifs) found in RNA-binding proteins from all three domains of life [14,15].
To date, the Cmr effector complex is the only CRISPR–Cas complex known to target RNA (rather than DNA) and is found in ~70% of archaea and ~30% of bacteria with CRISPR–Cas systems, suggesting that it plays an important role in the war against invaders [42]. In recent work, we demonstrated that the Cmr complex can be directed to cleave a desired target RNA in vitro (in a prokaryotic RNAi-like manner) by programming it with an appropriate crRNA [30], indicating that the site-specific cleavage function of crRNA–Cmr complexes could be exploited for gene knockdown.
Csa and Cst DNA-targeting defence systems
We have also isolated native P. furiosus crRNP complexes containing Cst and Csa proteins and distinct profiles of crRNAs (S. Majumdar, R.M. Terns and M.P. Terns, unpublished work) (Figure 3).We identified the Cas proteins by MS and the crRNAs by deep sequencing and Northern blot analysis. The crRNAs associated with the Cst and Csa proteins all include the 8-nt 5′ repeat sequence tag and possess various 3′ ends that often include a portion of the downstream repeat. Using plasmid challenge assays similar to those used to study the function of the Csa and Cst systems in Thermococcus kodakarensis in vivo [43], we have obtained evidence that CRISPR–Cas-mediated defences also target DNA in P. furiosus, presumably via the action of the Csa and/or Cst effector complexes (J.R. Elmore, R.M. Terns and M.P. Terns, unpublished work).
Conclusion
Studies of the CRISPR–Cas systems from P. furiosus are yielding fundamental information about mechanisms of crRNA biogenesis and silencing for three of the diverse CRISPR–Cas pathways (Figure 3). These studies reveal that organisms such as P. furiosus possess multiple RNA-guided mechanisms (including both RNA- and DNA-targeting systems) that provide layered protection against diverse invaders. Future studies are required to obtain a detailed understanding of how each of the individual RNA- and DNA-targeting systems function and are regulated as well as to determine the extent to which these coexisting systems influence the activity of one another. Detailed knowledge of the CRISPR–Cas pathways is also expected to lead to exciting new biomedical and biotechnological applications.
Acknowledgements
We thank the members of the Terns laboratory (present and past) for their contributions to our understanding of P. furiosus CRISPR-based immunity. We express our enthusiastic gratitude to Haruyuki Atomi, Claiborne Glover, Brenton Gravely, Hong Li, Lance Wells and Jan Westpheling for enjoyable and fruitful collaborations.
Funding
This work was supported by the National Institutes of Health [grant number R01 GM54682]
Abbreviations used
- Cas
CRISPR-associated
- Cmr
Cas module RAMP (repeat-associated mysterious proteins)
- CRISPR
clustered regularly interspaced short palindromic repeats
- crRNA
CRISPR RNA
- crRNP
CRISPR ribonucleoprotein
- Csa
Cas subtype Apern
- Cse
Cas-subtype Escherichia coli
- Cst
Cas subtype Tneap
References
- 1.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu. Rev. Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 2.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 3.Westra ER, Swarts DC, Staals RH, Jore MM, Brouns SJ, van der Oost J. The CRISPRs, they are a-changin’: how prokaryotes generate adaptive immunity. Annu. Rev. Genet. 2012;46:311–339. doi: 10.1146/annurev-genet-110711-155447. [DOI] [PubMed] [Google Scholar]
- 4.Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr. Opin. Microbiol. 2011;14:321–327. doi: 10.1016/j.mib.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhaya D, Davison M, Barrangou R. CRISPR–Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 6.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 7.Lillestøl RK, Redder P, Garrett RA, Brügger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mojica FJ, Díez-Villaseñor C, García-Martí nez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 9.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 10.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA–Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR–Cas systems. Nat. Rev. Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 19.Robb FT, Maeder DL, Brown JR, DiRuggiero J, Stump MD, Yeh RK, Weiss RB, Dunn DM. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 2001;330:134–157. doi: 10.1016/s0076-6879(01)30372-5. [DOI] [PubMed] [Google Scholar]
- 20.Norais C, Moisan A, Gaspin C, Clouet-d’Orval B. Diversity of CRISPR systems in the euryarchaeal Pyrococcales. RNA Biol. 2013;10:659–670. doi: 10.4161/rna.23927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farkas J, Chung D, DeBarry M, Adams MW, Westpheling J. Defining components of the chromosomal origin of replication of the hyperthermophilic archaeon Pyrococcus furiosus needed for construction of a stable replicating shuttle vector. Appl. Environ. Microbiol. 2011;77:6343–6349. doi: 10.1128/AEM.05057-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farkas J, Stirrett K, Lipscomb GL, Nixon W, Scott RA, Adams MW, Westpheling J. Recombinogenic properties of Pyrococcus furiosus strain COM1 enable rapid selection of targeted mutants. Appl. Environ. Microbiol. 2012;78:4669–4676. doi: 10.1128/AEM.00936-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipscomb GL, Stirrett K, Schut GJ, Yang F, Jenney FE, Jr, Scott RA, Adams MW, Westpheling J. Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 2011;77:2232–2238. doi: 10.1128/AEM.02624-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinf. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rousseau C, Gonnet M, Le Romancer M, Nicolas J. CRISPI: a CRISPR interactive database. Bioinformatics. 2009;25:3317–3318. doi: 10.1093/bioinformatics/btp586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geslin C, Le Romancer M, Erauso G, Gaillard M, Perrot G, Prieur D. PAV1, the first virus-like particle isolated from a hyperthermophilic euryarchaeote “Pyrococcus abyssi”. J. Bacteriol. 2003;185:3888–3894. doi: 10.1128/JB.185.13.3888-3894.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soler N, Marguet E, Cortez D, Desnoues N, Keller J, van Tilbeurgh H, Sezonov G, Forterre P. Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins. Nucleic Acids Res. 2010;38:5088–5104. doi: 10.1093/nar/gkq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA. 2010;16:2181–2188. doi: 10.1261/rna.2230110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CV, 3rd, Graveley BR, Terns RM, Terns MP. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol. Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2479. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure. 2011;19:257–264. doi: 10.1016/j.str.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gesner EM, Schellenberg MJ, Garside EL, George MM, Macmillan AM. Recognition and maturation of effector RNAs in a CRISPR interference pathway. Nat. Struct. Mol. Biol. 2011;18:688–692. doi: 10.1038/nsmb.2042. [DOI] [PubMed] [Google Scholar]
- 34.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calvin K, Li H. RNA-splicing endonuclease structure and function. Cell. Mol. Life Sci. 2008;65:1176–1185. doi: 10.1007/s00018-008-7393-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cocozaki AI, Ramia NF, Shao Y, Hale CR, Terns RM, Terns MP, Li H. Structure of the Cmr2 subunit of the CRISPR–Cas RNA silencing complex. Structure. 2012;20:545–553. doi: 10.1016/j.str.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu X, Ye K. Crystal structure of Cmr2 suggests a nucleotide cyclase-related enzyme in type III CRISPR–Cas systems. FEBS Lett. 2012;586:939–945. doi: 10.1016/j.febslet.2012.02.036. [DOI] [PubMed] [Google Scholar]
- 39.Shao Y, Cocozaki AI, Ramia NF, Terns RM, Terns MP, Li H. Structure of the Cmr2–Cmr3 subcomplex of the Cmr RNA silencing complex. Structure. 2013;21:376–384. doi: 10.1016/j.str.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osawa T, Inanaga H, Numata T. Crystal structure of the Cmr2–Cmr3 subcomplex in the CRISPR–Cas RNA silencing effector complex. J. Mol. Biol. 2013;425:3811–3823. doi: 10.1016/j.jmb.2013.03.042. [DOI] [PubMed] [Google Scholar]
- 41.Park JH, Sun J, Park SY, Hwang HJ, Park MY, Shin M, Kim JS. Crystal structure of Cmr5 from Pyrococcus furiosus and its functional implications. FEBS Lett. 2013;587:562–568. doi: 10.1016/j.febslet.2013.01.029. [DOI] [PubMed] [Google Scholar]
- 42.Shah SA, Garrett RA. CRISPR/Cas and Cmr modules, mobility and evolution of adaptive immune systems. Res. Microbiol. 2011;162:27–38. doi: 10.1016/j.resmic.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Elmore JR, Yokooji Y, Sato T, Olson S, Glover CV, 3rd, Graveley BR, Atomi H, Terns RM, Terns MP. Programmable plasmid interference by the CRISPR-Cas system in Thermococcus kodakarensis. RNA Biol. 2013;10:828–840. doi: 10.4161/rna.24084. [DOI] [PMC free article] [PubMed] [Google Scholar]