Abstract
Species are generally regarded as a fundamental unit of biodiversity. By contrast, higher taxa such as genera and families, while widely used as biodiversity metrics and for classification and communication, are generally not believed to be shaped by shared evolutionary processes in the same way as species. We use simulations to show that processes which are important for emergence of evolutionarily significant units (ESUs) at the species level, namely geographical isolation and ecological divergence, can generate evolutionary independence above the species level and thereby lead to emergence of discrete phylogenetic clusters (higher ESUs). Extending phylogenetic approaches for delimiting evolutionarily significant species to broader phylogenetic scales, we find evidence for the existence of higher ESUs in mammals. In carnivores, euungulates and lagomorphs the hierarchical level of units detected correspond, on average, to the level of family or genus in traditional taxonomy. The units in euungulates are associated with divergent patterns of body mass, consistent with occupation of distinct ecological zones. Our findings demonstrate a new framework for studying biodiversity that unifies approaches at species and higher levels, thus potentially restoring higher taxa to their historical status as natural entities.
Keywords: adaptive radiation, Artiodactyla, Carnivora, diversification, Lagomorpha, Perissodactyla
1. Introduction
Identifying units of biodiversity is an important step in understanding how diversity evolves. Species are generally regarded as real biological entities because their members not only share evolutionary ancestry, but also interact through shared evolutionary processes such as interbreeding and natural selection [1]. Numerous concepts have developed to define what species are [2] and their status as real, biological entities is supported by a rich theoretical and empirical literature [1,3,4]. Classically, higher taxa (e.g. those named as genera and families) were also viewed as natural entities [5–8], but the recent view has been that there are no shared processes above the species level that generate evolutionarily significant units (ESUs) equivalent to species [1,2]. Consequently, while named higher taxa are convenient for communication and classification they are not attributed evolutionary significance in the way that species are, beyond summarizing ancestry and approximate levels of relatedness. This limits the use of higher taxa as comparative units [9], although they continue to be used as biodiversity units in many disciplines (e.g. conservation biology, macroecology and palaeontology).
Mid-twentieth century evolutionary biologists held a view of higher taxa that encompassed both their processes of formation and resulting pattern of discreteness. The basic tenet was that acquisition of new, key adaptations allows occupation of and radiation into novel ecological or adaptive space leading over time to the emergence of distinct lineages of adaptively similar species, recognized by systematists as higher taxa [5,7]. With the rise of phylogenetic methods, there was a shift towards focusing instead on clades—groups of species sharing a common ancestor—but without consideration of processes that may earn some clades the status of a particular taxonomic level over others [10]. Higher taxa remain in usage as labels for higher groups, but units of diversity above the species are deemed to reflect only ancestry, not the action of evolutionary forces like those that maintain coherence within species and promote divergence between them [1,11]. Instead, diversity patterns above the species level are mostly explained in terms of shifts in rates of diversification [e.g. 12,13]. Recent work [14] has found phylogenetic evidence consistent with radiation of species into new adaptive zones but the emphasis has remained mainly on rates (but see [15]), rather than on evolutionary processes generating patterns of coherence and disparity in phylogenetic branching across higher clades. The processes involved in the formation of discrete entities at the species level and below are treated as distinct from those operating at broader scales (distinguished as ‘microevolution’ and ‘macroevolution’ [16]), where efforts to define groups are wholly divorced from efforts to understand the evolutionary processes affecting biodiversity patterns at those scales.
Here, we use a simulation model to show how ESUs above the species (higher ESUs) can evolve and a novel approach for analysing the phylogenetic branching pattern across broad clades to demonstrate their existence in mammals. Our simulations are designed to capture the phylogenetic signature of applying population genetic theory to higher clades. We simulate a metacommunity of species in which the total number of individuals is governed by ecological limits. The composition and diversity of the metacommunity fluctuate over time owing to species turnover, caused by chance birth and death of individuals. By splitting a single metacommunity into two, through separation in geographical or ecological space, we show that discrete, independently evolving entities can evolve above the species. Higher level phylogenetic clusters can arise if species in one area (or ecological guild) are independently limited [17,18] from species in other areas (or guilds): species turnover occurs within clusters but species in one cluster cannot replace species in another. This leads over time to the emergence of discrete phylogenetic clusters of closely related species.
To test for the presence of such entities in mammals, we extend the generalized mixed Yule coalescent (GMYC) method of species delimitation [11,19] to analysis of higher clades. In brief, the GMYC method uses a gene tree to test for the existence of phylogenetic clusters, separated by long internal branches, compared to a null model of evenly distributed branching across the tree. Because the method requires dated trees containing dense samples of species, we focused on three clades (encompassing four orders) for which genetic data are available in sufficient depths (cytochrome b (cytb) sequences for approx. 80% of species in Carnivora (cats, dogs, bears, seals and mustelids), Euungulata (odd-toed and even-toed ungulates and cetaceans) and Lagomorpha (rabbits, hares and pikas)). We also repeated the tests on a sparser sample across the entire Mammalia (46% of species). There is strong evidence for the existence of higher ESUs, corresponding to traditionally named families or genera. Analyses of trait variation suggest that they were established following both geographical and ecological shifts. Our study provides a new framework for studying biodiversity that unifies current approaches at species and higher levels and provides a connection between evolution and taxonomy, currently lacking above the species level.
2. Material and methods
(a). Simulation model
We use a simulation model to show how processes that cause emergence of ESUs at the species level—through the maintenance of coherence within species and divergence between species—can operate more broadly to generate discrete, ESUs of diversity above the species (higher ESUs; hESUs). The model first considers a metacommunity of a given number of individuals in a single ancestral region, as in neutral models of biodiversity [20]. New species arise by speciation events, which occur at random with an average per metacommunity rate. Species abundances fluctuate by drift over time because individuals are recruited at random with respect to species membership every generation. Species go extinct if their abundances dwindle to zero. The key features of the model are that species compete for resources (owing to fixed metacommunity size determined by the carrying capacity of the region) and there is species turnover owing to recurring speciation and extinction [21–23]. Such a view of diversity is gaining empirical support from analyses of phylogenetic patterns among living species [e.g. 23–25] and is consistent with patterns in the fossil record [e.g. 26,27]. We then split the initial metacommunity into two, which might represent two geographically isolated regions or two ecological guilds of species using different resources [28]. Then we compare the phylogenetic outcome of varying the level of isolation between the two metacommunities, by varying the rate of dispersal between regions or shifts between ecological guilds.
The model was devised as follows. Consider a number of coexisting species, S, with abundances, Ni, in a region that can support a finite number of individuals, K. Speciation occurs randomly at a rate b per metacommunity per generation. When a speciation event occurs, a parental species is chosen at random in proportion to Ni and the two descendant species are assigned abundances following a broken stick model. Extinction occurs when Ni = 0. Now consider the introduction of a barrier, splitting the metacommunity into R regions. Then let species disperse between regions randomly at a rate, D. Starting parameters were set to S = 100 and R = 2. Other parameters were sampled at random from a uniform distribution: the total number of individuals, Ntot, was sampled between 104 and 105; b between 10−4 and 10−2; D between log(10−6) and log(0.9), individuals (Ntot) were split evenly among regions to represent a region's K and species abundances, Ni, were set initially to K/S.
Each simulation was run for 10Ntot generations. Model output was recorded for the first 103 generations and then every 103 generations for a total of 103 individual simulations. At the end of each simulation, the phylogenetic pattern of the standing diversity was recorded and the influence of different parameters on the evolution of hESUs, measured as a high ratio of between to within region phylogenetic distances, was determined with regressions. All analyses were performed in R [29].
(b). Tree building
Cytb sequences for Carnivora (285 species; 82.5% sampled), Euungulata (346 species; 87.3% sampled) and Lagomorpha (92 species; 77.2% sampled) were obtained from previous studies [30,31] and GenBank (http://www.ncbi.nlm.nih.gov/genbank/) using International Union for the Conservation of Nature (IUCN) species lists ([32]; electronic supplementary material, table S1). We chose cytb because it is the best sampled gene for mammals and our analyses rely on dated trees encompassing as many species as possible. Sequences were aligned by eye in Geneious Pro v. 5.3.6 (2005–2006 Biomatters Ltd.). Trees were generated in BEAST [33,34] with two or three independent runs of 107 generations of Markov chain Monte Carlo sampling, a GTR + I + G substitution model, a birth–death (BD) tree prior and an uncorrelated lognormal relaxed clock prior. Three age priors [35] were set per tree (electronic supplementary material, table S2). Convergence among runs and mixing and sampling of parameters within runs were evaluated in Tracer and post-burnin samples of trees (−10%) summarized in LogCombiner and TreeAnnotator.
The resulting trees differed substantially from other recent, detailed studies [36–39]. Previous studies have shown that cytb can yield relationships not supported by other data [40] and that mitochondrial DNA in combination with only a few calibration points can inflate age estimates [36,37]. We therefore generated a second set of trees, employing numerous topological constraints and age priors based on empirical estimates from multigene studies (electronic supplementary material, table S3). Analyses were run as above but for 307 generations on the CIPRES Science Gateway [41].
The broadly sampled mammalian tree was constructed by searching GenBank for mammalian cytb sequences following the IUCN [32] species lists and a modified version of the database querying script from phyloGenerator [42]. Additional sequences were added from the detailed matrices (above) if these species were not found by the queries (e.g. owing to synonymy differences; electronic supplementary material, table S1). Sequences were aligned using the translation align algorithm of ClustalW v. 2.0.11 [43], followed by manual editing in Geneious Pro v. 5.3.6. A maximum-likelihood (ML) tree was generated using RAxML [44], run on the CIPRES Science Gateway [41], using 100 rapid bootstrap searches followed by a full ML search. Trees were inspected for rogue sequences (e.g. those on extremely long branches), which were removed before another search was performed with the backbone constrained according to the topology of Meredith et al. [45]. In addition, Chiroptera, Rodentia and Eulipotyphla and each of their sampled constituent families were constrained to be monophyletic. The resulting tree was made ultrametric in pathd8 [46] using 135 age constraints (electronic supplementary material, table S4) based on published empirical estimates [45]. The final tree was based on sequence data for 2538 species (46%), representing all orders and 89% of mammalian families [32].
(c). Inference of evolutionarily significant units above the species
To test for the signature of independent limitation above the species, we extended the GMYC method of species delimitation [11,19] to analysis of higher clades. At the species level, the GMYC method tests whether the gene tree of a sample of individuals conforms to a single population coalescent or has diversified into independently evolving species. The presence of independently evolving species is supported if one or more increases in the rate of phylogenetic branching, which cannot be explained by a single coalescent process alone, have occurred. Such shift(s) are interpreted as transitions from between (diversification) to within (coalescence) species processes. Analysis above the species using the GMYC method requires a densely sampled, time calibrated tree, with tips representing species rather than individuals. If all species belong to a single entity (the null expectation), then the pattern of phylogenetic branching will fit a constant rates process [47]. If species are differentiated into independently limited higher ESUs (the alternative expectation) one (single threshold (ST) model) or more (multiple threshold (MT) model) increases in the rate of branching towards the present should be apparent [19,48,49], denoting transition from between to within hESU branching. The MT model allows the transition to occur at different times in different lineages and may be important for analysis above the species, where heterogeneity in the mode and tempo of evolution is to be expected.
For analysis of higher clades in mammals, outgroup taxa were pruned from each constrained maximum clade credibility (MCC) tree and the fit of the null (single branching process) and alternative (one or more shifts in branching rate) GMYC models compared using Akaike information criterion (AIC) weights. Inferences were made from the 95% confidence set of models. Support for each hESU was obtained from the Akaike weights (GMYC support; GS [49]). The influence of incomplete sampling was taken into account by multiplying the GS by the robustness of each hESU to addition of missing species (see §2d). Finally, GMYC models were fitted to the large mammalian tree. Analyses were carried out using the R package splits [50] and code modified from Fujisawa & Barraclough [49].
(d). Investigating the effect of incomplete sampling
The equations behind the GMYC method assume complete sampling within and across hESUs [19]. The possibility that our sample—both incomplete and non-random—has biased results was quantified by adding missing species based on known genus or family membership (electronic supplementary material, table S5) [32], assuming a Yule branching process [51]. Tips were imputed at a randomly chosen phylogenetic position within their constituent group, attaching to a crown or the stem branch, at a randomly selected position along the chosen branch. The length of the imputed branch was determined as the depth of the position of attachment. For each dataset, this was repeated 500 times to generate a set of 500 simulated ‘completely sampled’ trees per clade. Null, ST and MT GMYC models were fitted to each of the simulated trees and ML solutions compared to those obtained for each empirical tree. To test the robustness of GMYC inferences to decreasing levels of sampling more generally, species were selected at random and removed incrementally from each MCC tree to generate 100 jackknifed trees, each with 70, 60 and 50% sampling (800 trees in total; lagomorph jackknifed trees were generated for 60 and 50% sampling only). GMYC inferences across jackknifed trees were compared to empirical inferences as above.
(e). The power of the generalized mixed Yule coalescent method applied above the species
To test the power of the GMYC method for detecting hESUs further, we simulated species trees of the same size as the empirical trees under a BD model of evolution [47]. Speciation (λ) and extinction (μ) parameters were randomly sampled from the range of estimates obtained across 500 empirical trees for each dataset. The estimate of extinction for carnivores was always zero so this set of trees was simulated with μ = 0, thus representing a pure birth (Yule) branching process [51]. We simulated 500 trees for each dataset. The GMYC models were fitted to each tree in turn and the fit of each compared using a likelihood ratio (LR) test with two degrees of freedom (d.f.). Finally, the difference in fit across simulated trees was compared to the difference in fit for each empirical dataset. Speciation and extinction parameters were estimated in the R package ape [52], Yule trees were simulated in diversitree [53] and BD trees in Geiger [54]. The expectation is that no hESUs will be found for trees simulated under a constant rates model and, indeed, the null model was not rejected for 88–93% (ST) or 68–79% (MT) of the trees. This suggests a Type I error rate of 7–12% (ST) or 21–32% (MT). We therefore used the difference in fit between null and alternative models across simulated trees to determine the critical value for significance for empirical trees.
(f). Testing for trait coherence within higher evolutionarily significant units
If hESUs reflect ecological divergence into different guilds, we might expect to observe shared ecological traits indicative of guild membership. For example, Uyeda et al. [55] found evidence for bounded evolution among close relatives but substantial shifts over longer timescales, which they argued reflected a signature of adaptive zones. Within hESU coherence is expected to be detectable as lower trait variance among species within than among hESUs, equivalent to a decrease in the rate of trait change under Brownian motion (BM) [56,57]. To test for such a pattern, the continuous traits with the highest data coverage (less than or equal to 25% missing data for sampled species) were extracted from PanTHERIA [58]. For non-marine carnivores, we also obtained a dataset of seven skull measurements from S. Meiri, representing 169 or 194 of the species in the phylogeny and with on average 82 skulls measured per species (see also [59]). This resulted in a database of 12 traits for Carnivora, seven for Euungulata and five for Lagomorpha (table 2). Because we were interested in testing for the expected pattern generally we analysed all these traits without any a priori hypotheses about their direct roles in influencing diversification dynamics.
Table 2.
Fit of single-rate and two-rate models of trait change for (a) Carnivora, (b) Euungulata and (c) Lagomorpha. AET, actual evapotranspiration; M1, length of lower carnassials; PM4, length of upper carnassials; CSL, maximum diameter of upper canine; ZYG, breadth of skull at widest part of zygomatic arches; SH (skull height), height of occipital triangle; CBL, condylo-basal length; SW, skull width.
| traitb | LL (single-rate) | LL (two-rate)c | within-hESU ML rate | between-hESU ML rate | within: between hESU ratio |
|---|---|---|---|---|---|
| (a) log adult body mass | 111.4 | 215.7*** | 0.20 | 0.0023 | 89.2 |
| litter size | −254.2 | −124.5*** | 1.41 | 0.0032 | 435.1 |
| Log AET (mean) | 182.1 | 336.3*** | 0.13 | 0.00011 | 1200.3 |
| latitude–longitudea | −1049463 | −66548.1*** | 7762.2 | 2017.6 | 3.85 |
| M1 | 608.9 | 676.2*** | 0.015 | 0.00065 | 23.6 |
| PM4 | 547.5 | 619.4*** | 0.022 | 0.00057 | 38.1 |
| CSL | 436.4 | 549.6*** | 0.037 | 0.00018 | 202.3 |
| Log SH | 557.7 | 662.8*** | 0.020 | 0.00014 | 142.8 |
| Log SW | 691.3 | 711.7*** | 0.014 | 0.00035 | 38.8 |
| Log ZYG | 358.8 | 462.6*** | 0.042 | 0.00024 | 175.2 |
| Log CBL | 555.2 | 655.7*** | 0.020 | 0.00021 | 94.3 |
| (b) log adult body massb | 536.3 | 594.5*** | 0.040 | 0.068 | 0.59 |
| litter size | 631.0 | 724.4*** | 0.029 | 0.000060 | 475.1 |
| diet breadth | 77.3 | 202.1*** | 0.25 | 0.0013 | 191.5 |
| Log AET (mean) | 475.6 | 510.3*** | 0.029 | 0.010 | 2.79 |
| latitude–longitudea | −384498.6 | −3371.8*** | 558113.9 | 87640.7 | 6.37 |
| (c) log adult body mass | 88.0 | 97.3*** | 0.049 | 0.023 | 2.1 |
| litter size | −81.1 | −29.7*** | 1.84 | 0.00000095 | 1931478.0 |
| Log AET (mean) | 59.4 | 104.9*** | 0.14 | 0.0000013 | 107680.7 |
| latitude–longitudea | −702428.2 | −71277.8*** | 9356.5 | 1443.6 | 6.5 |
aLatitudinal midrange and longitudinal midrange were fitted together using custom scripts modified from the ‘ace’ function of ape.
bLower rates within than between hESUs were found for body mass in euungulates (shown in bold).
cSignificance of the alternative model is denoted by asterisks (***p ≤ 0.001).
Fit of a single-rate model (BM) was compared with the fit of a two-rate model, where separate rates were optimized on branches within and between hESUs. This approach rules out the possibility that observed clumping of trait variation arose solely owing to chance Brownian walks [60]. Internal nodes were labelled based on the GMYC results with the highest support (figure 2; electronic supplementary material, table S6). Models were fitted using a customized version of the ‘ace’ functions in ape and the difference in fit was assessed with LR tests with 1 d.f.
Figure 2.
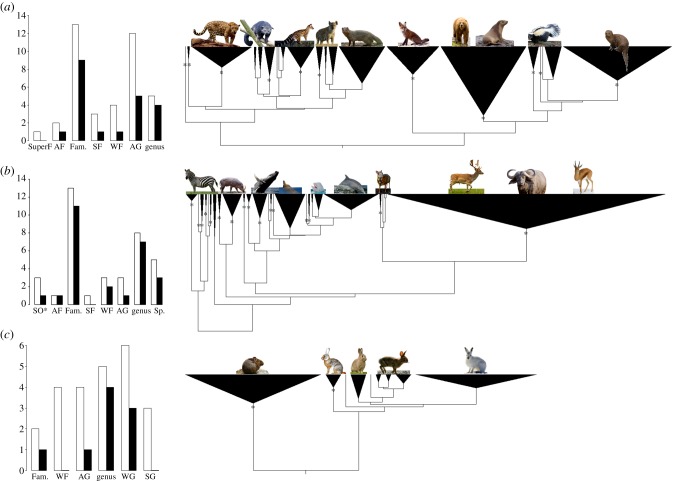
ESUs above the species (higher ESUs) in carnivores (a), euungulates (b) and lagomorphs (c). The solution with the highest GMYC support is shown (see also the electronic supplementary material, table S8 and figure S3). Stars denote GS ≥ 0.80 on a 0–1.0 scale. Left-hand panels: taxonomic rank (formal) or level (informal) of hESUs inferred from all models in the confidence set (white) or the solution with the highest support (black), arranged by decreasing taxonomic rank: SO*, suborder; AF, above family; SuperF, superfamily; Fam., family; WF, within family; SF, subfamily; AG, above genus; SG, subgenus; Sp., species. *SO comprises 1 suborder, 1 infraorder and 1 superfamily. Images: Wikipedia commons.
To allow for the possibility that trait evolution varies in different regions of the tree, we next fitted a model that searches for rate shifts anywhere on the tree, constrained to find only well-supported shifts (ΔAIC ≥ 8 for shifts within clades of at least five species; ΔAIC ≥ 6 for a minimum clade size of 10 [61]) as implemented in the ‘transformPhylo.ML’ function from the R package motmot [61]. Background rates were determined by excluding clades with exceptional rates and then estimating the average (BM) rate for the remaining sample. Rate shifts were scored as speed-ups or slowdowns based on the rate at the preceding node. Model fit was assessed using AIC with a correction for finite sample size (AICc). Shifts associated with hESUs were determined by eye.
3. Results
(a). Evolution of significant units of diversity above the species
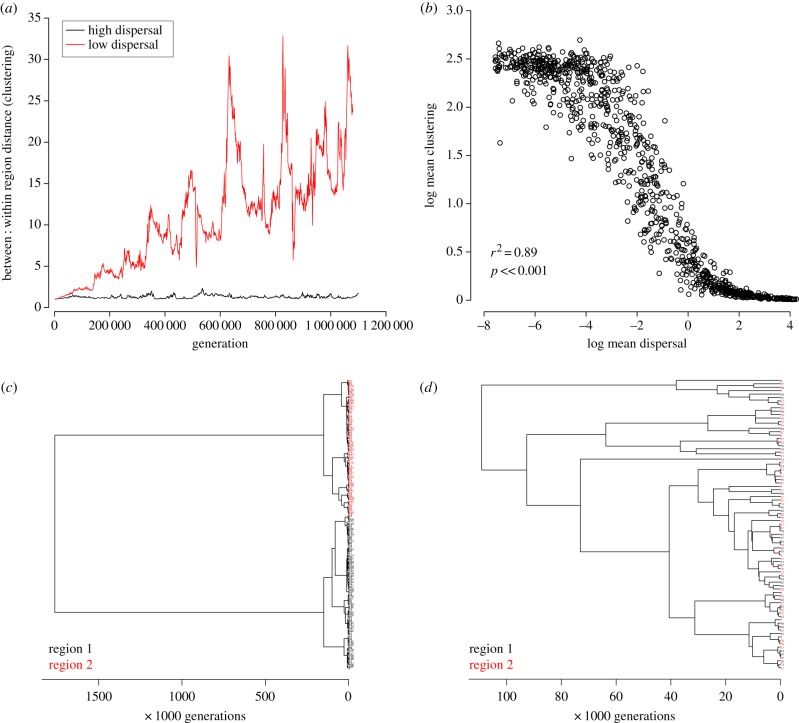
Our simulation model revealed that species turnover occurs separately within each region when rates of dispersal between regions (or shifts between ecological guilds) are low. This leads to the maintenance of low phylogenetic diversity within regions while divergence between regions increases over time (figure 1a), indicated by long internal branches separating species clusters occupying each region (or guild; figure 1c). Thus, higher ESUs evolve: two distinct clusters, each comprising closely related species that share evolutionary cohesion owing to ongoing turnover within each region (or guild). By contrast, when there is dispersal of species between regions, even at relatively low rates, between-region divergence remains low (figure 1c), formation of species clusters diminishes dramatically (figure 1b) and no phylogenetic separation of species in different regions is evident (figure 1d).
Figure 1.
The evolution of ESUs above the species level. Consider two sets of species corresponding to higher clades, separated geographically (or ecologically). (a) At low dispersal rates between regions the ratio of between to within region phylogenetic distance increases over time. At high dispersal rates the ratio between : within region distance remains low. (b) Relationship between dispersal rate and phylogenetic clustering, measured as between : within region distance. (c) Phylogenetic pattern under low dispersal rates: distinct clades of closely related species emerge in each region, separated from each other by long internal branches. (d) Phylogenetic pattern under high dispersal rates: no clustering beyond that expected under neutral coalescence occurs and there is no evidence of occupation of different regions. These results apply equally to separation into different ecological guilds, in which case degree of isolation of metapopulations refers to rate of shifts between guilds rather than rates of geographical dispersal.
(b). Evolutionarily significant units above the species in mammals
Based on analysis of constrained trees (electronic supplementary material, figure S1; available from TreeBase: http://purl.org/phylo/treebase/phylows/study/TB2:S15307), we found strong evidence for hESUs in all three clades: (p = 0.001 (carnivores and euungulates), p = 0.01 (lagomorphs), LR tests between null and MT models; table 1). These results remain significant using a critical value for significance determined through BD simulations of species trees (carnivores and euungulates at p = 0.01 and lagomorphs at p = 0.05; electronic supplementary material, figure S2). Inference across the 95% confidence set of GMYC models indicated 17–24 hESUs in carnivores, 18–29 in euungulates and 2–11 in lagomorphs (electronic supplementary material, tables S6–S7). In carnivores and euungulates, hESUs mostly correspond to traditionally named families and infra-family groups (figure 2; electronic supplementary material, table S8, figure S3). Lagomorph units correspond to genera and infrageneric units.
Table 1.
ML GMYC results for single (ST) and multiple (MT) thresholds models across (a) empirical, (b) complete and (c) jackknifed trees. Significance compared to the null model is determined with a likelihood ratio test with 2 d.f. and denoted by asterisks. For comparisons across multiple trees, asterisks denote median significance. Ranges reported for analyses across multiple trees denote median and 95% confidence interval. Lh, maximum likelihood; Ma, Mega annum. **p ≤ 0.01, ***p ≤ 0.001.
| ST |
MT |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| dataset | Lh (null) | Lh | hESUs | threshold (Ma) | Lh | hESUs | mean threshold (Ma) | lower threshold | upper threshold |
| (a) empirical trees | |||||||||
| Carnivora (82.5%) | 319.46 | 326.05*** | 19 | 19.0 | 328.17*** | 22 | 14.6 | 4.1 | 20.4 |
| Euungulata (87.3%) | 475.90 | 487.52*** | 18 | 20.5 | 491.87*** | 29 | 11.6 | 3.98 | 26.1 |
| Lagomorpha (77.2%) | 42.89 | 48.76** | 5 | 11.8 | 49.35** | 9 | 5.53 | 0.23 | 11.5 |
| Mammalia (46%) | 8279.6 | 8305.6*** | 100 | 40.4 | 8379.0*** | 224 | 17.0 | 5.84 | 37.9 |
| (b) completely sampled trees | |||||||||
| Carnivora (100%) | 472.4 (467.6–477.8) | 477.5 (473.4–481.5)** | 18 (18–19) | 19.5 (19.0–19.5) | 480.0 (475.7–483.6)*** | 22 (21–47.5) | 14.3 (8.19–14.6) | 6.40 (0.88–6.42) | 29.4 (17.3–29.6) |
| Euungulata (100%) | 623.3 (619.3–628.0) | 634.0 (630.8–637.6)*** | 12 (12–18) | 26.6 (20.5–26.6) | 636.3 (633.8–639.7)*** | 20 (16–21) | 13.6 (10.9–14.7) | 4.23 (1.29–4.30) | 28.0 (24.6–28.9) |
| Lagomorpha (100%) | 92.1 (88.4–95.3) | 97.4 (95.0–99.4)** | 5 (3–11) | 11.8 (9.5–13.4) | 97.8 (95.8–101.1)** | 10 (4–15.5) | 7.17 (5.38–10.1) | 4.31 (0.33–6.81) | 11.5 (11.3–14.6) |
| (c) jackknifed trees | |||||||||
| Carnivora (70%) | 225.0 (219.6–231.1) | 231.2 (226.5–238.2)** | 19 (17–26) | 19.0 (15.0–19.5) | 234.3 (228.7–242.3)*** | 23 (20–45.5) | 13.6 (9.73–15.0) | 6.16 (1.32–8.23) | 28.4 (16.2–30.2) |
| Carnivora (60%) | 154.6 (149.2–161.3) | 160.5 (155.0–167.9)** | 20 (17–27.5) | 17.3 (13.4–19.5) | 163.7 (158.9–171.4)*** | 37 (20–43) | 11.7 (10.0–14.8) | 5.87 (1.62–8.58) | 18.6 (16.3–30.1) |
| Carnivora (50%) | 94.9 (88.5–101.7) | 99.6 (94.0–107.8)** | 19 (16–26) | 17.4 (14.0–19.5) | 102.3 (97.3–110.7)*** | 31.5 (19.5–43) | 12.4 (10.0–14.8) | 5.89 (1.53–10.2) | 19.2 (16.2–30.7) |
| Euungulata (70%) | 305.6 (299.3–312.8) | 316.5 (310.6–324.1)*** | 17 (12–28) | 20.5 (15.3–26.6) | 321.3 (314.4–328.5)*** | 28 (19–37.5) | 11.6 (10.0–15.5) | 4.16 (0.97–4.89) | 23.4 (19.1–27.9) |
| Euungulata (60%) | 218.8 (210.6–228.8) | 230.1 (221.5–242.4)*** | 17 (10.5–28) | 20.5 (15.3–28.3) | 234.6 (226.2–246.5)*** | 27 (19–37.5) | 11.7 (10.1–15.5) | 4.11 (0.66–5.72) | 21.2 (18.4–27.8) |
| Euungulata (50%) | 138.0 (132.1–145.0) | 148.5 (141.4–158.3)*** | 16 (11.5–26) | 20.5 (15.1–25.9) | 152.9 (145.8–162.2)*** | 26 (17–37.5) | 12.2 (10.5–16.8) | 4.10 (0.21–9.37) | 20.5 (18.5–28.0) |
| Lagomorpha (60%) | 14.7 (11.5–19.3) | 19.8 (18.1–24.6)** | 5 (2–8.1) | 11.8 (10.4–15.2) | 20.6 (18.3–25.5)** | 6 (3–10) | 8.50 (6.23–11.4) | 5.28 (1.72–8.99) | 11.8 (11.4–14.8) |
| Lagomorpha (50%) | 2.47 (0.73–6.30) | 6.76 (4.61–10.9)** | 4 (2–5) | 11.8 (11.6–15.2) | 8.03 (5.00–11.2)** | 6 (2–9.5) | 8.50 (6.44–13.5) | 5.21 (1.75–11.9) | 11.8 (11.4–15.1) |
The MT version of the GMYC is computationally demanding and we were unable in its current implementation to run this analysis across the broadly sampled mammal tree to completion. The solution inferred under the local optimum reached after two months suggests the presence of 224 hESUs, with threshold times ranging from 5.8 to 38 million years (Ma; electronic supplementary material, table S7 and figure S4). This solution is much more likely than that inferred under the ST model (100 hESUs) but is still probably an underestimate of the true number of hESUs in mammals (electronic supplementary material, figure S5). To test this, we analysed the largest (more than 50 species) hESUs inferred under the ST model separately and found support for 78 more hESUs overall (electronic supplementary material, table S9).
(c). Robustness to incomplete sampling
Results for all three clades are generally robust to sampling differences, although the effect of adding missing species or jackknifing differs among the clades (table 1; electronic supplementary material, figures S6–S10). In carnivores and lagomorphs, more hESUs are inferred when missing species are added, suggesting that addition of missing species fills in branching within clades that are then separated as distinct hESUs. By contrast, in euungulates, fewer hESUs are inferred, caused by ‘filling of gaps’ between hESUs inferred from the sampled tree. Well-supported hESUs are generally less sensitive to sampling differences.
Jackknifing tended to reduce the significance of the clustering model and the correspondence with the empirical tree. For carnivores, more hESUs were inferred for jackknifed trees, as expected if removed taxa cause remaining clades to appear more distinct. For euungulates and lagomorphs, by contrast, fewer hESUs were inferred, as expected if clusters become less distinct from one another as species are removed or if clusters go completely unsampled. Once again, sampling biases mainly affected hESUs with low GMYC support.
(d). Trait coherence among species within higher evolutionarily significant units
In euungulates, adult body mass showed significantly lower rates of change within hESUs than between them (table 2). For all other morphological and ecological traits, rates were greater within than between hESUs. Other slowdowns corresponding to hESUs included geographical traits in the Madagascan family Eupleridae (fossa and relatives) and subgenera Conothoa and Ochotona (mountain pikas and shrub-steppe pikas, respectively), the New World family Procyonidae (racoons and relatives), litter size in Pinnipedia (seals, sea lions and walrus) and Balaenidae (right whales and bowhead whale) and dentition and skull traits for Herpestidae (mongooses) and Felidae (cats) (electronic supplementary material, tables S10, S11 and figure S11).
4. Discussion
Despite decades of research [e.g. 62,63] the uneven distribution of biodiversity among clades or higher taxa remains unexplained. We develop a new framework for studying these biodiversity patterns that integrates how such patterns may evolve. This framework rests on the assumption that there are limits to diversity [21,22,28] and that these limits operate independently in different clades, meaning that speciation and extinction are determined separately in different clades. Our results support the existence of phylogenetic clusters of species separated by longer internal branches than expected under an even process of branching through time (figure 1). This pattern is consistent with our predictions for independently evolving higher clades. The simulation model results apply equally to ecological specialization within the same geographical region. If shifts between niches are rare relative to the rate of species turnover, distinct ecological guilds of closely related species should emerge over time [28,64,65]. Note that strict neutrality is assumed for simplicity but is not required for hESUs to emerge, as long as limits are present that cause species turnover across the entire clade. For example, species might diverge in resource use or other traits permitting coexistence (i.e. alpha niche), but if the species in the clade are limited to particular abiotic conditions (i.e. beta niche), hESUs representing ecological guilds with shared beta niches could still evolve.
We found evidence for the existence of hESUs in mammals; 17–24 in carnivores, 18–29 in euungulates and 2–11 in lagomorphs (table 1). For comparison, around 16 families and 123 genera are currently recognized in carnivores, 24 families and 138 genera in euungulates and two families and 12 genera in lagomorphs [32]. Accordingly, most inferred carnivore and euungulates hESUs correspond to traditionally named or informal taxa of family or infra-family rank, while lagomorph hESUs correspond to taxa of generic or infrageneric rank (figure 2). Families and genera, delimited by taxonomists based primarily on patterns of morphological variation, might therefore reflect underlying evolutionary processes [5,62].
Several processes may have contributed to the evolution of hESUs. Geographical isolation could explain independent evolution of Old World lineages (e.g. pigs, Malayan tapir and the baiji) and their respective New World sisters (peccary, American tapirs and the two South American river dolphins). We found evidence for lower rates of change within hESUs than among them for adult body size in euungulates and lower rates in certain hESUs for tooth, skull and geographical traits in carnivores, life history in both carnivores and euungulates, and geographical traits in lagomorphs (electronic supplementary material, table S10 and figure S11). These traits are known to correlate with ecological differences and to habitat and dietary preferences, in particular [66]. Our results are thus consistent with hESUs being established following occupation of distinct ecological or dietary zones, and that (carnivore and euungulate) hESUs are specialized on different resources, thus representing major dietary or habitat ‘adaptive zones’. For other traits, rates of change tend to be greater within than between hESUs, consistent with adaptive radiation of alpha niche traits within evolutionary guilds [67]. Further understanding of the causes of separation into hESUs, however, remain to be determined: comprehensive tooth data are not available for euungulates and lagomorphs to our knowledge and no single trait can be associated with patterns of hESU clustering across all three clades. Few other groups have datasets with dense enough sampling to test for these patterns with accuracy at present. We note, however, that a recent analysis of all birds [14] found the same signature of a general increase in branching rate as was used to detect clusters here, although in the Eocene (at 50 Ma) compared to the Miocene (8–14 Ma) as found here.
There are other potential explanations for the patterns quantified here, including increased diversification rates owing to a recent shift in environmental conditions or gaps in the phylogenetic tree resulting from non-random and whole-clade extinction. For instance, the perissodactyl hESUs might be explained by extinction of entire stem lineages (e.g. the two extinct subfamilies of horses, Hyracotheriinae and Anchitheriinae [68]). The GMYC approach cannot distinguish long internal phylogenetic branches resulting from species turnover within an independently limited clade (as modelled here) from those resulting from extinction of whole clades. This alternative scenario is still consistent, however, with extinction operating differently on different higher level groups and therefore remains consistent with the existence of hESUs. The relative contribution of different processes might be distinguishable with analyses of fossil data [69]. In the meantime, our findings provide new theory and empirical support for Simpson's [5] idea that higher taxa represent biologically real entities which are genetically coherent and definable by the processes that underlie their emergence.
Acknowledgements
We are indebted to Graham Slater for detailed comments that improved the phylogenies analysed here. Shai Meiri supplied unpublished carnivore skull measurements. Fabian Leuenberger prepared the images in figure 2. Albert Phillimore, Andy Purvis, Daniel Rabosky, Gavin Thomas and one anonymous reviewer commented on an earlier draft. Several analyses used the Imperial College High Performance Computing Service (http://www.imperial.ac.uk/ict/services/teachingandresearchservices/highperformancecomputing).
Data accessibility
Phylogenies generated in this study are available from TreeBase: http://purl.org/phylo/treebase/phylows/study/TB2:S15307.
Funding statement
A.M.H. was funded by a Swiss NF Fellowship (grant PBZHP3-133420).
References
- 1.Coyne JA, Orr HA. 2004. Speciation. Sunderland, MA: Sinauer Associates Inc [Google Scholar]
- 2.Lee MSY. 2003. Species concepts and species reality: salvaging a Linnaean rank. J. Evol. Biol. 16, 179–188 (doi:10.1046/j.1420-9101.2003.00520.x) [DOI] [PubMed] [Google Scholar]
- 3.Butlin RK, et al. (The Marie Curie SPECIATION Network). 2011. What do we need to know about speciation? Trends Ecol. Evol. 27, 27–39 [DOI] [PubMed] [Google Scholar]
- 4.Rieseberg LH, Wood TE, Baack EJ. 2006. The nature of plant species. Nature 440, 524–527 (doi:10.1038/nature04402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson GG. 1953. The major features of evolution. New York, NY: Columbia University Press [Google Scholar]
- 6.Bentham GJDH. 1862–1883. Genera plantarum. London, UK: Lovell Reeve and Williams and Norgate [Google Scholar]
- 7.Mayr E. 1942. Systematics and the origin of species. New York, NY: Columbia University Press [Google Scholar]
- 8.Linnaeus C. 1751. Philosophia botanica (translated by S. Freer, 2003). Oxford, UK: Oxford University Press [Google Scholar]
- 9.Avise JC, Johns GC. 1999. Proposal for a standardised temporal scheme of biological classification for extant species. Proc. Natl Acad. Sci. USA 96, 7358–7363 (doi:10.1073/pnas.96.13.7358) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humphreys AM, Linder HP. 2009. Concept versus data in delimitation of plant genera. Taxon 58, 1054–1074 [Google Scholar]
- 11.Fontaneto D, Herniou EA, Boschetti C, Caprioli M, Melone G, Ricci C, Barraclough TG. 2007. Independently evolving species in asexual bdelloid rotifers. PLoS Biol. 5, e87 (doi:10.1371/journal.pbio.0050087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alfaro ME, Santini F, Brock C, Alamillo H, Dornburg A, Rabosky DL, Carnevale G, Harmon LJ. 2009. Nine exceptional radiations plus high turnover explain species diversity in jawed vertebrates. Proc. Natl Acad. Sci. USA 106, 13 410–13 414 (doi:10.1073/pnas.0811087106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Etienne RS, Haegeman B. 2012. A conceptual and statistical framework for adaptive radiations with a key role for diversity dependence. Am. Nat. 180, E75–E89 (doi:10.1086/667574) [DOI] [PubMed] [Google Scholar]
- 14.Jetz W, Thomas GH, Joy JB, Hartmann K, Mooers AO. 2012. The global diversity of birds in space and time. Nature 491, 444–448 (doi:10.1038/nature11631) [DOI] [PubMed] [Google Scholar]
- 15.Rabosky DL, Slater GJ, Alfaro ME. 2012. Clade age and species richness are decoupled across the eukaryotic tree of life. PLoS Biol. 10, e1001381 (doi:10.1371/journal.pbio.1001381) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldschmidt RB. 1940. The material basis of evolution. New Haven, CT: Yale University Press [Google Scholar]
- 17.Barraclough TG, Birky CW, Burt A. 2003. Diversification in sexual and asexual organisms. Evolution 57, 2166–2172 [DOI] [PubMed] [Google Scholar]
- 18.Templeton AR. 1989. The meaning of species and speciation: a genetic perspective. In Speciation and its consequences (eds Otte D, Endler JA.), pp. 3–27 Sunderland, MA: Sinauer Associates Inc [Google Scholar]
- 19.Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran DP, Hazell S, Kamoun S, Sumlin WD, Vogler AP. 2006. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst. Biol. 55, 595–609 (doi:10.1080/10635150600852011) [DOI] [PubMed] [Google Scholar]
- 20.Hubbel SP. 2001. The unified neutral theory of biodiversity and biogeography. Princeton, NJ: Princeton University Press [Google Scholar]
- 21.Ricklefs RE. 2007. Estimating diversification rates from phylogenetic information. Trends Ecol. Evol. 22, 601–610 (doi:10.1016/j.tree.2007.06.013) [DOI] [PubMed] [Google Scholar]
- 22.Rosenzweig ML. 1995. Species diversity in space and time. Cambridge, UK: Cambridge University Press [Google Scholar]
- 23.Rabosky DL. 2009. Ecological limits and diversification rate: alternative paradigms to explain the variation in species richness among clades and regions. Ecol. Lett. 12, 735–743 (doi:10.1111/j.1461-0248.2009.01333.x) [DOI] [PubMed] [Google Scholar]
- 24.Phillimore AB, Price TD. 2008. Density-dependent cladogenesis in birds. PLoS Biol. 6, 483–489 (doi:10.1371/journal.pbio.0060071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morlon H, Potts MD, Plotkin JB. 2010. Inferring the dynamics of diversification: a coalescent approach. PLoS Biol. 8, e1000493 (doi:10.1371/journal.pbio.1000493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alroy J. 2008. Dynamics of origination and extinction in the marine fossil record. Proc. Natl Acad. Sci. USA 105, 11 536–11 542 (doi:10.1073/pnas.0802597105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raup DM. 1972. Taxonomic diversity during the phanerozoic. Science 177, 1065–1071 (doi:10.1126/science.177.4054.1065) [DOI] [PubMed] [Google Scholar]
- 28.Barraclough TG. 2010. Evolving entities: towards a unified framework for understanding diversity at the species and higher levels. Phil. Trans. R. Soc. B 365, 1801–1813 (doi:10.1098/rstb.2009.0276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.R Development Core Team. 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 30.Agnarsson I, Kuntner M, May-Collado LJ. 2010. Dogs, cats, and kin: a molecular species-level phylogeny of Carnivora. Mol. Phylogenet. Evol. 54, 726–745 (doi:10.1016/j.ympev.2009.10.033) [DOI] [PubMed] [Google Scholar]
- 31.Agnarsson I, May-Collado LJ. 2008. The phylogeny of Cetartiodactyla: the importance of dense taxon sampling, missing data, and the remarkable promise of cytochrome b to provide reliable species-level phylogenies. Mol. Phylogenet. Evol. 48, 964–985 (doi:10.1016/j.ympev.2008.05.046) [DOI] [PubMed] [Google Scholar]
- 32.Wilson DE, Reeder DA. 2005. Mammal species of the world: a taxonomic and geographic reference, 3rd edn Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 33.Drummond A, Rambaut A. 2007. Beast: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7, 214 (doi:10.1186/1471-2148-7-214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUTi and the BEAST 1.7. Mol. Biol. Evol. 29, 1969–1973 (doi:10.1093/molbev/mss075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bininda-Emonds ORP, et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512 (doi:10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 36.Bibi F. 2013. A multi-calibrated mitochondrial phylogeny of extant Bovidae (Artiodactyla, Ruminantia) and the importance of the fossil record to systematics. BMC Evol. Biol. 13, 166 (doi:10.1186/1471-2148-13-166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fulton TL, Strobeck C. 2010. Multiple fossil calibrations, nuclear loci and mitochondrial genomes provide new insight into biogeography and divergence timing for true seals (Phocidae, Pinnipedia). J. Biogeogr. 37, 814–829 (doi:10.1111/j.1365-2699.2010.02271.x) [Google Scholar]
- 38.McGowen MR, Spaulding M, Gatesy J. 2009. Divergence date estimation and a comprehensive molecular tree of extant cetaceans. Mol. Phylogenet. Evol. 53, 891–906 (doi:10.1016/j.ympev.2009.08.018) [DOI] [PubMed] [Google Scholar]
- 39.Slater GJ, Harmon LJ, Alfaro ME. 2012. Integrating fossils with molecular phylogenies improves inference of trait evolution. Evolution 66, 3931–3944 (doi:10.1111/j.1558-5646.2012.01723.x) [DOI] [PubMed] [Google Scholar]
- 40.Gaubert P, Tranier M, Delmas A-S, Colyn M, Veron G. 2004. First molecular evidence for reassessing phylogenetic affinities between genets (Genetta) and the enigmatic genet-like taxa Osbornictis, Poiana and Prionodon (Carnivora, Civerridae). Zool. Scr. 33, 117–129 (doi:10.1111/j.1463-6409.2004.00140.x) [Google Scholar]
- 41.Miller MH, Holder MT, Vos R, Midford PE, Liebowitz T, Chan L, Hoover P, Warnow T. 2012. The CIPRES portals (CIPRES) See http://www.phylo.org/sub_sections/portal.
- 42.Pearse WD, Purvis A. 2013. Phylogenerator: an automated phylogeny generation tool for ecologists. Methods Ecol. Evol. 4, 692–698 (doi:10.1111/2041-210x.12055) [Google Scholar]
- 43.Thompson JD, Higgins DG, Gibson TJ. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (doi:10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (doi:10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 45.Meredith RW, et al. 2011. Impacts of the Cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524 (doi:10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 46.Britton T, Anderson CL, Jacquet D, Lundqvist S, Bremer K. 2007. Estimating divergence times in large phylogenetic trees. Syst. Biol. 56, 741–752 (doi:10.1080/10635150701613783) [DOI] [PubMed] [Google Scholar]
- 47.Nee S, May RM, Harvey PH. 1994. The reconstructed evolutionary process. Phil. Trans. R. Soc. Lond. B 344, 305–311 (doi:10.1098/rstb.1994.0068) [DOI] [PubMed] [Google Scholar]
- 48.Monaghan MT, et al. 2009. Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Syst. Biol. 58, 298–311 (doi:10.1093/sysbio/syp027) [DOI] [PubMed] [Google Scholar]
- 49.Fujisawa T, Barraclough TG. 2013. Delimiting species using single-locus data and the generalized mixed Yule coalescent (GMYC) approach: a revised method and evaluation on simulated datasets. Syst. Biol. 62, 707–724 (doi:10.1093/sysbio/syt033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ezard T, Fujisawa T, Barraclough TG.2009. Splits: species’ limits by threshold statistics. R package version 1.0–14/r31. See http://r-forge.r-project.org/projects/splits .
- 51.Yule GU. 1925. A mathematical theory of evolution, based on the conclusions of Dr JC Willis, FRS. Phil. Trans. R. Soc. B 213, 21–87 (doi:10.1098/rstb.1925.0002) [Google Scholar]
- 52.Paradis E, Claude J, Strimmer K. 2004. Ape: analyses of phylogenetics and evolution in R language. Bioinformatics 20, 289–290 (doi:10.1093/bioinformatics/btg412) [DOI] [PubMed] [Google Scholar]
- 53.FitzJohn RG. 2012. Diversitree: comparative phylogenetic analyses of diversification (with GeoSSE, ClaSSE by Emma W. Goldberg and BiSSE-ness by Karen Magnuson-Ford). R package version 0.9–3 See http://CRAN.R-project.org/package=diversitree.
- 54.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. 2008. Geiger: investigating evolutionary radiations. Bioinformatics 24, 129–131 (doi:10.1093/bioinformatics/btm538) [DOI] [PubMed] [Google Scholar]
- 55.Uyeda JC, Hansen TF, Arnold SJ, Pienaar J. 2011. The million-year wait for macroevolutionary bursts. Proc. Natl Acad. Sci. USA 108, 15 908–15 913 (doi:10.1073/pnas.1014503108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schluter D, Price T, Mooers AO, Ludwig D. 1997. Likelihood of ancestor states in adaptive radiation. Evolution 51, 1699–1711 (doi:10.2307/2410994) [DOI] [PubMed] [Google Scholar]
- 57.Harmon LJ, Schulte JA, Larson A, Losos JB. 2003. Tempo and mode of evolutionary radiation in iguanian lizards. Science 301, 961–964 (doi:10.1126/science.1084786) [DOI] [PubMed] [Google Scholar]
- 58.Jones KE, et al. 2009. Pantheria: a species-level database of life history, ecology, and geography of extant and recently extinct mammals. Ecology 90, 2648 (doi:10.1890/08-1494.1) [Google Scholar]
- 59.Meiri S, Dayan T, Simberloff D. 2005. Variability and correlations in carnivore crania and dentition. Funct. Ecol. 19, 337–343 (doi:10.1111/j.1365-2435.2005.00964.x) [Google Scholar]
- 60.Pie MR, Weitz JS. 2005. A null model of morphospace occupation. Am. Nat. 166, E1–E13 (doi:10.1086/430727) [DOI] [PubMed] [Google Scholar]
- 61.Thomas GH, Freckleton RP. 2011. MOTMOT: models of trait macroevolution on trees. Methods Ecol. Evol. 3, 145–151 (doi:10.1111/j.2041-210X.2011.00132.x) [Google Scholar]
- 62.Rabosky DL. 2010. Primary controls on species richness in higher taxa. Syst. Biol. 59, 634–645 (doi:10.1093/sysbio/syq060) [DOI] [PubMed] [Google Scholar]
- 63.Slowinski JB, Guyer C. 1993. Testing whether certain traits have caused amplified diversification: an improved method based on a model of random speciation and extinction. Am. Nat. 142, 1019–1024 (doi:10.1086/285586) [DOI] [PubMed] [Google Scholar]
- 64.Crisp MD, et al. 2009. Phylogenetic biome conservatism on a global scale. Nature 458, 754–758 (doi:10.1038/nature07764) [DOI] [PubMed] [Google Scholar]
- 65.Pennington RT, Cronk QCB, Richardson JA. 2004. Introduction and synthesis: plant phylogeny and the origin of major biomes. Phil. Trans. R. Soc. Lond. B 359, 1455–1464 (doi:10.1098/rstb.2004.1539) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cardillo M, Mace GM, Jones KE, Bielby J, Bininda-Emonds ORP, Sechrest W, Orme CDL, Purvis A. 2005. Multiple causes of high extinction risk in large mammal species. Science 309, 1239–1241 (doi:10.1126/science.1116030) [DOI] [PubMed] [Google Scholar]
- 67.Ackerly DD, Schwilk DW, Webb CO. 2006. Niche evolution and adaptive radiation: testing the order of trait divergence. Ecology 87, S50–S61 (doi:10.1890/0012-9658(2006)87[50:neaart]2.0.co;2) [DOI] [PubMed] [Google Scholar]
- 68.Janis CM. 2007. The horse series. In Icons of evolution (ed. Regal B.), pp. 251–280 Westport, CT: Greenwood Press [Google Scholar]
- 69.Ezard THG, Aze T, Pearson PN, Purvis A. 2011. Interplay between changing climate and species’ ecology drives macroevolutionary dynamics. Science 332, 349–351 (doi:10.1126/science.1203060) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Phylogenies generated in this study are available from TreeBase: http://purl.org/phylo/treebase/phylows/study/TB2:S15307.