Abstract
Mammalian olfaction comprises two chemosensory systems: the odorant-detecting main olfactory system (MOS) and the pheromone-detecting vomeronasal system (VNS). Mammals are diverse in their anatomical and genomic emphases on olfactory chemosensation, including the loss or reduction of these systems in some orders. Despite qualitative evidence linking the genomic evolution of the olfactory systems to specific functions and phenotypes, little work has quantitatively tested whether the genomic aspects of the mammalian olfactory chemosensory systems are correlated to anatomical diversity. We show that the genomic and anatomical variation in these systems is tightly linked in both the VNS and the MOS, though the signature of selection is different in each system. Specifically, the MOS appears to vary based on absolute organ and gene family size while the VNS appears to vary according to the relative proportion of functional genes and relative anatomical size and complexity. Furthermore, there is little evidence that these two systems are evolving in a linked fashion. The relationships between genomic and anatomical diversity strongly support a role for natural selection in shaping both the anatomical and genomic evolution of the olfactory chemosensory systems in mammals.
Keywords: olfaction, genomic evolution, anatomical evolution, mammalian evolution
1. Introduction
Olfaction is a key sensory adaptation of mammals [1]. Colloquially referred to as the ‘sense of smell’, mammalian olfaction actually derives from two chemosensory systems that are distinct at the anatomical, genomic and neurological levels—the main olfactory system (MOS) and vomeronasal system (VNS) (figure 1). Mammals are diverse in their anatomical and genomic development of these two olfactory chemosensory systems, ranging from ‘microsmatic’ cetaceans to ‘macrosmatic’ rodents and canines [3–5].
Figure 1.
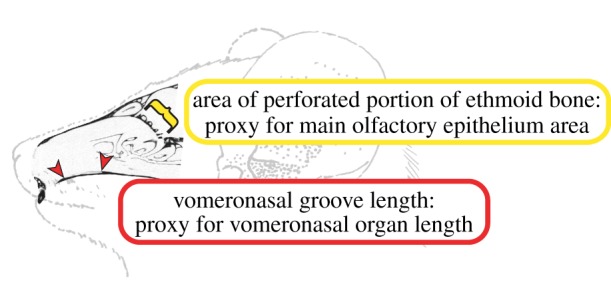
Schematic diagram of the anatomical components of the mammalian olfactory chemosensory systems. The diagram shows a line drawing of Microcebus murinus with a CT scan (AMNH 174535) of the nasal fossa in the sagittal plane. The red arrows indicate the length of the VNG, which forms on the palatal portion of the maxilla from the articulation of the cartilaginous capsule surrounding the VNO. When data on VNO length were not available, we measured the VNG length as an osteological proxy. The yellow bracket indicates the perforated portion of the ethmoid bone, through which nerves projecting from the MOE pass and then connect to the MOB. We measured ethmoid area as a proxy for MOE area when data were not available in the literature. Line drawing adapted from Smith et al. [2]. (Online version in colour.)
The MOS detects volatile stimuli using olfactory sensory neurons expressed in the olfactory epithelium (OE) of the nasal mucosa. Olfactory sensory neurons are encoded by the large and genomically scattered olfactory receptor (OR) superfamily [4–7] and expressed in the main OE (MOE). These nerves pass through the cribiform plate of the ethmoid bone and connect to the brain's main olfactory bulb (MOB) yielding information relating to diet, activity pattern, habitat, sociosexual signals and potentially navigation [4,8,9]. On the other hand, the VNS is specialized for the detection of non-volatile stimuli that may play a role in innate responses to odorants responsible for sociosexual behaviours and predator avoidance [10–12]. The peripheral sensory organ of the VNS is the vomeronasal organ (VNO), which comprises bilateral epithelial tubes situated at the anterior base of the nasal cavity [10]. In most mammals, vomeronasal sensory neurons are expressed in the VNO and their axons project to a distinct brain structure, the accessory olfactory bulb. Vomeronasal receptors are encoded by two gene families (V1R and V2R) that are distributed across most chromosomes [13].
Despite considerable variation in the olfactory chemosensory systems of mammals [3–5], little work has explored the relationship between the genomic and anatomical correlates of each olfactory system. In other words, do mammals with well-developed olfactory chemosensory anatomy have relatively well-developed receptor gene families? Hildebrand & Shepherd [14] suggested that receptor cell density (relating to epithelial distribution) would be greater in taxa with larger numbers of olfactory receptor genes. In addition, a positive relationship between the anatomic and genomic diversity has been hypothesized for both the MOS [15] and the VNS [16]. Qualitative studies support a relationship between VNO complexity and VNS gene family evolution [3,17–20]. In addition, quantitative studies of anatomy in birds [21] and ecological behaviour in mammals [4] have shown clear links between genomic and phenotypic evolution of the olfactory chemosensory systems.
Along the lines of these earlier studies, we predict that mammals with larger chemosensory gene family repertoires will have larger and more complex peripheral chemosensory organs. Conversely, in mammals with smaller chemosensory gene family repertoires, we predict a correspondent decrease in anatomical size and/or complexity of their sensory organs. In this study, we quantitatively test this prediction in a diverse set of mammals. We use phylogenetic least-squares regression to test whether anatomical features of the MOS and VNS, such as anatomical complexity and relative and absolute size of sensory organs, are related to features of the chemoreceptor gene families, such as the absolute and relative numbers of functional receptor genes. We interpret any correlation among these variables as evidence for natural selection's role in shaping the evolution of both anatomy and underlying genetics of these olfactory chemosensory systems.
In addition to the relationships within the MOS and VNS, there is considerable debate on the relationship between these two olfactory chemosensory systems. The dual olfactory hypothesis states that each system is adapted to detect specific types of stimuli, namely that the MOS is adapted to detect a broad range of ecological cues while the VNS is specifically adapted to detect pheromones [10,22–25]. Yet, other studies suggest that there is substantial functional overlap between the MOS and the VNS, and particularly that the VNS is not uniquely adapted for pheromone detection [26–29]. To address these questions, we use phylogenetic least-squares regression to test whether anatomical and genomic features of the MOS and VNS are correlated with one another.
2. Material and methods
(a). Anatomical data
We characterized anatomical diversity of the olfactory subsystems for 32 mammals, following the sampling of the genomic analysis of Young et al. [18]. We collected data on the VNO and the surface area of the MOE (the peripheral sensory organ of main olfaction) (figure 1). Owing to the destructive nature of histological methods, data on the sensory epithelium of the VNO or MOE have not been published for all taxa. When possible, epithelial structure of the VNO was recorded from the literature documenting histology and categorized using criteria from Takami [30] who described the VNO of a wide range of tetrapods and Smith et al. [31] who described it in primates (table 1). In this analysis, VNO category was treated as a continuous variable given that each character state represents decreasing complexity on a continuum rather than a discrete morphological trait. Length of the VNO was also recorded when presented in the literature. When these data were unavailable, we used an osteological proxy to estimate VNO length in cranial specimens, the vomeronasal groove (VNG) [32]. We used ethmoid area as a proxy for MOE surface area, following Pihlström et al. [33]. Ethmoid and skull areas (to correct for size) were recorded from Pihlström et al. [33] where available. Size-adjusted ethmoid area was calculated by dividing ethmoid area by skull area. Taxa without published data on VNO length or ethmoid area were CT scanned at the American Museum of Natural History Microimaging Facility in New York, NY. VNG length was collected using ImageJ software [34], and ethmoid and skull area data were extracted from three-dimensional rendered images using Avizo Standard software. Published data on head and body length (HBL) were used to correct for size of the VNO (see electronic supplementary material, table S1 and references therein). Size-adjusted VNO length was calculated by dividing VNO length by HBL. The anatomical variables include VNO category, VNO length (mm) and ethmoid area (mm2). We used both size-corrected and non-corrected values in the analysis and implemented the natural log transformation for non-corrected anatomical variables. When calculating the natural log transformation for VNO length, we added one to our variable prior to transformation so all values were positive.
Table 1.
VNO categories. Six defined character states of the mammalian VNO based on descriptions of epithelial distribution in the literature. VNNE, vomeronasal neuroepithelium; VNS, vomeronasal system; RFE, receptor free epithelium; VNO, vomeronasal organ.
| VNO category | description | reference |
|---|---|---|
| 1 | well-developed VNNE, connective tissue components penetrate VNNE, segregated VNS | [30] |
| 2 | well developed: VNNE medially with thin lateral RFE | [31] |
| 3 | VNNE only | [31] |
| 4 | VNNE interrupted by RFE | [31] |
| 5 | respiratory-like epithelium, VNO displaced superiorly | [31] |
| 6 | absent | [31] |
(b). Gene family data
We compiled data on the olfactory genomes by recoding published values reported in Hayden et al. [4] and Matsui et al. [35] for the OR gene family, and Young et al. [18] for the V1R gene family. From these data, we calculated the total number of genes, the total number of presumed functional/intact genes and the proportion of functional/intact genes for both gene families (electronic supplementary material, table S1). A second class of vomeronasal receptor genes, V2R, was not included in the analysis because functional V2R genes have only been identified in a small number of mammals [17,36].
(c). Quantitative methods
We tested for relationships within and between the anatomical and genetic components of the MOS and VNS using phylogenetically corrected generalized least-squares (PGLS) regression using the caper package [37] of R [38]. We used a pruned version of the mammalian supertree of Bininda-Emonds et al. [39] for all analyses. The integrated likelihood function of caper was used to estimate the phylogenetic scaling parameter λ [40]. To correct for Type I error when performing multiple tests, we used Benjamini & Yekutieli's [41] false discovery rate approach, as well as the Bonferroni correction to rescale significance levels (calculated in R using p.adjust) [38].
3. Results
Data for multiple variables were collected or compiled for 32 different species of mammals (electronic supplementary material, table S1). From these data, 11 variables were extracted for analysis. For the MOS, we analysed three genomic features of the mammalian OR superfamily: the total number of OR genes, the number of functional OR genes and the proportion of functional OR genes. As an anatomical index of the MOS, we examined the absolute and relative area of the cribiform plate of the ethmoid bone (referred to as ethmoid area), a predictor of the size and performance of the MOS [33].
Phylogenetically corrected regressions show that absolute ethmoid area is positively correlated with both the total and functional number of OR genes (table 2 and figure 2a; electronic supplementary material, table S2 and figure S1). There is no relationship between the proportion of functional OR genes and any other variable, though smaller bodied mammals (e.g. rodents) appear to have higher proportions of functional OR genes when compared with larger mammals (figure 2a).
Table 2.
Pairwise comparisons of variables using PGLS regression. The top left quadrant denotes VNS-related variables and comparisons; bottom right denotes MOS and top right/bottom left denotes comparisons between the VNS and MOS. Above diagonal: probability values for each of the pairwise PGLS regressions. Bold p-values are those significant after controlling for the false discovery rate using the method of Benjamini & Yekutieli [41]. Italic p-values are those significant after both the Bonferroni correction and the false discovery method. Below diagonal: R2 values for each pairwise comparison.
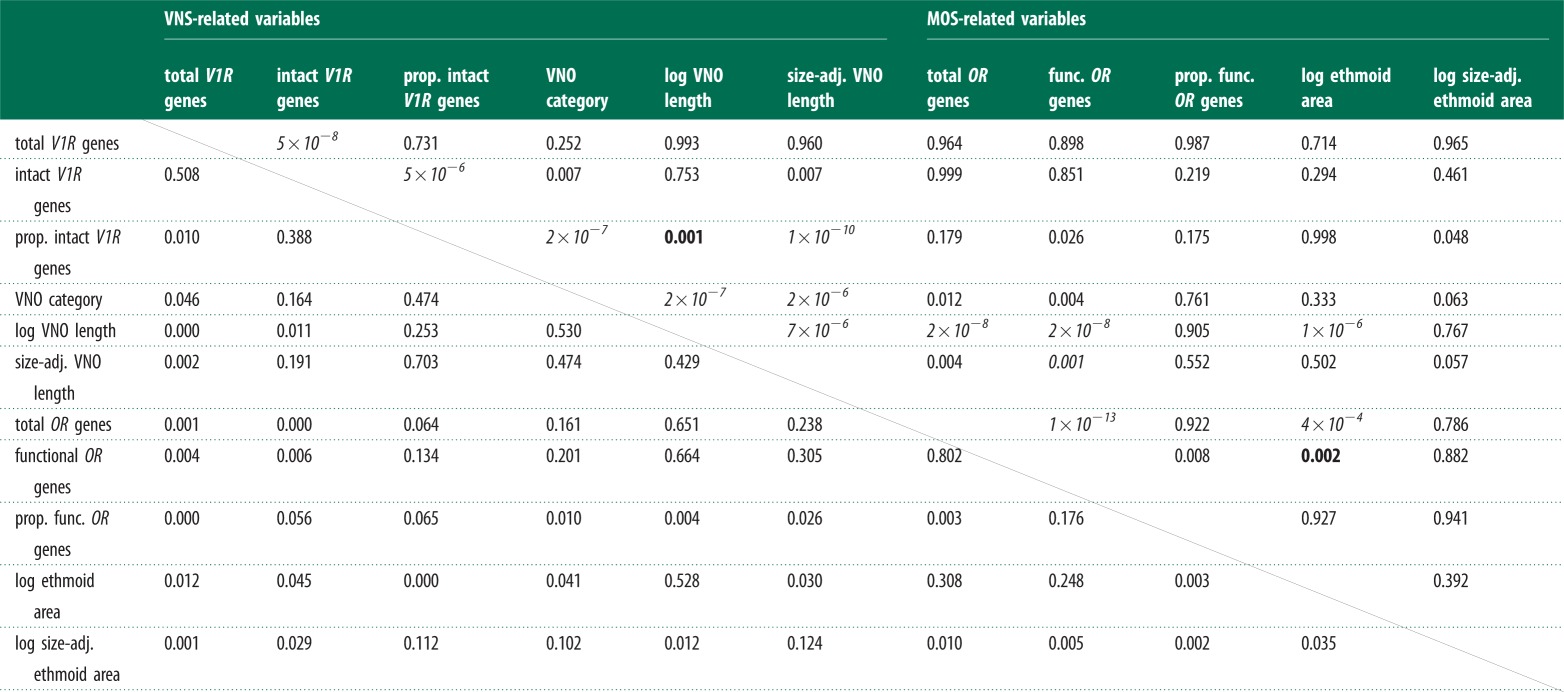 |
Figure 2.
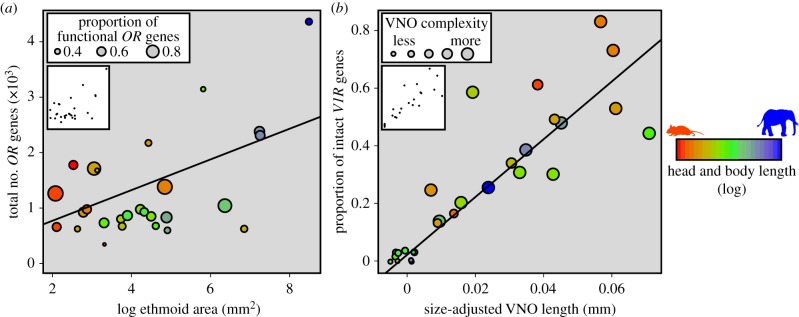
Relationships among selected VNS and MOS variables. Panel (a) shows the total number of OR genes on the y-axis and absolute ethmoid area on the x-axis. The size of each point reflects the proportion of functional OR genes, which is not significantly related to ethmoid area. The colour of each point indicates the overall size of each particular mammal (log head and body length). Panel (b) shows size-adjusted VNO length on the x-axis and the proportion of intact V1R genes on the y-axis. The size of each point reflects the morphological complexity of the VNO, which is also positively correlated with both size-adjusted VNO length and the proportion of intact V1R genes. The colour of each point indicates the overall size of each particular mammal (log head and body length). Superimposed points were ‘jittered’ for visualization purposes; inset plots show points in their original position. Scatterplots of all comparisons appear in electronic supplementary material, figures S1–S3 and all regression statistics appear in electronic supplementary material, table S2.
For the VNS, we examined three genomic features of the main family of receptor genes (V1R): the total number of V1R genes, the number of intact V1R genes and the proportion of intact V1R genes. We examined three of anatomical variables of the VNO: its overall complexity, as well as its absolute and relative length.
Within the VNS, the proportion of intact V1R genes is positively correlated with both the absolute and relative length of the VNO and its anatomical complexity (table 2 and figure 2b; electronic supplementary material, table S2 and figure S2) based on phylogenetically corrected regressions. Additionally, VNO complexity scales with absolute and relative VNO length. As in the MOS, there are highly significant positive correlations among variables, suggesting that natural selection has shaped the genomic and anatomical variation of the VNS together.
There are few significant relationships between the MOS and VNS, and those that exist could be explained by changes in body size (table 2; electronic supplementary material, table S2 and figure S3). Significant relationships exist between VNO length and several variables within the MOS, including ethmoid area, the total number of OR genes and the number of functional OR genes. These relationships likely reflect the relationship between overall size and the MOS (see Discussion), given the significant relationship between VNO length and body length (p-value < 0.01) (electronic supplementary material, figure S4).
4. Discussion
Within each olfactory chemosensory system, there are features of chemosensory anatomy that are strongly related to specific aspects of their constituent gene families. Interestingly, these relationships differ in the MOS and VNS. The VNS is modulated in proportional terms—the proportion of intact V1R genes relates to multiple aspects of VNO anatomy, including its relative size. This is supported by the large proportion of variance explained by the linear relationship between percentage of intact V1R genes and the relative size of the VNO (R2 = 0.70), which was comparatively larger than all other coefficients of determination among the VNS variables (table 2; electronic supplementary material, table S1). On the other hand, the MOS responds in absolute terms, both in anatomical size and numbers of OR receptors. Indeed, functional studies show that the absolute size of the olfactory organ is closely related to olfactory sensitivity, rather than its relative size [33]. The pattern is somewhat similar in birds, where the relative olfactory bulb size is related to the absolute number of OR genes [21]. Both our findings and those of Steiger et al. [21] support a link between absolute numbers of OR genes and olfactory ability [42].
Furthermore, there are fewer significant relationships between the MOS and VNS, except for a relationship between VNO length and ethmoid area, the total number of OR genes and the number of functional OR genes. These likely reflect the relationship between overall body size and the MOS, an interpretation in line with observations by Pihlström et al. [33] that the MOS is not under size constraints that might limit other sensory organs. In the VNS, on the other hand, we find that relative size of the VNO and relative number of intact V1R genes is positively correlated suggesting that the VNS is limited by size constraints. Neither sensory neuron bodies nor cell density scales isometrically with body size, therefore absolutely larger surface areas for olfactory sensory neurons have a positive relationship with olfactory sensitivity [33,43]. Alternatively, the positive relationship between VNO length and OR gene repertoires could be the result of both olfactory chemosensory systems evolving to be more complex in taxa that are more reliant on both systems. Additionally, selection for larger surface areas for main olfactory and vomeronasal neuroepithelium could result in larger nasal cavity size, potentially driving absolute sizes of the ethmoid area and VNO together.
We interpret these findings as evidence that natural selection has worked to shape the genomic and anatomical variation of both chemosensory systems, though selection may not act on them in a tightly correlated fashion. The MOS may respond to selective pressure in absolute terms, i.e. in absolute size and total number of functional gene copies. Indeed, it may be constrained to respond in an absolute size-dependent manner [33]. The VNS appears to respond to selection by varying relative size and complexity of its constituent features. Mammals that are more reliant on vomeronasal olfaction maintain larger proportions of functional V1R genes in a relatively longer and more complex VNO. Mammals that are less reliant on the VNS are under reduced selection to maintain complex VNOs and functional genes, thereby allowing for the accumulation of pseudogenes. This would account for lower percentages of functional genes in species with a less complex and relatively smaller VNO. These differences in the evolutionary patterns of the MOS and VNS can be interpreted to extend the differential tuning hypothesis [44], which states that each olfactory chemosensory system is under a distinct selective regime where the MOS is broadly tuned to detect a wide range of odorants, while the VNS is more finely tuned and under lineage-specific selection. Our results suggest that the differential tuning hypothesis [44] can be extended to include differences in the specific anatomical and genomic responses of the VNS and MOS to selective pressures. While our results do not bear on the distinct functions of each olfactory system, they support increasing evidence for the role of the VNS in innate responses to sociosexual and predator information, while the MOS may respond to a broader set of stimuli including some overlap in detecting sociosexual and predator odorants [11].
One noteworthy caveat is apparent when considering the MOS of the smallest and largest mammals. In figure 2a, the mouse (red) has a very high proportion of functional OR genes, yet its absolute number of functional OR genes is unremarkable. The elephant (blue) has an exceptionally large absolute number of functional OR genes, yet the proportion of functional genes is modest. This may be interpreted as two size-specific strategies. Smaller mammals may increase their proportion of functional OR genes as a size-limited ‘macrosmatic’ strategy constrained by their smaller available surface area for sensory neurons. Additionally, the significant relationship between relative VNO length and the number of functional OR genes reflect an enhanced olfactory ability in the largest mammals. As such, larger mammals may have greater capacity to pursue an overall ‘macrosmatic’ sensory strategy. Additional work is needed to determine how size affects the different olfactory chemosensory systems and sensory strategies of mammals, especially in the largest and smallest species.
These results also bear on the debate over the relative contributions of adaptive and random processes in the evolution of the MOS and VNS receptor gene families [45]. Interestingly, some genetic analyses provide evidence for adaptive evolution within the OR and V1R gene families [46–51], while others suggest that random processes also play a role [19,45,52–54]. Hayden et al. [4] recently argued for an adaptive interpretation of OR gene family evolution based on relationships between OR types and ecology in a wide array of mammalian species. Our analyses are similarly supportive of an adaptive interpretation for both the MOS and VNS, given the close relationship among multiple genomic and anatomical variables across mammals. Taken together, there is evidence for adaptive evolution in the mammalian olfactory chemosensory systems from evidence based on gene sequences [46,51], receptor copy numbers [18,52], anatomical diversity [18] and ecological parameters [4]. A challenge remains in reconciling the compelling evidence for neutral interpretations of olfactory chemoreceptor gene family evolution [19,45,52–54] with these multiple levels of evidence for the adaptive evolution in the olfactory chemosensory systems of mammals.
Acknowledgements
We thank J. Higham, C. Nunn, H. Pontzer, T. Smith and two anonymous reviewers for comments on the manuscript and T. Smith and J. Zichello for assistance with figure 1. We thank E. Westwig and N. Duncan for access to specimens at the American Museum of Natural History, and T. Smith for access to histological specimens. R. Rudolph, J. Thostenson and M. Hill of the AMNH Microimaging Facility were responsible for CT-scanning specimens. We thank E. Delson for providing additional CT scans, H. Pihlström for providing MatLab code, and L. Tallman and the 2011 AnthroTree Workshop participants for methodological assistance.
Funding statement
This research was supported by NSF grant nos. 0961964 (to E.G.) and 0966166 (NYCEP IGERT). The infrastructure of the Anthropological Genetics Laboratory at Hunter College was supported, in part, by grant no. RR003037 from the National Center for Research Resources, a component of the National Institutes of Health, its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR, NIMHD or NIH.
References
- 1.Rowe TB, Macrini TE, Luo ZX. 2011. Fossil evidence on origin of the mammalian brain. Science 332, 955–957 (doi:10.1126/science.1203117) [DOI] [PubMed] [Google Scholar]
- 2.Smith TD, Rossie JB, Bhatnagar KP. 2007. Evolution of the nose and nasal skeleton in primates. Evol. Anthropol. 16, 132–146 (doi:10.1002/evan.20143) [Google Scholar]
- 3.Grus WE, Shi P, Zhang YP, Zhang J. 2005. Dramatic variation of the vomeronasal pheromone receptor gene repertoire among five orders of placental and marsupial mammals. Proc. Natl Acad. Sci. USA 102, 5767–5772 (doi:10.1073/pnas.0501589102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC. 2010. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res. 20, 1–9 (doi:10.1101/gr.099416.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moulton DG. 1967. Olfaction in mammals. Am. Zool. 7, 421–429 [DOI] [PubMed] [Google Scholar]
- 6.Buck L, Axel R. 1991. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 (doi:10.1016/0092-8674(91)90418-X) [DOI] [PubMed] [Google Scholar]
- 7.Firestein S. 2001. How the olfactory system makes sense of scents. Nature 413, 211–218 (doi:10.1038/35093026) [DOI] [PubMed] [Google Scholar]
- 8.Baron G, Frahm HD, Bhatnagar KP, Stephan H. 1983. Comparison of brain structure volumes in Insectivora and Primates. III. Main olfactory bulb (MOB). J Hirnforsch 24, 551–568 [PubMed] [Google Scholar]
- 9.Jacobs LF. 2012. From chemotaxis to the cognitive map: the function of olfaction. Proc. Natl Acad. Sci. USA 109(Suppl. 1), 10 693–10 700 (doi:10.1073/pnas.1201880109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doving KB, Trotier D. 1998. Structure and function of the vomeronasal organ. J. Exp. Biol. 201, 2913–2925 [DOI] [PubMed] [Google Scholar]
- 11.Fortes-Marco L, Lanuza E, Martinez-Garcia F. 2013. Of pheromones and kairomones: what receptors mediate innate emotional responses? Anat. Rec. 296, 1346–1363 [DOI] [PubMed] [Google Scholar]
- 12.Papes F, Logan DW, Stowers L. 2010. The vomeronasal organ mediates interspecies defensive behaviors through detection of protein pheromone homologs. Cell 141, 692–703 (doi:10.1016/j.cell.2010.03.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roppolo D, Vollery S, Kan CD, Luscher C, Broillet MC, Rodriguez I. 2007. Gene cluster lock after pheromone receptor gene choice. EMBO J. 26, 3423–3430 (doi:10.1038/sj.emboj.7601782) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildebrand JG, Shepherd GM. 1997. Mechanisms of olfactory discrimination: converging evidence for common principles across phyla. Annu. Rev. Neurosci. 20, 595–631 (doi:10.1146/annurev.neuro.20.1.595) [DOI] [PubMed] [Google Scholar]
- 15.Issel-Tarver L, Rine J. 1997. The evolution of mammalian olfactory receptor genes. Genetics 145, 185–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawley EM. 1998. Species, sex, and seasonal differences in VNO size. Microsc. Res. Tech. 41, 506–518 (doi:10.1002/(SICI)1097-0029(19980615)41:6<506::AID-JEMT6>3.0.CO;2-K) [DOI] [PubMed] [Google Scholar]
- 17.Grus WE, Shi P, Zhang J. 2007. Largest vertebrate vomeronasal type 1 receptor gene repertoire in the semiaquatic platypus. Mol. Biol. Evol. 24, 2153–2157 (doi:10.1093/molbev/msm157) [DOI] [PubMed] [Google Scholar]
- 18.Young JM, Massa HF, Hsu L, Trask BJ. 2010. Extreme variability among mammalian V1R gene families. Genome Res. 20, 10–18 (doi:10.1101/gr.098913.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J. 2007. The drifting human genome. Proc. Natl Acad. Sci. USA 104, 20 147–20 148 (doi:10.1073/pnas.0710524105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H, Xu D, Zhang S, Zhang J. 2011. Widespread losses of vomeronasal signal transduction in bats. Mol. Biol. Evol. 28, 7–12 (doi:10.1093/molbev/msq207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steiger SS, Fidler AE, Valcu M, Kempenaers B. 2008. Avian olfactory receptor gene repertoires: evidence for a well-developed sense of smell in birds? Proc. R. Soc. B 275, 2309–2317 (doi:10.1098/rspb.2008.0607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aujard F. 1997. Effect of vomeronasal organ removal on male socio-sexual responses to female in a prosimian primate (Microcebus murinus). Physiol. Behav. 62, 1003–1008 (doi:10.1016/S0031-9384(97)00206-0) [DOI] [PubMed] [Google Scholar]
- 23.Powers JB, Winans SS. 1975. Vomeronasal organ: critical role in mediating sexual behavior of the male hamster. Science 187, 961–963 (doi:10.1126/science.1145182) [DOI] [PubMed] [Google Scholar]
- 24.Scalia F, Winans SS. 1975. The differential projections of the olfactory bulb and accessory olfactory bulb in mammals. J. Comp. Neurol. 161, 31–55 (doi:10.1002/cne.901610105) [DOI] [PubMed] [Google Scholar]
- 25.Smith TD, Siegel MI, Mooney MP, Burdi AR, Burrows AM, Todhunter JS. 1997. Prenatal growth of the human vomeronasal organ. Anat. Rec. 248, 447–455 (doi:10.1002/(SICI)1097-0185(199707)248:3<447::AID-AR18>3.0.CO;2-P) [DOI] [PubMed] [Google Scholar]
- 26.Baxi KN, Dorries KM, Eisthen HL. 2006. Is the vomeronasal system really specialized for detecting pheromones? Trends Neurosci. 29, 1–7 (doi:10.1016/j.tins.2005.10.002) [DOI] [PubMed] [Google Scholar]
- 27.Bhatnagar KP, Smith TD. 2001. The human vomeronasal organ. III. Postnatal development from infancy to the ninth decade. J. Anat. 199, 289–302 (doi:10.1046/j.1469-7580.2001.19930289.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martínez-García F, Martínez-Ricós J, Agustín-Pavón C, Martínez-Hernández J, Novejarque A, Lanuza E. 2009. Refining the dual olfactory hypothesis: pheromone reward and odour experience. Behav. Brain Res. 200, 277–286 (doi:10.1016/j.bbr.2008.10.002) [DOI] [PubMed] [Google Scholar]
- 29.Pause BM. 2004. Are androgen steroids acting as pheromones in humans? Physiol. Behav. 83, 21–29 (doi:10.1016/j.physbeh.2004.07.019) [DOI] [PubMed] [Google Scholar]
- 30.Takami S. 2002. Recent progress in the neurobiology of the vomeronasal organ. Microsc. Res. Tech. 58, 228–250 (doi:10.1002/jemt.10094) [DOI] [PubMed] [Google Scholar]
- 31.Smith TD, Siegel MI, Bhatnagar KP. 2001. Reappraisal of the vomeronasal system of catarrhine primates: ontogeny, morphology, functionality, and persisting questions. Anat. Rec. 265, 176–192 (doi:10.1002/ar.1152) [DOI] [PubMed] [Google Scholar]
- 32.Smith TD, Garrett EC, Bhatnagar KP, Bonar CJ, Bruening AE, Dennis JC, Kinznger JH, Johnson EW, Morrison EE. 2011. The vomeronasal organ of New World monkeys (Platyrrhini). Anat. Rec. 294, 2158–2178 (doi:10.1002/ar.21509) [DOI] [PubMed] [Google Scholar]
- 33.Pihlström H, Fortelius M, Hemilä S, Forsman R, Reuter T. 2005. Scaling of mammalian ethmoid bones can predict olfactory organ size and performance. Proc. R. Soc. B 272, 957–962 (doi:10.1098/rspb.2004.2993) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abramoff MD, Magalhaes PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 35.Matsui A, Go Y, Niimura Y. 2010. Degeneration of olfactory receptor gene repertories in primates: no direct link to full trichromatic vision. Mol. Biol. Evol. 27, 1192–1200 (doi:10.1093/molbev/msq003) [DOI] [PubMed] [Google Scholar]
- 36.Hohenbrink P, Mundy NI, Zimmermann E, Radespiel U. 2013. First evidence for functional vomeronasal 2 receptor genes in primates. Biol. Lett. 9, 20121006 (doi:10.1098/rsbl.2012.1006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orme D, Freckleton R, Thomas G, Petzholdt T, Fritz S, Isaac N. 2011. caper: comparative analyses of phylogenetics and evolution in R. R package v. 0.4 See http://cran.r-project.org/web/packages/caper/index.html
- 38.R Development Core Team. 2010. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 39.Bininda-Emonds OR, et al. 2007. The delayed rise of present-day mammals. Nature 446, 507–512 (doi:10.1038/nature05634) [DOI] [PubMed] [Google Scholar]
- 40.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884 (doi:10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (doi:10.1214/aos/1013699998) [Google Scholar]
- 42.Niimura Y, Nei M. 2006. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J. Hum. Genet. 51, 505–517 (doi:10.1007/s10038-006-0391-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith TD, Bhatnagar KP, Tuladhar P, Burrows AM. 2004. Distribution of olfactory epithelium in the primate nasal cavity: are microsmia and macrosmia valid morphological concepts? Anat. Rec. A Discov. Mol. Cell Evol. Biol. 281, 1173–1181 (doi:10.1002/ar.a.20122) [DOI] [PubMed] [Google Scholar]
- 44.Grus WE, Zhang J. 2008. Distinct evolutionary patterns between chemoreceptors of 2 vertebrate olfactory systems and the differential tuning hypothesis. Mol. Biol. Evol. 25, 1593–1601 (doi:10.1093/molbev/msn107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nei M, Niimura Y, Nozawa M. 2008. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9, 951–963 (doi:10.1038/nrg2480) [DOI] [PubMed] [Google Scholar]
- 46.Gilad Y, Bustamante CD, Lancet D, Pääbo S. 2003. Natural selection on the olfactory receptor gene family in humans and chimpanzees. Am. J. Hum. Genet. 73, 489–501 (doi:10.1086/378132). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilad Y, Man O, Pääbo S, Lancet D. 2003. Human specific loss of olfactory receptor genes. Proc. Natl Acad. Sci. USA 100, 3324–3327 (doi:10.1073/pnas.0535697100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hohenbrink P, Radespiel U, Mundy NI. 2012. Pervasive and ongoing positive selection in the vomeronasal-1 receptor (V1R) repertoire of mouse lemurs. Mol. Biol. Evol. 29, 3807–3816 (doi:10.1093/molbev/mss188) [DOI] [PubMed] [Google Scholar]
- 49.Mundy NI, Cook S. 2003. Positive selection during the diversification of class I vomeronasal receptor-like (V1RL) genes, putative pheromone receptor genes, in human and primate evolution. Mol. Biol. Evol. 20, 1805–1810 (doi:10.1093/molbev/msg192) [DOI] [PubMed] [Google Scholar]
- 50.Rouquier S, Blancher A, Giorgi D. 2000. The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc. Natl Acad. Sci. USA 97, 2870–2874 (doi:10.1073/pnas.040580197) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shi P, Bielawski JP, Yang H, Zhang YP. 2005. Adaptive diversification of vomeronasal receptor 1 genes in rodents. J. Mol. Evol. 60, 566–576 (doi:10.1007/s00239-004-0172-y) [DOI] [PubMed] [Google Scholar]
- 52.Niimura Y, Nei M. 2007. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS ONE 2, e708.. (doi:10.1371/journal.pone.0000708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nozawa M, Kawahara Y, Nei M. 2007. Genomic drift and copy number variation of sensory receptor genes in humans. Proc. Natl Acad. Sci. USA 104, 20 421–20 426 (doi:10.1073/pnas.0709956104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perry GH, et al. 2008. Copy number variation and evolution in humans and chimpanzees. Genome Res. 18, 1698–1710 (doi:10.1101/gr.082016.108) [DOI] [PMC free article] [PubMed] [Google Scholar]


