Abstract
The extent of female multiple mating (polyandry) can strongly impact on the intensity of sexual selection, sexual conflict, and the evolution of cooperation and sociality. More subtly, polyandry may protect populations against intragenomic conflicts that result from the invasion of deleterious selfish genetic elements (SGEs). SGEs commonly impair sperm production, and so are likely to be unsuccessful in sperm competition, potentially reducing their transmission in polyandrous populations. Here, we test this prediction in nature. We demonstrate a heritable latitudinal cline in the degree of polyandry in the fruitfly Drosophila pseudoobscura across the USA, with northern population females remating more frequently in both the field and the laboratory. High remating was associated with low frequency of a sex-ratio-distorting meiotic driver in natural populations. In the laboratory, polyandry directly controls the frequency of the driver by undermining its transmission. Hence we suggest that the cline in polyandry represents an important contributor to the cline in sex ratio in nature. Furthermore, as the meiotic driver causes sex ratio bias, variation in polyandry may ultimately determine population sex ratio across the USA, a dramatic impact of female mating decisions. As SGEs are ubiquitous it is likely that the reduction of intragenomic conflict by polyandry is widespread.
Keywords: polyandry, sexual selection, sex ratio distorter, sperm competition, meiotic drive, geographical cline
1. Introduction
Variation in female mating frequency in nature is profound. Females of some species mate only once in their life, whereas others may mate with hundreds of males [1]. Research in the last 30 years suggests that the frequency at which females remate is of key importance in the ecology and evolution of many animals [2]. The frequency of polyandry can affect the level of gene flow and the effective population size [3], the population viability [4,5], and the intensity of post-copulatory sexual selection and sexual conflict [6], giving rise to a range of adaptations and counter-adaptations for the manipulation of mates and rivals [2,7]. In addition, polyandry can reduce within-family relatedness and thereby inhibit within-family cooperation, thus affecting the intensity of parent–offspring conflict [8], and the evolution of cooperation and sociality [9,10]. However, theory and experimental evolution studies also indicate that polyandry can promote harmony within the genome, through undermining the spread of selfish genetic elements (SGEs) [11,12].
SGEs are genes that subvert the normal patterns of inheritance to increase their representation in subsequent generations [13,14]. SGEs are ubiquitous in living organisms [13] and can make up a large proportion of the genome, and the intragenomic conflicts they create are thought to have had major impacts on the evolution of sex and reproductive systems [13,15]. Recent discoveries of novel SGEs in well-studied species [16–18] suggest that a vast array of SGEs remain to be discovered. Models of the dynamics of many SGEs suggest that they should spread rapidly through populations, but most empirical studies have found their abundance to be stable in nature [13,14]. Although many mechanisms have been suggested to control the frequency of SGEs, in most cases we do not understand how their abundance is determined in natural populations [14,19]. Many SGEs, including meiotic drive elements, B chromosomes, endosymbionts and some transposons, often target male gametes and have been shown to impair male fertility through manipulation of spermatogenesis [20]. This reduces the success of males carrying the SGE during sperm competition [20]. Polyandry is therefore likely to reduce the transmission of any SGEs that reduce the sperm competitive ability of males [11]. Hence it is possible that differences in degree of polyandry are important in determining the frequency of many SGEs in the wild. However, this hypothesis has never been tested, because it requires intra-specific variation in the level of polyandry.
Sex ratio (SR), a meiotic driving X chromosome, has been studied in the fruitfly Drosophila pseudoobscura for more than 75 years [21]. The biology of this SGE indicates that its population frequency should be vulnerable to control by polyandry. SR has little consistent effect in females [22], but in males, SR causes the death of all Y-chromosome-bearing sperm during spermatogenesis [23], leaving all functional sperm carrying only the SR X chromosome. Hence all offspring of SR males are daughters, and inherit the SR X chromosome. As there is no genetic resistance to sex ratio drive in D. pseudoobscura [24], simple models predict that SR should spread rapidly through populations until it causes extinction owing to a lack of males [21,25]. In nature, the abundance of SR in D. pseudoobscura is broadly stable, having been historically common in populations in the southern USA, reaching frequencies of 30% at the US–Mexican border, and becoming rarer to the north, being absent in Canada [21,26]. This latitudinal cline in SR frequency has never been explained [27]. However, the loss of sperm by SR-carrying males makes them poor sperm competitors [28], raising the possibility that polyandry could regulate the frequency of SR by undermining its transmission, as predicted by Haig & Bergstrom [11]. Theory predicts that the transmission advantage of SR should be highest in monandrous populations, and that a sufficiently high frequency of sperm competition could prevent the spread of SR or eliminate it [29]. Experimental work has confirmed that allowing females just one additional mating is sufficient to prevent the spread of SR through laboratory populations, and that SR spreads rapidly and causes population extinction when female remating is prevented in as little as nine generations [4]. So polyandry can directly regulate SR in experimental populations, making polyandry a strong candidate for influencing the distribution of SR in nature.
To investigate the hypothesized link between polyandry and meiotic drive frequency in the wild, we determined the frequency of multiple paternity in seven natural populations of D. pseudoobscura. We then estimated the frequency of SR in these populations and tested for the predicted negative association between SR frequency and the rate of polyandry.
2. Material and methods
(a). Estimate of polyandry in wild females
We caught flies using standard banana baits [30] from seven locations across the USA in May–June 2008 (table 1; electronic supplementary material, figure S1). The seven locations were suitable forest habitat separated by areas of unsuitable habitat, such as desert or pasture. Most collections were carried out in National Forests, and no permits or licences were required. Two collection sites were on private land, and permission was given by the landowners for this. Baits were placed under trees, and emptied at dawn and dusk to reduce the likelihood that high densities at the bait would influence female mating frequency. Females were caught, isolated from males and sent to the laboratory, where they were maintained at 23°C, with a 14 L : 10 D cycle on standard Drosophila food [31]. We genotyped each wild-caught female and up to 22 of her randomly chosen offspring (range 9–22 offspring, median of 21 offspring per family, 189 families) using four highly polymorphic microsatellites (methods detailed in [32], microsatellites described in [33]). The number of sires was assayed by subtracting the maternal genotype from that of each offspring, and, for the most variable locus, dividing the number of remaining alleles by 2 to give a minimum number of fathers [34].
Table 1.
The locations of the seven populations and the percentage of wild-caught females found to have mated with more than one male.
| population | location | state | latitude north | longitude west | % multiple paternity |
|---|---|---|---|---|---|
| 1 | Chiricahua mountains | Arizona | 31°54′55″ | 109°15′95″ | 58 |
| 2 | Show Low | Arizona | 34°07′37″ | 110°07′37″ | 52 |
| 3 | Mount Lemmon | Arizona | 32°21′95″ | 110°41′66″ | 73 |
| 4 | Zion Forest | Utah | 37°25′91″ | 113°03′13″ | 59 |
| 5 | Panguitch | Utah | 37°55′87″ | 112°19′66″ | 73 |
| 6 | Fillmore | Utah | 38°55′86″ | 112°14′60″ | 73 |
| 7 | Lewistown | Montana | 47°04′47″ | 109°16′53″ | 92 |
One potential problem in assessing multiple paternity across populations is that allele frequencies typically differ between populations, meaning that the power of each locus to detect multiple paternity also differs between populations. If one population has few alleles, or one very common allele, then many males will share this allele, and detecting multiple paternity will be difficult, creating an artificially low rate of detected multiple paternity. To assess whether this could bias our results, we calculated the chance of failing to detect multiple paternity in each family, following Harshman & Clark [35]. This method calculates the probability of misidentifying multiple paternity as single paternity by combining the probabilities of the two males being identical in genotype, the two males sharing one common allele that is not represented in the offspring sampled, and the two males having no common alleles but only half the alleles being represented in the sample offspring. Using the population allele frequencies for each population derived from the maternal genotypes and samples of either nine offspring (the minimum sampled per real family) or 22 offspring (the majority of families sampled), the probability of misidentifying a multiply sired family was less than 0.00002 for every population. We also used the Gerudsim application of Gerud [36] to simulate families consisting of a mother, two fathers and nine offspring (three descended from one father, six from the other) using the allele frequencies found in each population. For each population, we ran 10 separate parental simulations with 1000 iterations (i.e. simulations of offspring genotypes). The mean probability of failing to detect multiple paternity owing to similar genotypes in two males was 0.4% per family, with the least successful population having a failure rate of 1.08% per family. Considering that this estimate is derived from the highly conservative assumption of only nine offspring per family (the fewest we were able to genotype for one of the families), rather than the 21 offspring used for most families, it is unlikely that these errors play a significant role affecting our results. Furthermore, the three southernmost populations, where we found the lowest rates of multiple paternity, had lower likelihoods of failing to detect multiple paternity than the three northernmost populations, so errors of this kind would mask the latitudinal cline we detected, rather than create it.
(b). Remating propensity of granddaughters in the laboratory
We collected one virgin daughter of 8–15 wild-caught females (see above) from each population (total 71 wild-caught females) and placed them in a standard Drosophila vial with a male sibling. Each pair was moved on to a new vial twice each week. We collected 8–10 virgin female offspring from each pair (the F2 granddaughters of the wild-caught females). At 3 days old, we placed each F2 granddaughter in an individual Drosophila vial, and left her overnight to acclimatize. The following day, we presented each F2 granddaughter with a single stock male for 2 h. These males were 3-day-old virgins collected from a stock mass population, all carrying the non-driving X chromosome standard (ST), derived from a collection carried out at Show Low, Arizona, USA, in 2004. We watched the vials continuously, and noted all copulations. Females that failed to mate were excluded from the experiment (264 of 748 females). Failure to mate with the first male did not correlate with the latitude of the population of origin (Spearman's correlation: n = 7 populations, coefficient = 0.286, p = 0.535), nor with the remating propensity of siblings (Spearman's correlation: n = 7 populations, coefficient = 0.071, p = 0.879). Hence we concluded that failure to mate with the initial male was stochastic, and could be disregarded in the rest of the analysis, as is standard in Drosophila remating trials. After a successful first copulation, we removed the male. Each day for the following 6 days, we presented the females with a new virgin stock male carrying ST. All vials were watched as before, and all rematings were noted. We blinded the experiment by labelling each female with a randomly assigned number, and the observers were not aware of the genotype of the individuals. All flies were moved by aspiration to avoid anaesthesia [37]. Remating was scored in two ways: first, whether or not the female remated at all over the 6 days; second, analysing only those females that remated, we examined mean number of days to remating. To test whether the results of the above experiments could have been due to the influence of maternal effects or differential response to the tester strain, we maintained isolines from two populations (Show Low, the southern population; and Lewistown, the northernmost population), and tested for remating propensity after 40 generations in the laboratory. At least 30 females from each of 23 isolines were tested as above, but were mated to males from their own population, and the mating assays were conducted simultaneously.
(c). Sperm competitive ability in northern and southern populations
Our previous estimate of the relative sperm competitive ability of SR and ST males was carried out using flies from a single southern population, collected at Show Low [28]. If ST males have lower success in sperm competition when competing against SR males, this would make polyandry less effective at reducing the transmission of SR in northern populations. However, if northern ST males are more successful in sperm competition against SR males than southern ST males, then this would increase the likelihood that polyandry prevents SR colonizing northern populations. To examine this, we established mass populations of SR and ST flies from the Show Low population by crossing individuals from 20 isolines from this population, and a mass Lewistown ST population by crossing 16 isolines from Lewistown. After two generations of free mating in each population, we collected homozygous ST/ST females from the Show Low population and mated them to both an SR male and an ST male, using the methodology described by Price et al. [28]. In brief, we mated 4-day-old virgin females to a male, and 3 days later she was mated to a second male, and then allowed to oviposit for 6 days. In half the trials, the ST male was from the Show Low population, whereas the SR males were always taken from the Show Low population, to represent the situation that occurs when SR and ST males compete in Show Low, and to represent an SR male immigrating into a northern population. The order of mating was randomized. Females that did not mate twice were discarded. After the second mating, females were allowed to oviposit for 12 days, and the resulting offspring were collected. The offspring were counted by sex, as all sons were fathered by the ST male, and 23 of the daughters were genotyped for SR [32]. We measured the success of the ST males in sperm competition by calculating the number of offspring that inherited SR by multiplying the number of daughters produced by the proportion of the 23 daughters that carried SR. However, as a conservative measure we also independently analysed the proportion of both sons and daughters inheriting SR.
(d). Correlation of polyandry and sex ratio frequency in nature
The distribution of SR across the USA was ascertained from collections between 1938 and 1957 by Dobzhansky [26], and our own collections in 2004 and 2008. We assayed SR frequency in our populations by genotyping at least 50 wild-caught males and females from each population. We detected SR and ST chromosomes using markers described by Price et al. [32]. The data from our collections in 2004 for Show Low and Lewistown were pooled with the data from 2008 for these locations. In 2004, we also carried out a collection at Flagstaff (35°05′00″ N, 111°44′10″ W), which was not repeated in 2008. This was used as a single data point. The data were analysed using a generalized linear model, with latitude as a fixed effect and survey as a random effect, using a normal error distribution and an identity link function. We used stepwise removal of factors from the full model to produce a final minimal model [38]. Analysis was conducted using R v. 2.13.1 [39].
3. Results
(a). Polyandry in wild females from populations across the USA
We first surveyed the frequency of multiple paternity in seven populations across the USA, ranging from southern Arizona to Montana (table 1). We observed that the mean number of sires per family was significantly correlated with latitude, with southern populations having fewer sires per brood than northern populations (figure 1). Carrying the SR chromosome does not affect the remating propensity of female carriers [27], and indeed the significance of the polyandry cline described above was not changed after re-analysis using only the non-SR females (figure 1).
Figure 1.
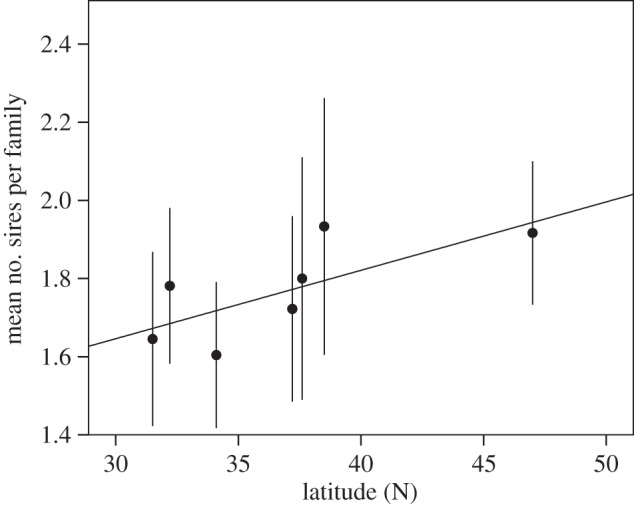
The mean number of sires detected in the broods of wild-caught females, with 95% confidence intervals. More sires are detected at higher latitudes (Spearman's correlation: all females: 12–40 broods from each of seven populations; n = 7, coefficient = 0.786, p = 0.036; analysing non-SR females only does not change the rank order: 12–34 broods from each of seven populations; n = 7, coefficient = 0.786, p = 0.036).
(b). Remating propensity of granddaughters in the laboratory
To investigate the genetic component of the cline in polyandry observed, we examined the remating propensity of the F2 granddaughters of the wild-caught females. The granddaughters of females from southern populations were significantly less likely to remate than the granddaughters of females from northern populations (figure 2a). Where remating occurred, the granddaughters of southern females also showed a significantly longer delay to remating than the granddaughters of northern females (figure 2b). This pattern was replicated 40 generations after collection, with females from a southern population having significantly lower remating propensities than females from the northernmost population (t test: n = 23, t = 2.326, p = 0.032), indicating the difference observed in the granddaughters was not because of maternal effects.
Figure 2.
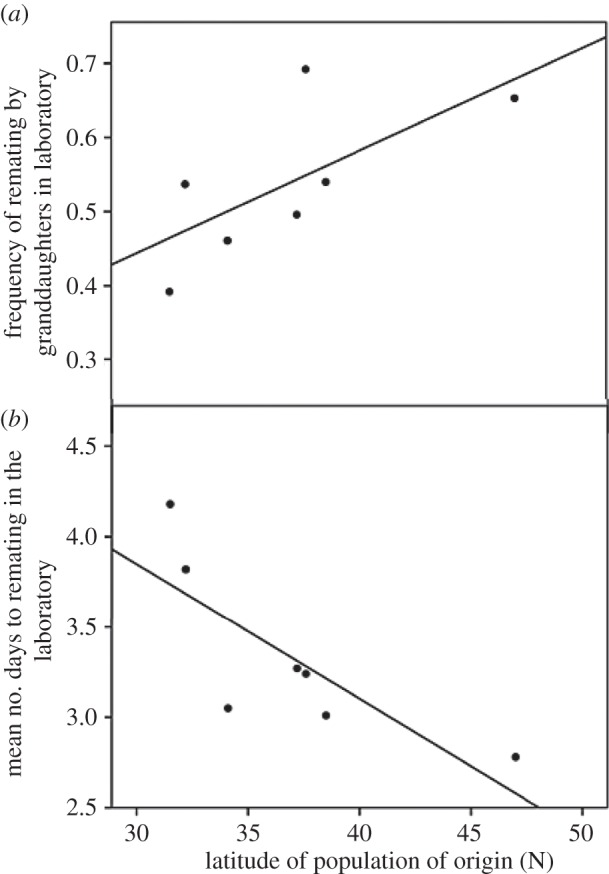
The remating propensity of females derived from seven populations across the USA. (a) The frequency of remating in the laboratory by the F2 granddaughters of each wild-caught female is positively correlated with the latitude of the population from which they were descended (10–12 families from each of seven populations, 484 females in total; Spearman's correlation: n = 7, coefficient = 0.786, p = 0.036). (b) The mean number of days to remate of the F2 granddaughters of each wild-caught female is negatively correlated with the latitude of the population from which they were descended (mean of 10–12 families from each of seven populations, 361 females total; Spearman's correlation: n = 7, coefficient = −0.893, p = 0.007).
(c). Sperm competitive ability in northern and southern populations
SR males were significantly worse sperm competitors than ST males from both populations, irrespective of mating order (proportion of offspring fathered by the SR male when competing with northern ST male: SR mated first: 0.06, n = 27; SR mated second: 0.31, n = 22; when competing with a southern ST male: SR mated first: 0.23, n = 20; SR mated second: 0.53, n = 26). SR males were significantly worse as sperm competition when mating in the first male role (F-test: F1,92 = 25.148, p < 0.001). ST males from the northern Lewistown population were significantly more successful in sperm competition than ST males from Show Low (F-test: F1,92 = 12.398, p < 0.001). However, there was no population difference in the sperm competitive success of non-SR males in relation to mating order (no interaction between non-SR male population of origin and mating order: F-test: F1,92 = 0.192, p = 0.662). The results were qualitatively the same when proportion of sons or proportion of daughters fathered by the non-SR male were analysed. The proportion of sons and daughters that were found to carry SR were significantly negatively correlated (Spearman's rank correlation of proportion of sons and proportion of daughters that inherited SR: n = 95, ρ = −0.619, p < 0.001), indicating that both measures were accurate, and that the combined estimate of the proportion of offspring inheriting SR was correct.
(d). Correlation of polyandry and frequency of sex ratio
We estimated the frequency of SR in our collections and tested whether SR frequency was correlated with latitude, both in our surveys and historical data from 1940 to 1958 [26]. The frequency of SR was negatively correlated with latitude across populations when both historical and contemporary data were pooled (figure 3; F-test: F1,23 = 16.284, p < 0.001), and in each dataset when analysed separately (F-tests: Dobzhansky data: F1,15 = 9.571, p = 0.007; our data: F1,7 = 19.474, p = 0.005). Furthermore, there was no evidence that the relationship between the frequency of SR and latitude had changed over the 70 years between the surveys (F-tests: effect of survey date—post-2004 versus pre-1958—on the relationship between latitude and SR frequency: F1,22 = 1.287, p = 0.269; effect of survey date per se: F1,21 = 2.728, p = 0.113), confirming that this latitudinal cline in SR frequency has remained broadly stable for at least 70 years.
Figure 3.
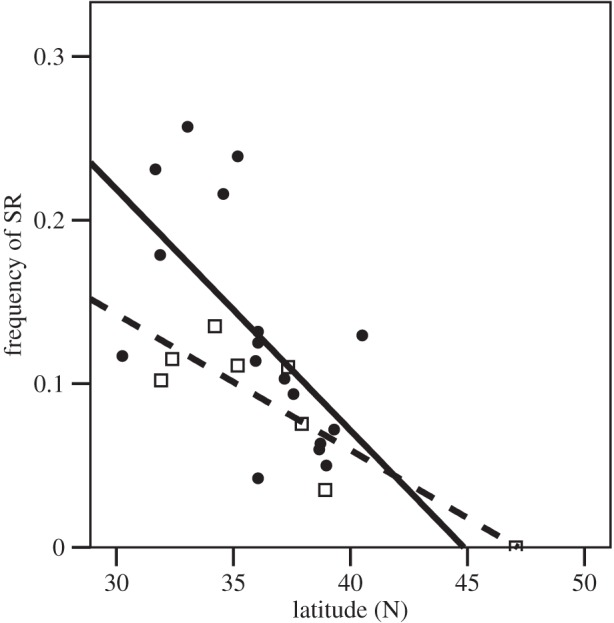
The frequency of SR and latitude across the USA. SR is more abundant in the southern USA, in surveys conducted by Dobzhansky [26] (solid circles, solid fit line) and in our eight populations (collected 2004–2008; hollow squares, dashed fit line).
The mean number of sires within the broods of field-caught females was negatively correlated with SR frequency across populations (figure 4; Spearman's correlation: n = 7, coefficient = −0.821, p = 0.023). Overall, the median number of sires per mother ranged from 1.62 (southern population, SR at 30% frequency) to 1.96 (northern population, SR near absent), a close match to the estimate that two mating opportunities is enough to cause the frequency of SR to decline in laboratory populations [4]. Moreover, the median number of fathers per brood was significantly lower than two in only the four southernmost populations, where SR was most common (one sample one-tailed Wilcoxon signed-ranks test: Chiricahua: n = 31, W = 16, p = 0.003; Show Low: n = 40, W = 56, p < 0.001; Mt Lemmon: n = 32, W = 12, p = 0.018; Zion Forest: n = 36, W = 52.5, p = 0.013; Panguitch: n = 19, W = 4, p = 0.09; Fillmore: n = 19, W = 16, p = 0.353; Lewistown: n = 12, W = 0, p = 0.159). Taken together, our results strongly support the hypothesis that variation in polyandry controls the frequency of SR in nature, as previously demonstrated in the laboratory [4].
Figure 4.
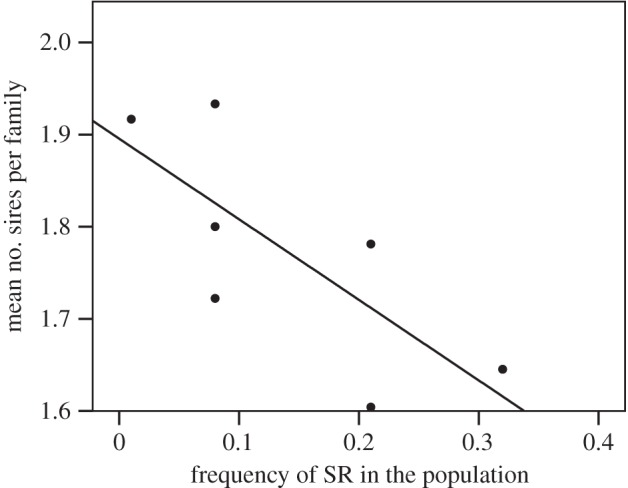
The number of fathers detected in the broods of wild-caught females and SR frequency. Mean number of sires is negatively correlated with the frequency of SR across natural populations (mean of 12–40 families from each of seven populations, total 189 families; Spearman's correlation: n = 7, coefficient = −0.821, p = 0.023).
4. Discussion
These results demonstrate first that there is a latitudinal cline in female remating frequency in D. pseudoobscura, and second that there is a genetic component to this cline. To the best of our knowledge, this is the first heritable geographical cline in female remating frequency to be discovered. The relatively high levels of gene flow between populations of D. pseudoobscura across the USA [40] make it unlikely that the genetic component of this cline is owing to historical factors, but rather is probably maintained by current selection. The cline is likely to be a result of selection on female mating behaviour, as the differences were expressed when females were reared under common garden conditions and mated with standard stock males. At present, the cause of this cline in polyandry in nature is not known.
Both methods of estimating population-level polyandry can be criticized. For example, the wild-caught females were of unknown age, and if females in the north live longer, this could result in higher numbers of brood sires. In addition, there are many well-known potential sources of error in genotyping the offspring of wild-caught females. Similarly, if northern females are adapted to cooler temperatures than southern females, then exposure to the standard 23°C laboratory temperature might lead to differences in female mating behaviour in the laboratory. However, using two independent methods to determine degree of polyandry makes it unlikely that our results are artefactual, which is further corroborated by the significant correlation of laboratory and field measures across populations (Spearman's correlation: n = 7 populations, rs = 0.821, p = 0.023). One additional risk is that an inability to detect SR sires owing to their poor sperm competitive ability might cause us to miss SR sires in some populations, hence generating the observed polyandry cline. However, SR is typically found at Hardy–Weinberg equilibrium [41–43], so SR is expected to occur at equal frequencies in males and females. We found no difference in frequencies of SR in fathers and mothers (overall SR frequency: males, 7.0%; females, 6.7%). As there is no overall deficit of SR in fathers, it is unlikely that an inability to detect SR fathers caused the observed cline. Taken together, our experiments demonstrate a north–south cline in frequency of polyandry across the USA. Furthermore, combined with previous evidence that female remating frequency in D. pseudoobscura is heritable [44], our results show that this cline in polyandry is genetically determined.
Our survey of SR frequency in eight populations and our formal analysis of previous data strongly support the conclusion by Dobzhansky [26] and others [27] that a latitudinal cline in SR exists across the USA. This cline appears to have remained stable for the past 75 years. The cline in polyandry will be an important driver of this cline. Theoretical work predicts that meiotic drivers could have their transmission inhibited by reduced sperm competitive success, and so variation in rates of polyandry could generate variation in the abundance of meiotic drive [11,45]. For this hypothesis to apply in nature, the driver must significantly reduce the success of male carriers in sperm competition, and sperm competition should occur at a high enough rate to be important. In addition, females should not be able to discriminate against meiotic drive males prior to mating, otherwise mate discrimination against drive-carrying males would be expected to be more important than sperm competition. There is considerable evidence that SR in D. pseudoobscura fulfils all these conditions. Males carrying SR are poor sperm competitors, both in the experiments presented here and as shown in previous studies [28]. This study suggests that SR males are particularly poor sperm competitors when competing against males from the northern, more highly polyandrous populations. Females are often polyandrous in nature [32,46], and show no evidence of being able to discriminate between SR and ST males prior to mating [47,48]. Laboratory experimental evolution studies have shown that in populations where females were allowed to remate, SR declined in frequency, whereas in populations where polyandry was prevented, SR spread rapidly [4]. As polyandry directly regulates the frequency of SR in laboratory populations, the evidence presented here that SR is rare or absent in populations with a high level of polyandry, and common in populations where polyandry is more rare, strongly suggests that polyandry also determines the frequency of SR in natural populations.
Alternative explanations for the distribution of SR have been suggested. Meta-population dynamics is one possibility, with subpopulations carrying high frequencies of drive repeatedly going extinct [49]. However, this would predict that very high frequencies of drive should be observed in local populations, and local extinctions should be common, but at present there is only one tentative report of this [50]. It is also notable that this dynamic would create a checkerboard pattern of presence and absence, rather than the observed clinal variation. [14]. A second suggestion is decreased fitness in females, particularly homozygotes, owing to the accumulation of deleterious mutations within the inversions on the SR chromosome [27,43]. Such decreased fitness is seen in the SR chromosome of Drosophila recens [51], and low fitness of homozygotes is found in t haplotypes in mice [52]. However, the evidence for a cost to females of carrying SR in D. pseudoobscura is poor [14,22,27,53]. Moreover, the one study that found low fitness of female SR carriers found that SR females were more successful at lower temperatures [53], and so cannot explain why SR is rare in the north. Alternatively, SR might decrease the fertility of males more at lower temperatures, thereby preventing SR persistence in colder areas. However, although male fertility does interact with temperature, SR males are less fertile at higher temperatures, making this an unlikely explanation of the observed cline in SR [22]. Further suggestions, such as increased vulnerability to parasites or lower overwinter survival of SR-bearing flies, have been put forward [22,27]. However, at present none are supported by data from either the laboratory or the field.
It is currently not possible to completely eliminate the possibility that the observed correlation of SR and polyandry is due to some unknown additional factor that directly influences both, or arose simply by chance. However, there is strong theoretical [11,29] and empirical evidence [4,28] that polyandry reduces the success of SR in populations. There is also a lack of experimental support for the major competing theories for the control of SR frequency in natural populations [14,22,27], and little evidence that these alternative factors correlate with latitude in a way that could create the observed stable cline in SR. Hence, by far the most parsimonious explanation for the clinal distribution of SR is that it is maintained by the underlying cline in polyandry reported here, providing a potential solution to a 75-year-old puzzle in population genetics [21].
Polyandry may play a major role in controlling the abundance of many SGEs in nature [13]. Evidence from mice supports this hypothesis [52]. The population dynamics of autosomal meiotic driving t alleles in mice have been investigated for decades [54]. Models commonly predict t allele frequencies 10 times higher than those found in natural populations (the ‘t frequency paradox’) [55]. Recent work using models parametrized with extensive laboratory and field experiments found that the dynamics of t alleles in a natural population of house mice could only be explained by the transmission disadvantage owing to sperm competition resulting from polyandry [52]. Clinal distributions may also be common in SGEs. Several other sex chromosome meiotic drivers are distributed along latitudinal clines, being commoner in southern populations than northern ones. These include drivers in Drosophila persimilis [27], D. subobscura [56] and D. recens [51]. The endosymbiont Wolbachia also seems to be clinally distributed in the weevil Curculio sikkimensis [57]. It is possible that these clines may also be caused by underlying clines in polyandry, although this has not been investigated. Our conclusion that polyandry can determine the distribution of an SGE in nature is supported by recent work in another Drosophila species, D. neotestacea [50]. In D. neotestacea, another X-chromosome meiotic driver shows a latitudinal cline across North America, being rare in Canada [58]. A similar cline in polyandry has been found in this species, with northern females remating more frequently, and degree of polyandry covaries with the frequency of meiotic drive in natural populations. Drosophila neotestacea is only distantly related to D. pseudoobscura, and the meiotic drivers evolved independently. The discovery of similar patterns in both species is strong evidence that polyandry can protect populations from SGEs in nature. Hence it is likely that geographical differences in degree of polyandry are generally important in determining the frequency of many SGEs in the wild. Meiotic drivers may be particularly likely to be controlled in this way, because damaging sperm is an essential part of the drive mechanism [14].
If the abundance of sex chromosome drivers is partly determined by the level of polyandry, this could have an impact on the whole population. The presence of meiotic drivers can cause females to evolve increased rates of polyandry [44], which can in turn promote the evolution of male counter-adaptations that suppress female remating [59], increasing the level of sexual conflict throughout a population. Most obviously, sex ratio distorters can influence population sex ratio [14]. Population sex ratio is a key ecological parameter, influencing factors such as effective population size [60], population growth rates [61], mate competition and mate choice [62]. High frequencies of SR result in female-biased population sex ratios, and the population sex ratio in D. pseudoobscura can be directly determined by the frequency of SR [63]. Laboratory studies tracking the spread of SR through experimental evolution populations also show that the frequency of SR can control population sex ratio [4]. Here we argue that, although correlational, our results strongly suggest that the level of polyandry also determines the frequency of SR in natural populations, and the population sex ratio in D. pseudoobscura is therefore ultimately determined by the frequency of polyandry. This is a remarkably powerful impact of individual female mating decisions on the ecology of populations [64].
Our results demonstrate the existence of a stable cline in polyandry across the USA. This cline is heritable, acts through female mating behaviour and may be maintained by current selection on the frequency of polyandry. Furthermore, these differences in polyandry across populations seem to control the frequency of a sex-ratio-distorting SGE in nature, ultimately determining population sex ratio at a landscape scale. Many other SGEs can control population sex ratio [13,62,65] and impair sperm production [20], making them vulnerable to control through polyandry [11]. Hence, although polyandry can increase conflict between individuals [2,7] and repress the evolution of sociality [9,10], it also has the potential to promote harmony within the genome by suppressing the spread of SGEs.
Acknowledgements
We thank Mari-Wyn Burley and the UCL Centre for Comparative Genomics for genotyping support, and Teri Markow and Doug Emlen for field support. We thank Karin Pfennig and two anonymous reviewers for useful comments on the manuscript.
Data accessibility
No large datasets are involved in this work. However, isolines and material will be made available on request.
Funding statement
This work was funded by NERC (NE/F003781/1 to N.W., T.A.R.P. and G.D.D.H., NE/I027711/1 to N.W., NE/I025905/1 to T.A.R.P. and G.D.D.H., and NE/H015604/1 to T.A.R.P.), and by the Genetics Society (summer studentship to A.C.). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Reguera P, Pomiankowski A, Fowler K, Chapman T. 2004. Low cost of reproduction in female stalk-eyed flies, Cyrtodiopsis dalmanni. J. Insect Physiol. 50, 103–108 (doi:10.1016/j.jinsphys.2003.10.004) [DOI] [PubMed] [Google Scholar]
- 2.Simmons LW. 2001. Sperm competition and its evolutionary consequences in the insects. Princeton, NJ: Princeton University Press [Google Scholar]
- 3.Jaffe R, Moritz RFA, Kraus FB. 2009. Gene flow is maintained by polyandry and male dispersal in the army ant Eciton burchellii. Popul. Ecol. 51, 227–236 (doi:10.1007/s10144-008-0133-1) [Google Scholar]
- 4.Price TAR, Hurst GDD, Wedell N. 2010. Polyandry prevents extinction. Curr. Biol. 20, 1–5 (doi:10.1016/j.cub.2010.01.050) [DOI] [PubMed] [Google Scholar]
- 5.Rankin D, Dieckmann U, Kokko H. 2011. Sexual conflict and the tragedy of the commons. Am. Nat. 177, 780–791 (doi:10.1086/659947) [DOI] [PubMed] [Google Scholar]
- 6.Hosken DJ, Stockley P, Tregenza T, Wedell N. 2009. Monogamy and the battle of the sexes. Annu. Rev. Entomol. 54, 361–378 (doi:10.1146/annurev.ento.54.110807.090608) [DOI] [PubMed] [Google Scholar]
- 7.Arnqvist G, Rowe L. 2005. Sexual conflict. Princeton, NJ: Princeton University Press [Google Scholar]
- 8.Trivers RL. 1974. Parent–offspring conflict. Am. Zool. 14, 249–264 [Google Scholar]
- 9.Cornwallis CK, West SA, Davis KE, Griffin AS. 2010. Promiscuity and the evolutionary transition to complex societies. Nature 466, 969–972 (doi:10.1038/nature09335) [DOI] [PubMed] [Google Scholar]
- 10.Hughes WO, Oldroyd BP, Beekman M, Ratnieks LW. 2008. Ancestral monogamy shows kin selection is key to the evolution of eusociality. Science 320, 1213–1216 (doi:10.1126/science.1156108) [DOI] [PubMed] [Google Scholar]
- 11.Haig D, Bergstrom CT. 1995. Multiple mating, sperm competition and meiotic drive. J. Evol. Biol. 8, 265–282 (doi:10.1046/j.1420-9101.1995.8030265.x) [Google Scholar]
- 12.Zeh JA, Zeh DW. 1996. The evolution of polyandry I: intragenomic conflict and genetic incompatibility. Proc. R. Soc. B 263, 1711–1717 (doi:10.1098/rspb.1996.0250) [Google Scholar]
- 13.Burt A, Trivers R. 2006. Genes in conflict: the biology of selfish genetic elements. Cambridge, MA: Harvard University Press [Google Scholar]
- 14.Jaenike J. 2001. Sex chromosome meiotic drive. Annu. Rev. Ecol. Syst. 32, 25–49 (doi:10.1146/annurev.ecolsys.32.081501.113958) [Google Scholar]
- 15.Hurst G, Werren J. 2001. The role of selfish genetic elements in eukaryotic evolution. Nat. Rev. Genet. 2, 597–606 (doi:10.1038/35084545) [DOI] [PubMed] [Google Scholar]
- 16.Axelsson E, et al. 2010. Segregation distortion in chicken and the evolutionary consequences of female meiotic drive in birds. Heredity 105, 290–298 (doi:10.1038/hdy.2009.193) [DOI] [PubMed] [Google Scholar]
- 17.Clark KA, Howe DK, Gafner K, Kusuma D, Ping S, Estes S, Denver DR. 2012. Selfish little circles: transmission bias and evolution of large deletion-bearing mitochondrial DNA in Caenorhabditis briggsae Nematodes. PLoS ONE 7, e41433 (doi:10.1371/journal.pone.0041433) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Corbett-Detig RB, Hartl DL. 2012. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8, e1003056 (doi:10.1371/journal.pgen.1003056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson GS, Johns PM, Kelleher ES, Muscedere ML, Lorsong A. 2006. Fitness effects of X chromosome drive in the stalk-eyed fly, Cyrtodiopsis dalmanni. J. Evol. Biol. 19, 1851–1860 (doi:10.1111/j.1420-9101.2006.01169.x) [DOI] [PubMed] [Google Scholar]
- 20.Price TAR, Wedell N. 2008. Selfish genetic elements and sexual selection: their impact on male fertility. Genetica 132, 295–307 (doi:10.1007/s10709-007-9173-2) [DOI] [PubMed] [Google Scholar]
- 21.Sturtevant AH, Dobzhansky T. 1936. Geographical distribution and cytology of ‘sex ratio’ in Drosophila pseudoobscura and related species. Genetics 21, 473–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Price TAR, Hoskyns RA, Rapley H, Evans J, Wedell N. 2012. Temperature effects on male fertility cannot explain the distribution of a selfish genetic element. Func. Ecol. 26, 657–665 (doi:10.1111/j.1365-2435.2012.01971.x) [Google Scholar]
- 23.Policansky D, Ellison J. 1970. ‘Sex ratio’ in Drosophila pseudoobscura: spermiogenic failure. Science 169, 888–889 (doi:10.1126/science.169.3948.888) [DOI] [PubMed] [Google Scholar]
- 24.Policansky D, Dempsey B. 1978. Modifiers and ‘sex-ratio’ in Drosophila pseudoobscura. Evolution 32, 922–924 (doi:10.2307/2407507) [DOI] [PubMed] [Google Scholar]
- 25.Hamilton WD. 1967. Extraordinary sex-ratios. Science 156, 477–488 (doi:10.1126/science.156.3774.477) [DOI] [PubMed] [Google Scholar]
- 26.Dobzhansky T. 1958. Genetics of natural populations. XXVII. The genetic changes in populations of Drosophila pseudoobscura in the American Southwest. Evolution 12, 385–401 (doi:10.2307/2405860) [Google Scholar]
- 27.Powell J. 1997. Progress and prospects in evolutionary biology: the Drosophila model. Oxford, UK: Oxford University Press [Google Scholar]
- 28.Price TAR, Bretman AJ, Avent TD, Snook RR, Hurst GDD, Wedell N. 2008. Sex ratio distorter reduces sperm competitive ability in an insect. Evolution 62, 1644–1652 (doi:10.1111/j.1558-5646.2008.00386.x) [DOI] [PubMed] [Google Scholar]
- 29.Taylor DR, Jaenike J. 2002. Sperm competition and the dynamics of X chromosome drive: stability and extinction. Genetics 160, 1721–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Markow T, O'Grady P. 2006. Drosophila: a guide to species identification and use. Amsterdam, The Netherlands: Academic Press [Google Scholar]
- 31.Shorrocks B. 1972. Invertebrate types: Drosophila. London, UK: Ginn & Co [Google Scholar]
- 32.Price TAR, Lewis Z, Smith DT, Hurst GDD, Wedell N. 2011. Remating in the laboratory reflects rates of polyandry in the wild. Anim. Behav. 82, 1381–1386 (doi:10.1016/j.anbehav.2011.09.022) [Google Scholar]
- 33.Noor M, Schug M, Aquadro C. 2000. Microsatellite variation in populations of Drosophila pseudoobscura and Drosophila persimilis. Genet. Res. Camb. 75, 25–35 (doi:10.1017/S0016672399004024) [DOI] [PubMed] [Google Scholar]
- 34.Bretman A, Tregenza T. 2005. Measuring polyandry in wild populations: a case study using promiscuous crickets. Mol. Ecol. 14, 2169–2179 (doi:10.1111/j.1365-294X.2005.02556.x) [DOI] [PubMed] [Google Scholar]
- 35.Harshman L, Clark A. 1998. Inference of sperm competition from broods of field-caught Drosophila. Evolution 52, 1334–1341 (doi:10.2307/2411303) [DOI] [PubMed] [Google Scholar]
- 36.Jones A. 2001. GERUD1.0: a computer program for the reconstruction of parental genotypes from progeny arrays using multilocus DNA data. Mol. Ecol. Notes 1, 215–218 (doi:10.1046/j.1471-8278.2001.00062.x) [Google Scholar]
- 37.Barron AB. 2000. Anaesthetising Drosophila for behavioural studies. J. Insect Physiol. 46, 439–442 (doi:10.1016/S0022-1910(99)00129-8) [DOI] [PubMed] [Google Scholar]
- 38.Grafen A, Hails R. 2002. Modern statistics for the life sciences. Oxford, UK: Oxford University Press [Google Scholar]
- 39.Ihaka R, Gentleman R. 1996. R: a language for data analysis and graphics. J. Comp. Graph. Stat. 5, 299–314 [Google Scholar]
- 40.Schaeffer S, Miller E. 1992. Estimates of gene flow in Drosophila pseudoobscura determined from nucleotide sequence analysis of the alcohol dehydrogenase region. Genetics 132, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beckenbach AT. 1991. Sex ratio polymorphism in Drosophila pseudoobscura. Am. Nat. 137, 340–343 (doi:10.1086/285166) [Google Scholar]
- 42.Beckenbach AT. 1996. Selection and the ‘sex-ratio’ polymorphism in natural populations of Drosophila pseudoobscura. Evolution 50, 787–794 (doi:10.2307/2410851) [DOI] [PubMed] [Google Scholar]
- 43.Curtsinger JW, Feldman MW. 1980. Experimental and theoretical analyses of the ‘sex ratio’ polymorphism in Drosophila pseudoobscura. Genetics 94, 445–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price TAR, Hodgson DJ, Lewis Z, Hurst GDD, Wedell N. 2008. Selfish genetic elements promote polyandry in a fly. Science 322, 1241–1243 (doi:10.1126/science.1163766) [DOI] [PubMed] [Google Scholar]
- 45.Lorch PD, Chao L. 2003. Selection for multiple mating in females due to mates that reduce female fitness. Behav. Ecol. 14, 679–686 (doi:10.1093/beheco/arg045) [Google Scholar]
- 46.Turner M. 1986. Multiple mating, sperm competition and the fertility component of fitness in Drosophila pseudoobscura. Florida Entomologist 69, 121–128 (doi:10.2307/3494750) [Google Scholar]
- 47.Price TAR, Lewis Z, Smith DT, Hurst GDD, Wedell N. 2012. No evidence of mate discrimination against males carrying a sex ratio distorter in Drosophila pseudoobscura. Behav. Ecol. Sociobiol. 66, 561–568 (doi:10.1007/s00265-011-1304-1) [Google Scholar]
- 48.Wu C-I. 1983. Virility deficiency and the sex-ratio trait in Drososphila pseudoobscura. I. Sperm displacement and sexual selection. Genetics 105, 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carvalho AB, Vaz SC. 1999. Are Drosophila SR drive chromosomes always balanced? Heredity 83, 221–228 (doi:10.1038/sj.hdy.6886100) [DOI] [PubMed] [Google Scholar]
- 50.Pinzone C, Dyer K. 2014. Association of polyandry and sex-ratio drive prevalence in natural populations of Drosophila neotestacea. Proc. R. Soc. B 280, 20131397 (doi:10.1098/rspb.2013.1397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dyer K, Charlesworth B, Jaenike J. 2007. Chromosome-wide linkage disequilibrium as a consequence of meiotic drive. Proc. Natl Acad. Sci. USA 104, 1587–1592 (doi:10.1073/pnas.0605578104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manser A, Lindholm A, König B, Bagheri H. 2011. Polyandry and the decrease of a selfish genetic element in a wild house mouse population. Evolution 65, 2435–2447 (doi:10.1111/j.1558-5646.2011.01336.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wallace B. 1948. Studies on ‘sex-ratio’ in Drosophila pseudoobscura. I. Selection and ‘sex-ratio’. Evolution 2, 189–217 (doi:10.2307/2405380) [PubMed] [Google Scholar]
- 54.Silver LM. 1993. The peculiar journey of a selfish chromosome: mouse t haplotypes and meiotic drive. Trends Genet. 9, 250–254 (doi:10.1016/0168-9525(93)90090-5) [DOI] [PubMed] [Google Scholar]
- 55.Carroll L, Meagher S, Morrison L, Penn D, Potts W. 2004. Fitness effects of a selfish gene (the mus t complex) are revealed in an ecological context. Evolution 58, 1318–1328 [DOI] [PubMed] [Google Scholar]
- 56.Solé E, Balanyà J, Sperlich D, Serra L. 2002. Long-term changes in the chromosomal inversion polymorphism of Drosophila subobscura. I. Mediterranean populations from southwestern Europe. Evolution 56, 830–835 [DOI] [PubMed] [Google Scholar]
- 57.Toju H, Fukatsu T. 2011. Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: relevance of local climate and host plants. Mol. Ecol. 20, 853–868 (doi:10.1111/j.1365-294X.2010.04980.x) [DOI] [PubMed] [Google Scholar]
- 58.Dyer K. 2012. Local selection underlies the geographic distribution of sex-ratio drive in Drosophila neotestacea. Evolution 66, 973–984 (doi:10.1111/j.1558-5646.2011.01497) [DOI] [PubMed] [Google Scholar]
- 59.Price T, Lewis Z, Smith D, Hurst G, Wedell N. 2010. Sex ratio drive promotes sexual conflict and sexual coevolution in the fly Drosophila pseudoobscura. Evolution 64, 1504–1509 [DOI] [PubMed] [Google Scholar]
- 60.Engelstädter J. 2010. The effective size of populations infected with cytoplasmic sex-ratio distorters. Genetics 186, 309–320 (doi:10.1534/genetics.110.120014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jenouvrier S, Caswell H, Barbraud C, Weimerskirch H. 2010. Mating behaviour, population growth, and the operational sex ratio: a periodic two-sex model approach. Am. Nat. 175, 739–752 (doi:10.1086/652436) [DOI] [PubMed] [Google Scholar]
- 62.Charlat S, Reuter M, Dyson E, Hornett E, Duplouy A, Davies N, Roderick G, Wedell N, Hurst GDD. 2007. Male-killing bacteria trigger a cycle of increasing male fatigue and female promiscuity. Curr. Biol. 17, 273–277 (doi:10.1016/j.cub.2006.11.068) [DOI] [PubMed] [Google Scholar]
- 63.Bryant SH, Beckenbach AT, Cobbs G. 1982. Sex ratio, sex composition, and relative abundance in Drosophila pseudoobscura. Evolution 36, 27–34 (doi:10.2307/2407963) [DOI] [PubMed] [Google Scholar]
- 64.Schoener T. 2011. The newest synthesis: understanding the interplay of evolutionary and ecological dynamics. Science 331, 426–429 (doi:10.1126/science.1193954) [DOI] [PubMed] [Google Scholar]
- 65.James A, Jaenike J. 1990. ‘Sex ratio’ meiotic drive in Drosophila testacea. Genetics 126, 651–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No large datasets are involved in this work. However, isolines and material will be made available on request.


