Abstract
Post-copulatory sexual selection in the form of sperm competition is known to influence the evolution of male reproductive proteins in mammals. The relationship between sperm competition and regulatory evolution, however, remains to be explored. Protamines and transition nuclear proteins are involved in the condensation of sperm chromatin and are expected to affect the shape of the sperm head. A hydrodynamically efficient head allows for fast swimming velocity and, therefore, more competitive sperm. Previous comparative studies in rodents have documented a significant association between the level of sperm competition (as measured by relative testes mass) and DNA sequence evolution in both the coding and promoter sequences of protamine 2. Here, we investigate the influence of sexual selection on protamine and transition nuclear protein mRNA expression in the testes of eight mouse species that differ widely in levels of sperm competition. We also examined the relationship between relative gene expression levels and sperm head shape, assessed using geometric morphometrics. We found that species with higher levels of sperm competition express less protamine 2 in relation to protamine 1 and transition nuclear proteins. Moreover, there was a significant association between relative protamine 2 expression and sperm head shape. Reduction in the relative abundance of protamine 2 may increase the competitive ability of sperm in mice, possibly by affecting sperm head shape. Changes in gene regulatory sequences thus seem to be the basis of the evolutionary response to sexual selection in these proteins.
Keywords: sexual selection, sperm competition, protamine, transition nuclear protein, sperm
1. Introduction
When females mate promiscuously, the sperm of rival males compete for the fertilization of available ova [1]. Post-copulatory sexual selection mediated by sperm competition has a profound influence on male reproductive traits across a wide range of taxa (reviewed in [2–4]). In mammals, key traits affected by sperm competition include sperm quality parameters [5], processes that prepare sperm to interact with the oocyte [6], sperm design (e.g. overall size, head shape and dimensions [7–10]) and sperm swimming velocity [10–12]. Several lines of evidence suggest that sperm head shape is particularly important in competitive situations. For example, the size and curvature of the apical hook of rodent sperm heads is thought to be associated with levels of sperm competition ([9], but see [13]). Likewise, head shape may affect the hydrodynamic efficiency of spermatozoa. Head elongation, which may reduce drag, associates with faster sperm swimming velocity [10]. Faster sperm are more likely to succeed in fertilization [14].
To date, most work on the molecular evolution of male reproductive genes has focused on protein-coding regions [15,16]. A number of studies have found a positive relationship between sequence divergence of these genes and levels of sperm competition, and several such genes show evidence of positive selection in coding regions ([17–21], but see [22,23]). However, a positive correlation between sequence divergence in the promoter region of protamine 2 and relative levels of sperm competition in house mice and their close relatives [24] suggests that regulatory changes may also contribute to species differences in sperm competitive ability. Surprisingly, despite order of magnitude differences in the absolute and relative expression levels of protamines and associated transition nuclear proteins across eutherian mammals [25], the relationship between sperm competition and gene expression remains largely unexplored.
Protamines and transition nuclear proteins are integral to chromatin remodelling and condensation during the final stages of spermatogenesis. This nuclear reshaping in postmeiotic spermatids affects the overall shape of the sperm head which, in turn, may influence hydrodynamic efficiency, resulting in an increase in sperm swimming speed and more competitive sperm. Notably, sperm from transition nuclear protein-deficient mice perform poorly in some competitive assays [26]. Whereas protamines (PRM1 and PRM2 in most eutherian mammals) bind directly to DNA in the nucleus of elongating spermatids and mature spermatozoa [27], transition nuclear proteins (TNP1 and TNP2) are involved in intermediate stages in the replacement of histones by protamines [28,29]. Protamines remain associated with sperm chromatin in the oocyte and influence the rate of nuclear decondensation, a trait associated with embryonic survival [30–33].
Protamine and transition protein mRNAs are highly co-expressed in round spermatids [34–37], and the protein products of both gene families exhibit significant overlap in elongating spermatid nuclei [28,38]. TNP1 and TNP2 seem to perform partially redundant functions: only double TNP1/TNP2 mouse knockouts are completely sterile [28]. However, deletion of either transition protein results in incomplete PRM2 processing and defective chromatin condensation [29,39]. This, together with the co-localization of mRNAs and mature proteins, strongly suggests that functional interactions between protamines and transition proteins are necessary for normal sperm development.
In mice and humans, both PRM1 and PRM2 are essential for male fertility [40]. Strikingly, although the relative abundance of PRM1 and PRM2 proteins differs widely across mammals (from 0 to 77% PRM2) [41], disruption of species-specific protamine ratios causes fertility defects comparable with gene knockouts [40,42]. In human males, for example, protamine imbalance can result in reduced sperm concentration and motility, and in abnormal head morphology, an indicator of deficits in chromatin condensation [43–45]. In particular, incomplete processing of the PRM2 precursor is associated with sperm dysfunction [45,46], and PRM2-deficient sperm are characterized by incomplete nuclear condensation and increased DNA damage [40,46,47], defects that can lead to embryonic mortality [31]. Thus, protamine ratios play a large role in sperm head morphology, a phenotype important for competitive ability both before and during fertilization. This suggests that sexual selection mediated by sperm competition should act on protamine ratios, resulting in an association between species differences in levels of sperm competition and protamine expression.
Here, we investigate the influence of sexual selection on protamine and transition nuclear protein mRNA expression in the testes of eight closely related species in the genus Mus. These species exhibit a wide range of relative testes mass, a robust proxy for different levels of sperm competition [2,4], and differ in sperm traits associated with competitive ability [5,7,24,48]. Moreover, evolution of the Prm2 promoter in seven of the same species is consistent with stronger selection in taxa with higher inferred levels of sperm competition [24]. This provides specific motivation for studying the relationship between protamine expression and sperm competition in Mus. Given the functional relationship between protamines and transition proteins, and the role of transition proteins in PRM2 processing, we expected that transition nuclear protein expression should covary with species differences in protamine expression. Because protamines and transition nuclear proteins are involved in the condensation of sperm chromatin and are expected to affect the shape of the sperm head, we also assessed the relationship between gene expression and sperm head shape.
2. Material and methods
(a). Species
This study included eight species in the genus Mus: M. caroli, M. castaneus, M. domesticus, M. macedonicus, M. musculus, M. pahari, M. spicilegus and M. spretus (four to five males per species). This group of species shows diverse levels of sperm competition, as inferred from their differences in relative testes mass (table 1). Large testes in relation to body mass (relative testes mass) is a strong predictor of high sperm competition levels in many taxa (reviewed in [2,4,50]), and relative testes mass is correlated with genetic paternity (i.e. percentages of multiple paternity) in mammals in general [51], and rodents in particular [52]. Therefore, relative testes mass is used in this study as a robust proxy for sperm competition levels.
Table 1.
Relative testes mass was calculated as described by Kenagy & Trombulak [49] followed by a calculation of the median for the species. Gene expression data are normalized, transformed median values. Species were ordered by relative testes mass (ascending).
| species | relative testes mass | Prm1 (ΔCT) | Prm2 (ΔCT) | Tnp1 (ΔCT) | Tnp2 (ΔCT) | Tnp1/Tnp2 | Prm1/Prm2 | Prm/Tnp | Prm2/Tnp | Prm2/Prm | Prm2/(Prm + Tnp) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mus castaneus | 0.27 | 3.16 | 4.29 | 2.96 | 3.38 | 0.83 | 0.67 | 1.24 | 0.75 | 0.60 | 0.33 |
| Mus pahari | 0.27 | 3.80 | 3.69 | 3.01 | 2.26 | 1.13 | 1.01 | 1.38 | 0.68 | 0.50 | 0.29 |
| Mus domesticus | 0.32 | 2.11 | 3.22 | 1.88 | 2.32 | 0.81 | 0.65 | 1.27 | 0.77 | 0.61 | 0.34 |
| Mus musculus | 0.44 | 2.87 | 3.72 | 3.27 | 2.98 | 1.14 | 0.76 | 1.06 | 0.61 | 0.57 | 0.29 |
| Mus caroli | 0.46 | 5.83 | 7.28 | 6.71 | 6.18 | 1.07 | 0.78 | 1.00 | 0.56 | 0.56 | 0.28 |
| Mus spretus | 0.87 | 1.90 | 2.31 | 2.32 | 1.70 | 1.38 | 0.82 | 1.05 | 0.58 | 0.55 | 0.28 |
| Mus macedonicus | 0.95 | 3.54 | 3.49 | 2.48 | 2.14 | 1.05 | 0.99 | 1.49 | 0.74 | 0.50 | 0.30 |
| Mus spicilegus | 1.51 | 4.61 | 4.60 | 4.76 | 3.92 | 1.16 | 0.98 | 1.05 | 0.53 | 0.50 | 0.26 |
| CV | 0.69 | 0.37 | 0.36 | 0.46 | 0.46 | 0.18 | 0.17 | 0.15 | 0.14 | 0.08 | 0.09 |
Individuals were purchased from the Institut des Sciences de l'Evolution-Montpellier, CNRS-Universite de Montpellier II. Males were kept in our animal facilities in individual cages under standard laboratory conditions in environmentally controlled rooms (20–24°C) on a 14 L : 10 D photoperiod and were provided with food and water ad libitum. All animal handling was done following Spanish Animal Protection Regulation RD1201/2005, which conforms to European Union Regulation 2003/65.
(b). Testes collection and relative testes mass
Animals were sacrificed at an age of two to four months by cervical dislocation and were immediately weighed and dissected. Testes were removed, weighed, flash-frozen in liquid nitrogen and stored at −80°C. All dissection instruments and areas were cleaned with RNase AWAY (Molecular BioProducts, Thermo Fisher Scientific, San Diego, CA, USA) before use. Relative testes mass was calculated based on the rodent power function, following the method in Kenagy & Trombulak [49].
(c). RNA extraction and cDNA synthesis
RNA was extracted in a sterile vertical laminar flow hood using either the RNeasy Plus kit (Qiagen) or the E.Z.N.A Total RNA kit I (Omega, Madrid, Spain) following the manufacturer's recommendations. All instruments and surface areas were cleaned with RNase AWAY. RNA concentration and purity were determined using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Madrid, Spain), and cDNA was synthesized the same day from 10 μg of RNA, using the Superscript III First Strand Synthesis Kit with oligo(dT) (Invitrogen, Barcelona, Spain) according to the manufacturer's recommendations. cDNA concentration and purity were determined using a NanoDrop1000 spectrophotometer, and samples were stored at −20°C.
(d). Quantitative PCR
Expression levels for M. musculus, M. spretus, M. spicilegus and M. pahari were determined at the University of Arizona in Tucson using a MyiQ2 light cycler (Bio-Rad), and expression levels for M. domesticus, M. castaneus, M. macedonicus and M. caroli were determined at the Museo Nacional de Ciencias Naturales in Madrid using a CFX96 Real Time System/C1000 Thermal Cycler (Bio-Rad). To check the consistency of results obtained using different cyclers, assays for the standard gene (see below) were run by the same person (L.L.) with a set of testes samples taken from the same individuals used in both Tucson and Madrid, using exactly the same protocol. Results were consistent across locations (e.g. M. musculus individual 1 (Tucson, right testis): average CT (±s.d.) = 12.94 (0.02); M. musculus individual 1 (Madrid, left testis): average CT (±s.d.) = 12.89 (0.07)).
Primers were designed in Primer3 (v. 0.4.0) to amplify a product between 70 and 150 bases across an exon–exon junction. Protamine primers were placed in sequences that are invariant across all species in this analysis. Transition protein primers were placed in sequences that are conserved between Mus and Rattus, and therefore are unlikely to vary among closely related Mus species. Primer sequences and amplicon sizes are provided in the electronic supplementary material, table S1. Each quantitative PCR (qPCR) run included one individual of each species with three technical replicates for the four experimental genes (Prm1, Prm2, Tnp1 and Tnp2) and two technical replicates for the standard gene (18SrRNA). qPCR reactions were run in 96-well plates with an end volume of 16 μl per sample containing 8 μl SYBR green Master Mix (Invitrogen), 15 ng of each primer and 50 ng μl−1 of cDNA. The conditions of the thermocycler program consisted of an initial denaturation of 95°C for 10 min, 40 cycles of 95°C for 15 s and an annealing and elongation stage of 62°C for 1 min. Melt curve analysis was performed at the end of each run to check for multiple peaks, indicative of non-specific amplification.
(e). Analysis of expression data
Cycle threshold data (CT) were normalized relative to 18SrRNA for each plate (ΔCT). To avoid statistical analysis using a dataset of mixed negative and positive values, data were transformed by adding a constant based on the lowest ΔCT value. Expression ratios and percentages were calculated from transformed individual ΔCT values (M. domesticus n = 4, all other species n = 5), and median values were obtained for each species. Because of the expectation that relative expression levels may be of greater functional significance than absolute expression levels (see above), we calculated ratios (Prm1/Prm2, Tnp1/Tnp2, Prm/Tnp, Prm2/Tnp) and proportions (Prm2/Prm, Prm2/(Prm + Tnp), Prm1/(Prm + Tnp)), where Prm refers to the combined expression of Prm1 and Prm2, and Tnp refers to the combined expression of Tnp1 and Tnp2. To obtain a measure of variability between individuals and species, as well as for individual genes, the coefficient of variation (CV = s.d./mean) was calculated.
(f). Phylogenetic generalized least-squares analysis
Species data may not be free of phylogenetic association because shared character values may result from common ancestry rather than independent evolution, and thus may not be truly independent. To control for this phylogenetic inertia, we used phylogenetic generalized least-squares (PGLS) analyses [53] to test for relationships between species differences in total and relative protamine and transition protein expression, and relative testes mass. PGLS analysis was implemented in COMPARE 4.6b [54], using a phylogenetic tree based on Lundrigan et al. [55] and Gómez Montoto et al. [5] (electronic supplementary material, figure S1).
(g). Geometric morphometrics analysis of sperm head shape
Geometric morphometrics methods were used to quantify head shape variation based on a set of landmarks that correspond to the spatial position of particular anatomical traits [56,57]. A total of 20 bidimensional landmark coordinates were gathered from spermatozoa of seven of the eight species used in the gene expression analysis (M. caroli, M. castaneus, M. domesticus, M. macedonicus, M. musculus, M. spicilegus, M. spretus; n = 5 males/species). Landmark data were processed as described previously [58]. All morphometric analyses were conducted with MorphoJ [59]. An independent contrast for morphometric shape data [60] was conducted to check for phylogenetic signal in the sperm head shape dataset. This test simulates the null hypothesis of total absence of phylogenetic signal by a permutation procedure. The p-value was not significant (p = 0.102) for the null hypothesis of independence, which indicates a lack of phylogenetic signal and, therefore, that phylogenetic correction was not needed for this analysis.
Canonical variate analysis (CVA) [61] was used to explore the relationship between sperm head shape and relative protamine expression. Species were grouped into three categories based on well-defined differences in relative protamine expression: low, intermediate and high expression ratios (table 1; electronic supplementary material, table S2; see Results section for details). The CVA produces a set of canonical variates that are uncorrelated within and among groups and account for the maximum amount of among-group variance relative to within-group variance. As a result of the CVA, distances in the original space are transformed to Procrustes distances. These Procrustes distances for between-category comparisons were used to test for significant differences in sperm head shapes between species with low, intermediate and high protamine expression ratios.
3. Results
(a). Expression of protamines and transition nuclear proteins
Median expression levels for each gene and species are shown in table 1. The ranges of expression medians and the CV for each gene and species are provided in the electronic supplementary material, table S2. Within species, expression levels were positively correlated in all pairwise comparisons among genes (electronic supplementary material, figure S2 and table S3), suggesting that there may be functional constraints to maintain consistent relative expression levels of these genes and/or common regulatory control. The median expression level for individual genes varied by a factor of approximately threefold among species (electronic supplementary material, table S2). Tnp1 was expressed at a slightly higher level than Tnp2 although both showed the same CV. Likewise, Prm2 was expressed at a slightly higher level than Prm1 but there was no difference in CV (table 1; electronic supplementary material, table S2).
The ratios and proportions of expression levels for different genes are shown in the electronic supplementary material, table S4. These relative levels of expression were much more constant among species (electronic supplementary material, table S4) than expression levels of individual genes (cf. electronic supplementary material, table S2). The ratio of total protamines to total transition nuclear proteins was close to one in half the species and above one in the other four species, revealing higher overall expression levels of protamines. Ratios between Tnp1 and Tnp2 were generally above one, in agreement with higher expression levels of Tnp1 in comparison to Tnp2 (see above). The reverse was true for protamines, with ratios of Prm1/Prm2 below one (electronic supplementary material, table S4).
(b). Relationships between relative testes mass and gene expression
We tested for associations between relative testes mass and patterns of protamine and transition protein expression, both for individual genes and for ratios of expression levels among genes.
The correlation between relative testes mass and Prm1/Prm2 or Prm2/Prm was not significant when all eight species were considered (figure 1a and table 2). However, we noted that M. pahari appears to be an outlier in this analysis. Mus pahari is basal to the other species included in this study and belongs to a different subgenus (Coelomys) [62]. When the analysis was restricted to the seven species in the subgenus Mus, there was a significant positive relationship between relative testes mass and Prm1/Prm2 (α = 15.5, CI 95% (slope) = 1.67–4.25, correlation = 0.89; figure 1a and table 2) and a significant negative relationship between relative testes mass and Prm2/Prm (α = 15.5, CI 95% (slope) = −0.14 to −0.06, correlation = 0.80; table 2). By contrast, there was no relationship between testes mass and transition protein ratios (data not shown).
Figure 1.
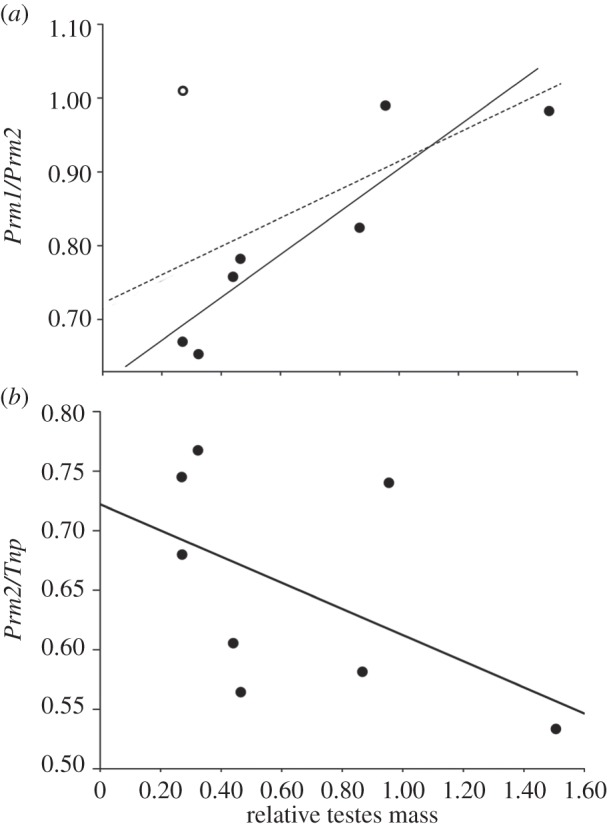
Relationships between relative testes mass and relative protamine 2 expression. (a) Protamine ratio (Prm1/Prm2): the dashed line corresponds to analyses with N = 8 mouse species, and the correlation is not significant. The open circle identifies M. pahari, a species that behaves as an outlier in these analyses. The solid line corresponds to analyses with n = 7 species in which M. pahari is not included, and this correlation is statistically significant. (b) Ratio of Prm2 to total Tnp (Prm2/Tnp). Results of statistical analyses are given in table 2.
Table 2.
Relationship between relative testes mass and relative protamine or transition protein expression. Analyses were carried out with all species and excluding Mus pahari (see text). CI− and CI+ indicate the confidence intervals for the regression slope, lnL = log likelihood estimate of alpha, alpha = measure of evolutionary constraints acting on phenotypes, corr = the correlation value (r). Bold CI values indicate statistical significance.
| excluding Mus pahari (n = 7) |
relationships for all species (n = 8) |
|||||
|---|---|---|---|---|---|---|
| Prm1/Prm2 | Prm2/Prm | Prm2/Tnp | Prm2/(Prm + Tnp) | Prm1/Prm2 | Prm2/Prm | |
| CI− | 1.67 | −0.14 | −4.64 | −0.19 | −0.47 | −0.12 |
| CI+ | 4.25 | −0.06 | −0.03 | −0.02 | 3.68 | 0.01 |
| lnL | 8.15 | 8.14 | 5.20 | 6.05 | 4.52 | 4.63 |
| alpha | 15.50 | 15.50 | 1.56 | 1.66 | 5.62 | 5.37 |
| corr | 0.89 | −0.80 | −0.63 | −0.72 | 0.53 | −0.54 |
Significant negative associations with relative testes mass were found for Prm2/Tnp (α = 1.56, CI 95% (slope) = −4.64 to −0.03, correlation = −0.63; figure 1a and table 2) and Prm2/(Prm + Tnp) (α = 6.05, CI 95% (slope) = −0.19 to −0.02, correlation = −0.72; table 2). By contrast, there was no association between relative testes mass and Prm1/Tnp, Prm1/(Prm + Tnp) or Prm/Tnp (data not shown), or between relative testes mass and any of the four genes when analysed separately (electronic supplementary material, table S5). Thus, significant relationships between testes mass and the expression of sperm condensation proteins are driven mainly by the relative expression of Prm2.
Together, these results indicate that species with higher inferred levels of sperm competition express proportionately less Prm2 in relation to total transition protein and in relation to total protamine and transition protein combined. Within the subgenus Mus, species with higher sperm competition have a higher Prm1/Prm2 expression ratio and therefore a lower Prm2/Prm proportion.
(c). Relationships between protamine expression and sperm head shape
Geometric morphometrics was employed to quantify differences in head shape between the seven species in the subgenus Mus. Species were categorized as having high, intermediate or low protamine expression ratios, and Procrustes distances (D) calculated from CVA were used to test for between-category differences in sperm head shape.
Sperm head shapes were significantly different between species with high, intermediate and low Prm1/Prm2 ratios (high versus intermediate: D = 0.08, p = 0.0002; high versus low: D = 0.1, p = 0.0001; intermediate versus low: D = 0.05, p = 0.05; figure 2). The same between-category differences in sperm head shape were obtained for Prm2/Prm ratio (high versus intermediate: D = 0.05, p = 0.0001; high versus low: D = 0.1, p = 0.0001; intermediate versus low: D = 0.08, p = 0.0001). These results support the idea that sperm head shape is influenced by relative protamine expression.
Figure 2.
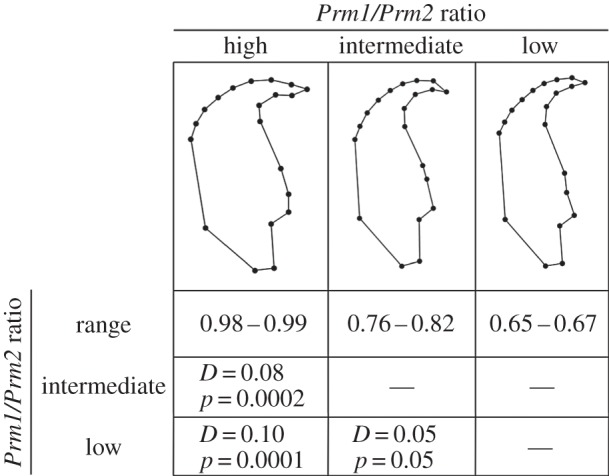
Procrustes distances (D) and p-values for canonical variate analyses examining head shape in relation to Prm1/Prm2 ratio. Three groups of species were defined according to their ratios of protamine expression: high (M. macedonicus and M. spicilegus), intermediate (M. musculus, M. caroli and M. spretus) and low (M. castaneus and M. domesticus) (see table 1). Morphometric data were taken from 35 individuals of seven species. Procrustes distances different from zero indicate shape differences between groups. Wireframe graphics show the shape associated with each group categorized according to its Prm1/Prm2 ratio.
4. Discussion
Despite the long-standing debate over the relative contribution of coding versus regulatory changes to adaptive evolution [63–65], mounting empirical evidence demonstrates that regulatory evolution can play a major role in adaptive divergence, particularly between closely related lineages [65–71]. In this study, we compared protamine and transition nuclear protein mRNA expression in the testes of eight species in the genus Mus that share recent common ancestry but differ widely in inferred levels of sperm competition. We found that species that experience higher levels of sperm competition express less protamine 2 in relation to both transition nuclear proteins and to protamine 1. This strongly suggests that species differences in relative expression levels of these key spermiogenesis genes are influenced by variation in the strength of post-copulatory sexual selection. The fact that this pattern is driven by the relative expression of protamine 2 is consistent with evidence that the promoter region of this gene is evolving under sexual selection in Mus [24]. Importantly, we found that species that differ in ratios of protamine 2 expression, both in relation to protamine 1 and in relation to total protamines, also differ in sperm head shape. This suggests that regulatory changes contribute to modifications of sperm phenotype that could, ultimately, influence sperm's competitive ability. Taken together, the results of this study support the proposition that selection on regulatory regions can fine-tune adaptive phenotypes on short evolutionary timescales [72]. We discuss these results in relation to previous work on the evolution of sperm chromatin condensation genes in mammals and the genetics and functional consequences of sperm competition in rodents.
(a). Protamines and sperm competition: evolution at two levels
Sperm chromatin condensation genes, including protamines, are thought to be among the fastest evolving male reproductive proteins in eutherian mammals [73,74]. There is ample evidence from primates and rodents that selection contributes to this rapid rate of change [16,21,75,76] and sperm competition is often invoked as the driving force [15]. However, how particular substitutions might enhance sperm competitiveness remains untested, and it has been suggested that selection for protein stability is an equally parsimonious explanation for protamine-coding sequence evolution in primates [77]. Notably, in case–control studies of human males, associations between infertility and coding region SNPs in either Prm1 or Prm2 are rare [78–80], whereas men with imbalanced PRM1/PRM2 ratios are consistently subfertile or sterile (reviewed in [81]). Thus, while the functional consequences of protamine-coding sequence substitutions are largely unknown, changes in protamine expression have a demonstrated impact on male fertility, and therefore might covary with the strength of post-copulatory sexual selection across species.
In the Mus clade comprising house mice and their close relatives, there is evidence for weak positive selection on Prm2-coding sequence in the three species with the highest inferred levels of sperm competition (M. spicilegus, M. spretus and M. macedonicus), whereas divergence in the promoter region is positively correlated with relative testes mass, and with sperm swimming speed, across the entire clade [24]. Here, using a subset of the same species, we show that the relative abundance of Prm2 mRNA in the testes is negatively correlated with relative testes mass. These findings suggest that nucleotide substitutions in the Prm2 promoter region influence expression and that high levels of sperm competition act to decrease the relative abundance of Prm2 in the testes.
We emphasize, however, that our understanding of the relationship between protamine 2 regulation and sperm competition in Mus is far from complete. First, the functional relationship between promoter evolution and expression is not straightforward: species with higher Prm2 promoter divergence express less Prm2 only in relation to transition nuclear proteins and Prm1. Despite substantial interspecific differences in the expression levels of all four genes, there was no relationship between relative testes mass and individual gene expression. Likewise, although the Prm1 promoter region is highly variable in Mus, there is no relationship between divergence and levels of sperm competition [24]. A plausible explanation for these patterns is that sexual selection for reduced PRM2 is counterbalanced by natural selection to maintain the relative proportions of protamines and transition nuclear proteins within a functional range. Potential mechanisms include compensatory evolution in the promoter regions of interacting sperm chromatin condensation proteins or a single regulatory modifier shared among genes. In mice, as in humans, Prm1, Prm2 and Tnp2 are tightly clustered in the genome. Thus, an enhancer element common to all three genes is a formal possibility. Comparative analysis of intergenic regions in the Prm1/Prm2/Tnp2 cluster, together with the Tnp1 and Tnp2 promoter regions, will help to discriminate these non-mutually exclusive alternatives.
Second, the correlation between mRNA expression levels and protein abundance is often imperfect [82]. Quantification of sperm chromatin condensation proteins in mature spermatozoa will provide a direct measure of species differences in their relative abundance. Finally, evidence for selection on the Prm2-coding sequence in M. spicilegus, M. spretus and M. macedonicus is intriguing, because it suggests that high levels of sperm competition can drive coding and regulatory evolution in tandem [24]. However, whether positively selected Prm2 amino acid substitutions in these species affect sperm phenotypes related to competitive ability remains to be determined.
(b). The relative abundance of protamine 2: functional implications for sperm phenotypes
Why should high levels of sperm competition favour reduction in the relative abundance of PRM2? While the phenotypic effects of interspecific differences in protamine ratios are largely unstudied, there is some evidence that sperm from species that either lack PRM2, or produce very little PRM2 relative to PRM1, exhibit slower DNA decondensation in the oocyte [32,41]. Sperm with more compact heads may have higher competitive ability [10], and sperm with incomplete DNA compaction often have over-sized or less streamlined heads [34]. Thus, it is plausible that high levels of sperm competition select for higher DNA compaction, and thus proportionately less PRM2. Evaluation of this hypothesis will require comparative analyses of sperm chromatin compaction in relation to head morphology, the proportion of PRM2 and the strength of sexual selection mediated by sperm competition. Notably, the finding that relative abundance of Prm2 is associated with differences in sperm head shape is an important first step towards revealing the functional relationship between protamine expression and sperm head morphology. Future studies will investigate the hydrodynamic consequences of these Prm2-associated differences in sperm head shape.
5. Conclusion
An important role of comparative studies such as this is to identify patterns that generate testable hypotheses [83]. Here, we show that species of mice with higher inferred levels of sperm competition express less protamine 2 in relation to protamine 1 and transition nuclear proteins. Based on this pattern, together with evidence for sexually selected divergence in the promoter region of protamine 2 [24], we propose that reduction in the relative abundance of protamine 2 enhances sperm competitive ability in mice by influencing sperm head shape and that regulatory evolution plays a key role in this evolutionarily rapid response to selection.
Acknowledgements
We thank François Bonhomme and Annie Orth for facilitating access to the animals used in this study.
Funding statement
L.L. holds a studentship from the Spanish Research Council (JAEpre-CSIC) co-financed by the European Social Fund (ESF). This work was supported by the Spanish Ministry of Economy and Competitiveness (grant no. CGL2011-26341) and was developed in part during a short stay at the University of Arizona with funding from CSIC (‘Estancia Breve’). This work was also financially supported by NSF and NIH grants to M.W.N.
References
- 1.Parker GA. 1970. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. 45, 525–567 (doi:10.1111/j.1469-185X.1970.tb01176.x) [Google Scholar]
- 2.Birkhead TR, Møller AP. 1998. Sperm competition and sexual selection. London, UK: Academic Press [Google Scholar]
- 3.Simmons LW. 2001. Sperm competition and its evolutionary consequences in the insects. Princeton, NJ: Princeton University Press [Google Scholar]
- 4.Birkhead TR, Hosken DJ, Pitnick S. 2009. Sperm biology: an evolutionary perspective. Oxford, UK: Academic Press [Google Scholar]
- 5.Gómez Montoto L, Varea Sánchez M, Tourmente M, Martín-Coello J, Luque-Larena JJ, Gomendio M, Roldan ERS. 2011. Sperm competition differentially affects swimming velocity and size of spermatozoa from closely related muroid rodents: head first. Reproduction 142, 819–830 (doi:10.1530/REP-11-0232) [DOI] [PubMed] [Google Scholar]
- 6.Gomendio M, Martin-Coello J, Crespo C, Magaña C, Roldan ERS. 2006. Sperm competition enhances functional capacity of mammalian spermatozoa. Proc. Natl Acad. Sci. USA 103, 15 113–15 117 (doi:10.1073/pnas.0605795103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roldan ERS, Gomendio M, Vitullo AD. 1992. The evolution of eutherian spermatozoa and underlying selective forces: female selection and sperm competition. Biol. Rev. 67, 551–593 (doi:10.1111/j.1469-185X.1992.tb01193.x) [DOI] [PubMed] [Google Scholar]
- 8.Breed WG, Taylor J. 2000. Body mass, testes mass, and sperm size in murine rodents. J. Mammal. 81, 758–768 (doi:10.1644/1545-1542(2000)081<0758:BMTMAS>2.3.CO;2) [Google Scholar]
- 9.Immler S, Moore HD, Breed WG, Birkhead TR. 2007. By hook or by crook? Morphometry, competition and cooperation in rodent sperm. PLoS ONE 2, e170 (doi:10.1371/journal.pone.0000170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tourmente M, Gomendio M, Roldan ERS. 2011. Sperm competition and the evolution of sperm design in mammals. BMC Evol. Biol. 11, 12 (doi:10.1186/1471-2148-11-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomendio M, Roldan ERS. 1991. Sperm competition influences sperm size in mammals. Proc. R. Soc. Lond. B 243, 181–185 (doi:10.1098/rspb.1991.0029) [DOI] [PubMed] [Google Scholar]
- 12.Gomendio M, Roldan ERS. 2008. Implications of diversity in sperm size and function for sperm competition and fertility. Int. J. Dev. Biol. 52, 439–447 (doi:10.1387/ijdb.082595mg) [DOI] [PubMed] [Google Scholar]
- 13.Firman RC, Simmons LW. 2009. Experimental evolution of sperm quality via postcopulatory sexual selection in house mice. Evolution 64, 1245–1256 [DOI] [PubMed] [Google Scholar]
- 14.Cummins JM, Yanagimachi R. 1982. Sperm–egg ratios and the site of the acrosome reaction during in vivo fertilization in the hamster. Gamete Res. 5, 239–256 (doi:10.1002/mrd.1120050304) [Google Scholar]
- 15.Swanson WJ, Vacquier VD. 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144 (doi:10.1038/nrg733) [DOI] [PubMed] [Google Scholar]
- 16.Turner LM, Hoekstra HE. 2008. Causes and consequences of the evolution of reproductive proteins. Int. J. Dev. Biol. 52, 769–780 (doi:10.1387/ijdb.082577lt) [DOI] [PubMed] [Google Scholar]
- 17.Dorus S, Evans PD, Wyckoff GJ, Choi SS, Lahn BT. 2004. Rate of molecular evolution of the seminal protein gene SEMG2 correlates with levels of female promiscuity. Nat. Genet. 36, 1326–1329 (doi:10.1038/ng1471) [DOI] [PubMed] [Google Scholar]
- 18.Finn S, Civetta A. 2010. Sexual selection and the molecular evolution of ADAM proteins. J. Mol. Evol. 71, 231–240 (doi:10.1007/s00239-010-9382-7) [DOI] [PubMed] [Google Scholar]
- 19.Kingan SB, Tatar M, Rand DM. 2003. Reduced polymorphism in the chimpanzee semen coagulating protein, semenogelin I. J. Mol. Evol. 57, 159–69 (doi:10.1007/s00239-002-2463-0) [DOI] [PubMed] [Google Scholar]
- 20.Ramm SA, Oliver PL, Ponting CP, Stockley P, Emes RD. 2008. Sexual selection and the adaptive evolution of mammalian ejaculate proteins. Mol. Biol. Evol. 25, 207–219 (doi:10.1093/molbev/msm242) [DOI] [PubMed] [Google Scholar]
- 21.Wyckoff GJ, Wang W, Wu CI. 2000. Rapid evolution of male reproductive genes in the descent of man. Nature 403, 304–309 (doi:10.1038/35002070) [DOI] [PubMed] [Google Scholar]
- 22.Lüke L, Vicens A, Serra F, Luque-Larena JJ, Dopazo H, Roldan ERS, Gomendio M. 2011. Sexual selection halts the relaxation of protamine 2 among rodents. PLoS ONE 6, e29247 (doi:10.1371/journal.pone.0029247) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walters JR, Harrison RG. 2011. Decoupling of rapid and adaptive evolution among seminal fluid proteins in Heliconius butterflies with divergent mating systems. Evolution 65, 2855–2871 (doi:10.1111/j.1558-5646.2011.01351.x) [DOI] [PubMed] [Google Scholar]
- 24.Martin-Coello J, Dopazo H, Arbiza L, Ausió J, Roldan ERS, Gomendio M. 2009. Sexual selection drives weak positive selection in protamine genes and high promoter divergence, enhancing sperm competitiveness. Proc. R. Soc. B 276, 2427–2436 (doi:10.1098/rspb.2009.0257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kleene KC, Bagarova J. 2008. Comparative genomics reveals gene-specific and shared regulatory sequences in the spermatid-expressed mammalian Odf1, Prm1, Prm2, Tnp1 and Tnp2 genes. Genomics 92, 101–106 (doi:10.1016/j.ygeno.2008.05.001) [DOI] [PubMed] [Google Scholar]
- 26.Nayernia K, Drabent B, Adham IM, Moschner M, Wolf S, Meinhardt A, Engel W. 2003. Mice lacking three germ cell expressed genes are fertile. Biol. Reprod. 69, 1973–1978 (doi:10.1095/biolreprod.103.018564) [DOI] [PubMed] [Google Scholar]
- 27.Brewer LR. 1999. Protamine-induced condensation and decondensation of the same DNA molecule. Science 286, 120–123 (doi:10.1126/science.286.5437.120) [DOI] [PubMed] [Google Scholar]
- 28.Meistrich ML, Mohapatra B, Shirley CR, Zhao M. 2003. Roles of transition nuclear proteins in spermiogenesis. Chromosoma 111, 483–488 (doi:10.1007/s00412-002-0227-z) [DOI] [PubMed] [Google Scholar]
- 29.Yu YE, Zhang Y, Unni E, Shirley CR, Deng JM, Russell LD, Weil MM, Behringer RR, Meistrich ML. 2000. Abnormal spermatogenesis and reduced fertility in transition nuclear protein 1 deficient mice. Proc. Natl Acad. Sci. USA 97, 4683–4688 (doi:10.1073/pnas.97.9.4683) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aoki VW, Carrell DT. 2003. Human protamines and the developing spermatid: their structure, function, expression and relationship with male infertility. Asian J. Androl. 5, 315–324 [PubMed] [Google Scholar]
- 31.Cho C, Jung-Ha H, Willis WD, Goulding EH, Stein P, Xu Z, Schultz RM, Hecht NB, Eddy EM. 2003. Protamine-2 deficiency leads to sperm DNA damage and embryo death in mice. Biol. Reprod. 69, 211–217 (doi:10.1095/biolreprod.102.015115) [DOI] [PubMed] [Google Scholar]
- 32.Perreault SD, Barbee RR, Elstein KH, Zucker RM, Keefer CL. 1988. Interspecies differences in the stability of mammalian sperm nuclei assessed in vivo by sperm microinjection and in vitro by flow cytometry. Biol. Reprod. 39, 157–167 (doi:10.1095/biolreprod39.1.157) [DOI] [PubMed] [Google Scholar]
- 33.McLay DW, Clarke HJ. 2003. Remodelling the paternal chromatin at fertilization in mammals. Reproduction 125, 625–633 (doi:10.1530/rep.0.1250625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balhorn R. 2007. The protamine family of sperm nuclear proteins. Genome Biol. 8, 227 (doi:10.1186/gb-2007-8-9-227) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hecht NB. 1993. Gene expression during male germ cell development. In Cell and molecular biology of the testis (eds Desjardins C, Ewing LL.), pp. 400–432 New York, NY: Oxford University Press [Google Scholar]
- 36.Mali P, Kaipia A, Kangasniemi M, Toppari J, Sandberg M, Hecht NB, Parvinen M. 1989. Stage-specific expression of nucleo-protein mRNAs during rat and mouse spenniogenesis. Reprod. Fertil. Dev. 1, 369–382 (doi:10.1071/RD9890369) [DOI] [PubMed] [Google Scholar]
- 37.Oliva R. 2006. Protamines and male infertility. Hum. Reprod. Update 12, 417 (doi:10.1093/humupd/dml009) [DOI] [PubMed] [Google Scholar]
- 38.Wu JY, Ribar TJ, Cummings DE, Burton KA, McKnight GS, Means AR. 2000. Spermiogenesis and exchange of basic nuclear proteins are impaired in male germ cells lacking Camk4. Nat. Genet. 25, 448–452 (doi:10.1038/78153) [DOI] [PubMed] [Google Scholar]
- 39.Zhao M, et al. 2001. Targeted disruption of transition protein 2 gene subtly affects spermatogenesis in mice. Mol. Cell Biol. 21, 7243–7255 (doi:10.1128/MCB.21.21.7243-7255.2001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cho C, Willis WD, Goulding EH, Haesook JH, Choi YC, Hecht NB, Eddy EM. 2001. Haploinsufficiency of protamine-1 or 2 causes infertility in mice. Nat. Genet. 28, 82–86 (doi:10.1038/ng0501-82) [DOI] [PubMed] [Google Scholar]
- 41.Corzett M, Mazrimas J, Balhorn R. 2002. Protamine 1, protamine 2 stoichiometry in the sperm of eutherian mammals. Mol. Reprod. Dev. 61, 519–527 (doi:10.1002/mrd.10105) [DOI] [PubMed] [Google Scholar]
- 42.Haueter S, Kawsumi M, Asner I, Brykczynska U, Cinelli P, Moisyadi S, Burki K, Peters AHFM, Pelczar P. 2010. Genetic vasectomy—overexpression of Prm1–EGFP fusion protein in elongating spermatids causes dominant male sterility in mice. Genesis 48, 151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aoki VW, Liu L, Carrell DT. 2005. Identification and evaluation of a novel sperm protamine abnormality in a population of infertile males. Hum. Reprod. 20, 1298–1306 (doi:10.1093/humrep/deh798) [DOI] [PubMed] [Google Scholar]
- 44.Carrell DT, Liu L. 2001. Altered protamine 2 expression is uncommon in donors of known fertility, but common among men with poor fertilizing capacity, and may reflect other abnormalities of spermiogenesis. J. Androl. 22, 604–610 [PubMed] [Google Scholar]
- 45.de Yebra L, Ballescá JL, Vanrell JA, Corzett M, Balhorn R, Oliva R. 1998. Detection of P2 precursors in the sperm cells of infertile patients who have reduced protamine P2 levels. Fertil. Steril. 69, 755–759 (doi:10.1016/S0015-0282(98)00012-0) [DOI] [PubMed] [Google Scholar]
- 46.Torregrosa N, Dominguez-Fandos D, Camejo MI, Shirley CR, Meistrich ML, Ballesca JL, Oliva R. 2006. Protamine 2 precursors, protamine 1/protamine 2 ratio, DNA integrity and other sperm parameters in infertile patients. Hum. Reprod. 21, 2084–2089 (doi:10.1093/humrep/del114) [DOI] [PubMed] [Google Scholar]
- 47.Carrell DT, Emery BR, Liu L. 1999. Characterization of aneuploidy rates, protamine levels, ultrastructure, and functional ability of round-headed sperm from two siblings and implications for intracytoplasmic sperm injection. Fertil. Steril. 71, 511–516 (doi:10.1016/S0015-0282(98)00498-1) [DOI] [PubMed] [Google Scholar]
- 48.Gómez Montoto L, Magaña C, Tourmente M, Martín-Coello J, Crespo C, Luque-Larena JJ, Gomendio M, Roldan ERS. 2011. Sperm competition, sperm numbers and sperm quality in muroid rodents. PLoS ONE 6, e18173 (doi:10.1371/journal.pone.0018173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kenagy GJ, Trombulak SC. 1986. Size of mammalian testes in relation to body size. J. Mammal. 67, 1–22 (doi:10.2307/1380997) [Google Scholar]
- 50.Gomendio M, Harcourt H, Roldan ERS. 1998. Sperm competition in mammals. In Sperm competition and sexual selection (eds Birkhead TR, Møller AP.), pp. 667–751 London, UK: Academic Press [Google Scholar]
- 51.Soulsbury CD. 2010. Genetic patterns of paternity and testes size in mammals. PLoS ONE 5, e9581 (doi:10.1371/journal.pone.0009581) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramm SA, Parker GA, Stockley P. 2005. Sperm competition and the evolution of male reproductive anatomy in rodents. Proc. R. Soc. B 272, 949–955 (doi:10.1098/rspb.2004.3048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Felsenstein J. 1985. Phylogenies and the comparative method. Am. Nat. 125, 1–15 (doi:10.1086/284325) [Google Scholar]
- 54.Martins EP. 2004. COMPARE, version 46b: computer programs for the statistical analysis of comparative data. Bloomington, IN: Department of Biology, Indiana University; (http://compare.bio.indiana.edu/) [Google Scholar]
- 55.Lundrigan BL, Jansa S, Tucker PK. 2002. Phylogenetic relationships in the genus Mus, based on paternally, maternally, and biparentally inherited characters. Syst. Biol. 51, 410–431 (doi:10.1080/10635150290069878) [DOI] [PubMed] [Google Scholar]
- 56.Kendall D. 1986. The diffusion of shape. Adv. Appl. Probab. 9, 428–430 (doi:10.2307/1426091) [Google Scholar]
- 57.Goodall C. 1991. Procrustes methods in the statistical analysis of shape. J. R. Stat. Soc. B 53, 285–339 [Google Scholar]
- 58.Varea Sanchez M, Bastir M, Roldan ERS. 2013. Geometric morphometrics of rodent sperm head shape. PLoS ONE 8, e80607 (doi:10.1371/journal.pone.0080607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Klingenberg CP. 2011. MorphoJ: an integrated software package for geometric morphometrics. Mol. Ecol. Res. 11, 353–357 (doi:10.1111/j.1755-0998.2010.02924.x) [DOI] [PubMed] [Google Scholar]
- 60.Klingenberg CP, Gidaszewski A. 2010. Testing and quantifiying phylogenetic signals and homoplasy in morphometric data. Syst. Biol. 59, 245–261 (doi:10.1093/sysbio/syp106) [DOI] [PubMed] [Google Scholar]
- 61.Campbell NA, Atchley WR. 1981. The geometry of canonical variate analysis. Syst. Zool. 30, 268–280 (doi:10.2307/2413249) [Google Scholar]
- 62.Veyrunes F, Dobigny G, Yang F, O'Brien PCM, Catalan J, Robinson TJ, Britton-Davidian J. 2006. Phylogenomics of the genus Mus (Rodentia; Muridae): extensive genome repatterning is not restricted to the house mouse. Proc. R. Soc. B 273, 2925–2934 (doi:10.1098/rspb.2006.3670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carroll SB. 2005. Evolution at two levels: on genes and form. PLoS Biol. 3, 1159–1166 (doi:10.1371/journal.pbio.0030245) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hoekstra HE, Coyne JA. 2007. The locus of evolution: evo devo and the genetics of adaptation. Evolution 61, 995–1016 (doi:10.1111/j.1558-5646.2007.00105.x) [DOI] [PubMed] [Google Scholar]
- 65.King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188, 107–188 (doi:10.1126/science.1090005) [DOI] [PubMed] [Google Scholar]
- 66.Abzhanov A, Winston PK, Hartmann C, Grant R, Grant PR, Tabin CJ. 2006. The calmodulin pathway and evolution of elongated beak morphology in Darwin's finches. Nature 442, 563–567 (doi:10.1038/nature04843) [DOI] [PubMed] [Google Scholar]
- 67.Abzhanov A, Protas M, Grant R, Grant PR, Tabin CJ. 2004. Bmp4 and morphological variation of beaks in Darwin's finches. Science 305, 1462–1465 (doi:10.1126/science.1098095) [DOI] [PubMed] [Google Scholar]
- 68.Carleton KL, Kocher TD. 2001. Cone opsin genes of African cichlid fishes: tuning spectral sensitivity by differential gene expression. Mol. Biol. Evol. 18, 1540–1550 (doi:10.1093/oxfordjournals.molbev.a003940) [DOI] [PubMed] [Google Scholar]
- 69.Jones FC, et al. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484, 55–61 (doi:10.1038/nature10944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Manceau M, Domingues VS, Mallarino R, Hoekstra HE. 2011. The developmental role of agouti in color pattern evolution. Science 331, 1062–1065 (doi:10.1126/science.1200684) [DOI] [PubMed] [Google Scholar]
- 71.Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereg KS, Jónsson B, Schluter D, Kingsley DM. 2004. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428, 717–723 (doi:10.1038/nature02415) [DOI] [PubMed] [Google Scholar]
- 72.Wray GA. 2007. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Gen. 8, 206–216 (doi:10.1038/nrg2063) [DOI] [PubMed] [Google Scholar]
- 73.Queralt R, Adroer R, Oliva R, Winkfein RJ, Retief JD, Dixon GH. 1995. Evolution of protamine P1 genes in mammals. J. Mol. Evol. 40, 601–607 (doi:10.1007/BF00160507) [DOI] [PubMed] [Google Scholar]
- 74.Su Y, Wu D, Zhou W, Irwin DM, Zhang Y. 2013. Rapid evolution of the mammalian HILS1 gene and the nuclear condensation process during mammalian spermiogenesis. J. Genet. Genomics 40, 55–59 (doi:10.1016/j.jgg.2012.10.003) [DOI] [PubMed] [Google Scholar]
- 75.Rooney AP, Zhang J. 1999. Rapid evolution of a primate sperm protein: relaxation of functional constraint or positive Darwinian selection? Mol. Biol. Evol. 16, 706–710 (doi:10.1093/oxfordjournals.molbev.a026153) [DOI] [PubMed] [Google Scholar]
- 76.Torgerson DG, Kulathinal RJ, Singh RS. 2002. Mammalian sperm proteins are rapidly evolving: evidence of positive selection in functionally diverse genes. Mol. Biol. Evol. 19, 1973–1980 (doi:10.1093/oxfordjournals.molbev.a004021) [DOI] [PubMed] [Google Scholar]
- 77.Clark AG, Civetta A. 2000. Evolutionary biology: protamine wars. Nature 403, 261–263 (doi:10.1038/35002236) [DOI] [PubMed] [Google Scholar]
- 78.Aoki VW, Christensen GL, Atkins JF, Carrell DT. 2006. Identification of novel polymorphisms in the nuclear protein genes and their relationship with human sperm protamine deficiency and severe male infertility. Fertil. Steril. 86, 1416–1422 (doi:10.1016/j.fertnstert.2006.04.033) [DOI] [PubMed] [Google Scholar]
- 79.He XJ, Ruan J, Du WD, Chen G, Zhou Y, Xu S, Zuo XB, Cao YX, Zhang XJ. 2012. Prm1 variant rs35576928. Arg > Ser is associated with defective spermatogenesis in the Chinese Han population. Reprod. Biomed. Online 25, 627–634 (doi:10.1016/j.rbmo.2012.09.005) [DOI] [PubMed] [Google Scholar]
- 80.Schlicker M, Schnulle V, Schneppel L, Vorob'ev VI, Engel W. 1994. Disturbances of nuclear condensation in human spermatozoa: search for mutations in the genes for protamine 1, protamine 2 and transition nuclear protein 1. Hum. Reprod. 9, 2313–2317 [DOI] [PubMed] [Google Scholar]
- 81.Carrell DT, Emery BR, Hammoud S. 2007. Altered protamine expression and diminished spermatogenesis: what is the link? Hum. Reprod. Update 3, 313–327 (doi:10.1093/humupd/dml057) [DOI] [PubMed] [Google Scholar]
- 82.Greenbaum D, Colangelo C, Williams K, Gerstein M. 2003. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 4, 117 (doi:10.1186/gb-2003-4-9-117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harvey PH, Pagel MD. 1991. The comparative method in evolutionary biology. New York, NY: Oxford University Press [Google Scholar]


