Abstract
Pathogen evasion of the host immune system is a key force driving extreme polymorphism in genes of the major histocompatibility complex (MHC). Although this gene family is well characterized in structure and function, there is still much debate surrounding the mechanisms by which MHC diversity is selectively maintained. Many studies have investigated relationships between MHC variation and specific pathogens, and have found mixed support for and against the hypotheses of heterozygote advantage, frequency-dependent or fluctuating selection. Few, however, have focused on the selective effects of multiple parasite types on host immunogenetic patterns. Here, we examined relationships between variation in the equine MHC gene, ELA-DRA, and both gastrointestinal (GI) and ectoparasitism in plains zebras (Equus quagga). Specific alleles present at opposing population frequencies had antagonistic effects, with rare alleles associated with increased GI parasitism and common alleles with increased tick burdens. These results support a frequency-dependent mechanism, but are also consistent with fluctuating selection. Maladaptive GI parasite ‘susceptibility alleles’ were reduced in frequency, suggesting that these parasites may play a greater selective role at this locus. Heterozygote advantage, in terms of allele mutational divergence, also predicted decreased GI parasite burden in genotypes with a common allele. We conclude that an immunogenetic trade-off affects resistance/susceptibility to parasites in this system. Because GI and ectoparasites do not directly interact within hosts, our results uniquely show that antagonistic parasite interactions can be indirectly modulated through the host immune system. This study highlights the importance of investigating the role of multiple parasites in shaping patterns of host immunogenetic variation.
Keywords: major histocompatibility complex, DRA, selection, zebra, parasites, pleiotropy
1. Introduction
Pathogens and hosts engage in coevolutionary cycles that shape the genetic variability of populations [1,2]. Pathogens may evolve increased virulence and host recognition avoidance mechanisms, challenging their hosts to respond by evolving a diversity of innate and adaptive immune defences [3]. This ongoing evolutionary ‘arms race’ may influence the molecular diversity of both pathogen and host genomes, particularly within immunological genes [4]. There has been increasing focus on this selective molecular interplay in wildlife populations [5] that has come along with recognizing the importance of immune system function not only to host–parasite coevolution and infectious disease emergence, but also population dynamics and life-history evolution.
The major histocompatibility complex (MHC), a gene family composed of immune-related genes, has been of particular interest due to its exceptional diversity and significance in mate choice, kin recognition and host immunity in vertebrates [6]. This gene complex encodes the molecules responsible for initiating host immune response, by delivering foreign peptides derived from pathogens to helper T cells. Given this fundamental role in immune function, observations of extreme MHC polymorphism have been attributed to pathogen-induced balancing selection [7–10]. For example, a global study on humans found that populations with increased pathogen diversity also exhibited elevated MHC diversity [11]. Additionally, associations between specific MHC alleles and pathogen resistance have been demonstrated in humans [12], non-human primates [13], ungulates [14], rodents [15,16], bats [17], birds [18,19], amphibians [20] and fish [21].
The mechanisms driving MHC diversity are the subject of much debate (reviewed in [9,22]), with three primary hypotheses being (i) heterozygote advantage [7], (ii) negative frequency-dependent selection [8], and (iii) fluctuating selection over time and space [12,23]. The heterozygote advantage hypothesis is based on the theory that heterozygous individuals are capable of recognizing a more diverse suite of antigenic peptides from pathogens than homozygotes [7], and is supported by empirical evidence from experimental co-infection [24] and natural system studies [25,26]. Negative frequency-dependent selection suggests that the advantage of specific host alleles varies with their frequency as the result of pathogen evasion [8]. This mode of selection has been particularly difficult to demonstrate given the long time scales needed to detect such effects when in play. Finally, under fluctuating selection, external environmental factors or demographic stochasticity may drive oscillations in pathogen communities over space and time, resulting in corresponding changes in host allele/genotype fitness values [12]. Spatio-temporally heterogeneous allele fitness generated through fluctuating selection has been theoretically shown to be capable of maintaining high levels of MHC polymorphism [23].
Resolving the relative importance of the aforementioned hypotheses in any given study system is a challenging task, particularly because these mechanisms are not mutually exclusive and, even when acting in isolation, often can produce similar effects on MHC patterns [22]. Also, heterozygote advantage may depend on the degree of molecular divergence at overlapping peptide-binding regions (i.e. divergent allele advantage) [27]. Further complications arise in disentangling the relative importance of different mechanisms due to antagonistic pleiotropic effects of multiple parasites on a single locus, such that resistance to different pathogens requires different MHC alleles or genotypes. Under pleiotropy, increased resistance to one pathogen may be accompanied by a cost through decreased resistance to another, resulting in perplexing associations between MHC alleles and increased susceptibility to infection [17,19,21,25,28,29]. Given that diverse parasites may act in concert or opposition to mediate selection on host MHC genes, incorporating knowledge about multiple parasite infections may lead to conclusions that would have otherwise gone undetected.
Gastrointestinal (GI) parasites may have negative fitness consequences on wild populations [14], and many studies to date have examined relationships between MHC genes and helminths in vertebrates [13–15]. The vertebrate immune system is also important in responding to ectoparasite infections. For example, host antibodies can bind proteins in tick saliva, interfering with tick engorgement and nutrient absorption, thereby inhibiting ovum production and viability [30]. Also, immune-triggered inflammatory response and increase in host skin temperature may result in tick detachment [31]. Several studies have found associations between the MHC and ectoparasite prevalence and intensities [17,26,32]. Others have assessed bacterial [21,25], protist [18,28] and fungal infections [20], but few have investigated the relationships of multiple parasite types with MHC diversity (but see [29,32]). To our knowledge, only one study has investigated host immunogenetic relationships with both endo- and ectoparasites concurrently in a natural system [32].
The plains zebra (Equus quagga) population of Etosha National Park (ENP), Namibia, provides an excellent natural system in which to elucidate the mechanisms by which pathogens shape MHC variation. ENP is a eutrophic savannah ecosystem [33] with low annual precipitation (rainfall less than 650 mm yr−1). This aridity probably plays a role in limiting E. quagga parasite diversity relative to that found in zebra inhabiting ecosystems with higher annual rainfall (e.g. Kruger National Park—KNP, South Africa) [34]. Nonetheless, ENP zebra are susceptible to GI parasites, with nearly all individuals in the population having nematode infections [35]. Additionally, hard-bodied ticks have been observed on most individuals examined in the field, though the ecology of the host–ectoparasite relationship has yet to be characterized in this system. Here, we take advantage of the high prevalence of both parasite groups (nematodes and ticks) in E. quagga to investigate the relationships between multiple parasite infections and MHC diversity.
In domestic equids, Strongylidae nematodes can cause significant damage to the intestinal mucosa and arterial system, and larvae, in some species, can migrate through host tissues, occlude small arteries, and cause arteritis, thrombosis, embolism and fatal infarction of the bowel [36]. Ticks are one of the chief vectors for infectious disease agents, often causing illness and even death to their hosts. For example, in equids, ticks can transmit the lethal African horse sickness virus [37] and the protozoan pathogen babesiosis [38]. Beyond harbouring disease agents, they may decrease host fitness through dermatoses (inflammation, itching and swelling) and envenomization (delivered through tick saliva). We recognize that other pathogens besides the macroparasites investigated here may also affect MHC diversity in E. quagga. For example, zebras in ENP are the main host of anthrax, a deadly bacterial disease caused by Bacillus anthracis [39], and the role of this pathogen in shaping host immunogenetic diversity is a subject that warrants further study.
The equid MHC, or equine lymphocyte antigen (ELA), has been molecularly characterized, and earlier work provides evidence for selection on class II ELA loci [40,41]. The DR alpha chain (ELA-DRA) is of particular importance as it encodes the antigen-binding domain responsible for recognition of foreign peptides. This locus is considered to be much less diverse relative to other classical MHC genes in vertebrates [42,43]. However, recent evidence has proven equids to be an exception, exhibiting uniquely high levels of DRA polymorphism [40,41]. In E. quagga populations of southern Africa, low DRA differentiation among populations has been reported, typical of a locus under balancing selection [44].
In this study, we investigated the relationships between ELA-DRA variation and parasitism in the ENP zebra population. We focused on the effects of DRA heterozygosity and specific alleles in predicting both GI and ectoparasite burdens in zebra, while accounting for ecological and demographic predictors (season, sex and age) known to influence parasitism in ungulate hosts of this system [35]. Our goal was to examine the effects of immunogenetic variables on parasite intensity, while addressing the role of the following selective mechanisms: (i) ‘heterozygote advantage’ in terms of heterozygote fitness at the population level; (ii) ‘heterozygote advantage’ under the divergent allele advantage hypothesis, assuming fitness increases with the number of heterozygous bases in an individual's genotype; (iii) frequency dependence in terms of the non-additive (i.e. dominance) effects of alleles of a particular frequency class; and (iv) frequency-dependent or fluctuating selection through examination of specific allele effects. We also considered that the pathogen-mediated selection mechanisms might not be mutually exclusive. Finally, we examined whether these variables concurrently predict multiple types of parasitism to elucidate possible pleiotropic effects of the MHC in modulating parasite resistance.
2. Material and methods
(a). Study population
This study focused on the plains zebra population in ENP, a large (22 915 km2) fenced nature reserve in northern Namibia. In 2012, the population size was estimated to be 16 174 (95% CI: 13 310 – 19 038) individuals. Both aerial survey and genetic data suggest that the population has recently and historically been stable (Namibian Ministry of the Environment and Tourism 2012, unpublished data) [44]. ENP is classified as semi-arid mopane savannah [45], with annual rainfall totals around 500 mm. Rainfall exhibits a seasonal pattern with the majority of precipitation between the months of October and April (electronic supplementary material, figure S1).
(b). Data collection
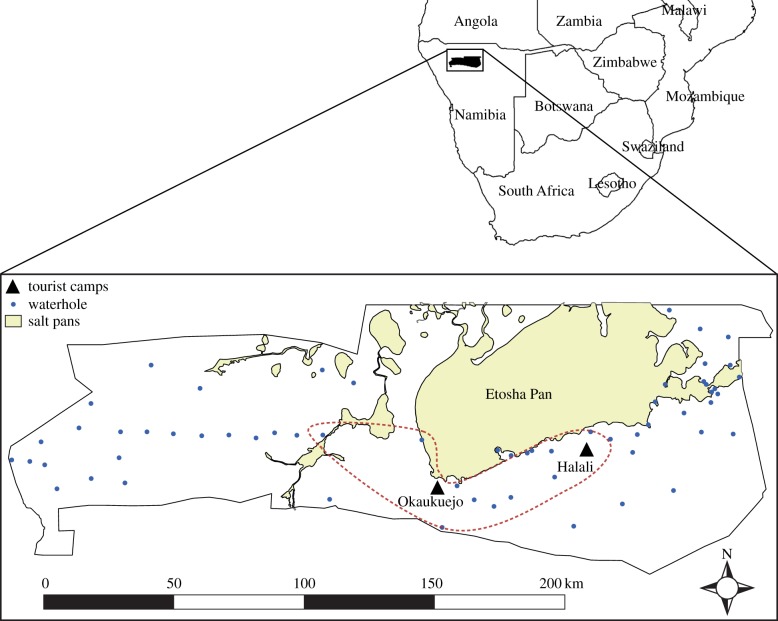
Data were collected from adult zebras (n = 70; females = 60, males = 10) during a series of captures taking place between March 2008 and August 2010 (three rainy and dry seasons) on the Okaukuejo and Halali plains of ENP (figure 1). During captures, zebras were anaesthetized and VHF- or GPS-collared, which enabled a subset to be re-captured. There were 173 total sampling observations, and each individual was sampled one to seven times. We collected faecal samples for GI parasite quantification (n = 140) and ectoparasites were picked from the animal (n = 140). Of these, we collected both parasite types for only a portion of the data (n = 107). Age was estimated from dental wear patterns of permanent incisors [46]. We also collected blood samples for immunogenetic characterization, stored in ethylenediaminetetraacetic acid tubes and preserved at −20°C. Faecal samples were stored at 4°C, and ticks preserved in 70% ethanol at room temperature.
Figure 1.
Map of Etosha National Park, Namibia, showing the area from which zebras were captured (dashed line) in the central plains region of the park. Tourist camps, where weather stations are located, are indicated by the black triangles. (Online version in colour.)
(c). Quantification of gastrointestinal parasite burden
GI parasite burden was measured in terms of faecal egg counts (FECs) in eggs per gram (EPG) of faeces of strongylid nematodes, following a modification of the McMaster flotation procedure [47,48] as described by Turner & Getz [35]. This approach provided an appropriate non-invasive means for quantifying relative parasite burdens among individuals and has proven to be valuable for assessing parasitism in wild ungulate hosts [14,49]. Previous work revealed nearly 100% prevalence of nematodes from the order Strongylida in ENP zebras [35], which accorded with our results. Microscopic observation did not allow us to resolve nematode taxa beyond the order level. However, nematodes of equid hosts are generally from the Strongylidae family, and necropsies identified 15 species of this family in ENP plains zebra [50]. A comparative study of intestinal helminth parasites in African equids found no single species infections, with a minimum of five strongylid species in any given individual [34]. These findings suggest that the nematodes in our study also fall within this taxonomic group and represent multiple strongylid species.
(d). Quantification of ectoparasite burden
We assessed tick burden during capture by collecting all visible arthropods of all life stages and focusing on the peri-anal (base of tail) and groin areas. Although this method does not ensure complete quantification and characterization of an individual's tick burden, we assumed that it provides a consistent relative estimate of tick abundance that is comparable among individuals. Five hard-bodied tick species (Family: Ixodidae) were observed from the genera Hyalomma and Rhipicelphalus: H. rufipes, H. truncatum, R. evertsi mimeticus, R. sulcatus and R. turanicus (I. Horak 2011, personal communication). We evaluated infection by ticks in terms of overall abundance, not distinguishing among species or life stages, owing to low statistical power associated with small sample sizes at these sub-levels.
(e). Major histocompatibility complex genotyping
Genomic DNA was extracted from blood samples using DNeasy extraction kits (Qiagen, Valencia, CA). We examined the diversity of the ELA-DRA exon 2, which encompassed the antigen-binding sites (ABS) known for their role in foreign peptide recognition. We amplified 246 bp of DRA exon 2 in ENP plains zebra following the protocols outlined by Kamath & Getz [41]. In E. quagga, the DRA locus exists as a single copy [41], and this was verified here by observations of no more than two alleles in any given individual. Individual DRA genotypes were determined through direct sequencing, and heterozygous nucleotide positions were confirmed by sequencing in both forward and reverse directions. Sequence chromatograms were aligned and edited manually using Geneious v. 5 [51]. Allele sequences were inferred using the haplotype phase determination algorithm, implemented in PHASE v. 2.1 [52]. A threshold posterior probability of 0.8 was upheld, and an allele was required to be observed at least twice before being considered a ‘true’ allele (i.e. in a minimum of one homozygote or two heterozygotes). We identified eight DRA alleles (DRA*01,*03–*05,*07,*09–*11; GenBank accession numbers AJ575299, EU930126, EU930121, EU930118, HQ637394–HQ637396), which were previously reported in this population [41]. This locus exhibited a total of six single nucleotide polymorphisms (SNPs) and four amino acid replacements, two of which occurred at ABS.
(f). Statistical analyses
Relationships between DRA variation and parasitism were assessed using generalized estimation equations (GEE), or marginal models analysed within the generalized linear model framework [53], and the following parasitological response variables: (i) GI parasite intensity defined as the square root (applied to reduce overdispersion) of the estimated number of nematode EPG of faeces, and (ii) ectoparasite intensity in terms of the number of ticks. We further assessed effects of co-infection by testing for a direct relationship between parasite types in an independent GEE analysis. Model estimates were determined by incorporating zebra identification as a random effect, following an exchangeable working correlation structure that accounts for repeated measurements from individuals [54,55]. A Poisson error distribution with log-link function was specified; with the GEE approach, however, no specific error distribution was directly assumed.
Previous research has shown corresponding patterns in the peaks of rainfall and GI parasitism, revealing season and age as significant predictors of GI parasitism in our study population [35]. Therefore, we accounted for season, sex and age in all statistical models. Season was defined based on rainfall observed during the study period (2007–2010), and a one-month lag was applied due to a known time lag in parasite egg shedding behind rainfall [56]. Thus, the wet season was defined as November to May and dry season as June to October (electronic supplementary material, figure S1). Individual age was represented as a continuous variable.
We fitted models with genetic explanatory variables that allowed for testing the hypotheses of heterozygote advantage, frequency-dependent and fluctuating selection (electronic supplementary material, table S1). We defined heterozygote advantage as heterozygotes having higher fitness than the average of homozygotes in the population, which can be explained by the ‘dominance’ of resistant alleles [24]. Heterozygosity was included as a binary fixed effect (i.e. heterozygote or homozygote). We also included heterozygosity as the number of SNPs observed within an individual's genotype to address the divergent allele advantage hypothesis. To address the frequency-dependent hypothesis, we included the presence or absence of rare (less than 5%: DRA*07,*10,*11), mid-frequency (5–10%: DRA*01*09,*05) and common alleles (more than 15%: DRA*03,*04) as explanatory variables. Allele frequencies were determined from our population data (electronic supplementary material, table S2). Finally, we fitted an allele model for each parasite response variable that included all DRA alleles (presence or absence of allele) as fixed effects, to assess whether specific alleles were behind observed associations between allele frequency classes and parasitism.
In choosing model parameters, we evaluated pairwise scatterplots, correlation coefficients and variance inflation factors (VIFMAX < 3) among explanatory variables to identify outliers and assess collinearity. Heterozygosity was highly correlated with SNPs, and thus we fitted our models by including these variables separately. We contrasted genetic with null models that included season, sex and age as covariates. In total, 49 models were examined (electronic supplementary material, tables S3 and S4) per response variable.
Model selection was conducted using the quasi-likelihood information criterion (QIC) [57], with best-fit models indicated by the smallest QIC values. Interactions among genetic variables were considered to account for the combined effects of heterozygosity and allele frequency. Model fit was evaluated by the difference from the best-fit model (ΔQIC = QICi – QICmin), improvement over the best-fit null model (ΔQIC/QICnull) and QIC weights (w) [58]. The effect of model structure was evaluated by marginal R2 (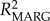 ) [59] or the proportion of variance in the response variable explained by the fitted model [55]. Finally, we validated candidate models by plotting Pearson's residuals against model-fitted values to assess homogeneity, examined residual histograms to assess normality and plotted residuals against each explanatory variable to test for homogeneity of error variances. Significance of parameter estimates was determined using a Wald test [54] and coefficients (β) were exponentiated (eβ) for interpretation as incidence rate ratios. In allele models, we controlled for multiple testing of allele effects by using a modified false discovery rate (FDR) procedure [60] to adjust the critical p-value (α). All computations were carried out using R v. 2.14.2 [61] with GEE fitting performed in geepack [62], assessment of collinearity using the corvif function [63] and calculation of Pan's QIC [57] in yags [64].
) [59] or the proportion of variance in the response variable explained by the fitted model [55]. Finally, we validated candidate models by plotting Pearson's residuals against model-fitted values to assess homogeneity, examined residual histograms to assess normality and plotted residuals against each explanatory variable to test for homogeneity of error variances. Significance of parameter estimates was determined using a Wald test [54] and coefficients (β) were exponentiated (eβ) for interpretation as incidence rate ratios. In allele models, we controlled for multiple testing of allele effects by using a modified false discovery rate (FDR) procedure [60] to adjust the critical p-value (α). All computations were carried out using R v. 2.14.2 [61] with GEE fitting performed in geepack [62], assessment of collinearity using the corvif function [63] and calculation of Pan's QIC [57] in yags [64].
3. Results
(a). Ecological patterns in parasitism
Mean (±s.e.) strongylid nematode egg count was 2524 (±129) EPG, ranging from 100 to 8050 EPG. We sampled an average of 4.7 (±0.3) ticks per individual (range: 0–29 ticks/individual). Of these, the majority of individuals (91%) were infected by the tick species R. e. mimeticus, supporting previous research on ticks from zebra of this region [65]. Fewer individuals sampled were infected by H. truncatum (20%), H. rufipes (11.4%), R. sulcatus (1%) or R. turanicus (1%). GI parasitism was significantly higher in the wet than the dry season (electronic supplementary material, figure S2): mean nematode FEC was 2893(±167) EPG and 1816(±152) EPG in the wet and dry seasons, respectively. By contrast, there was no apparent relationship with sex or age. The null model of GI parasitism (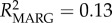 ) also revealed a significant effect of season (0.217 ± 0.051, p < 0.001; electronic supplementary material, table S5), predicting increased parasitism in the wet season. Initial data exploration exposed one high-leverage outlier within the ectoparasite data; therefore, we removed this observation from our data prior to model selection. There was no obvious effect of any of the evaluated ecological variables on tick abundance (electronic supplementary material, figure S2), although the null model for ectoparasitism revealed marginal significance for season (−0.204 ± 0.105, p = 0.052; electronic supplementary material, table S5), indicating decreased ectoparasite load in the wet season compared with the dry season. This model, however, provided a poor fit to the data (
) also revealed a significant effect of season (0.217 ± 0.051, p < 0.001; electronic supplementary material, table S5), predicting increased parasitism in the wet season. Initial data exploration exposed one high-leverage outlier within the ectoparasite data; therefore, we removed this observation from our data prior to model selection. There was no obvious effect of any of the evaluated ecological variables on tick abundance (electronic supplementary material, figure S2), although the null model for ectoparasitism revealed marginal significance for season (−0.204 ± 0.105, p = 0.052; electronic supplementary material, table S5), indicating decreased ectoparasite load in the wet season compared with the dry season. This model, however, provided a poor fit to the data (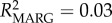 ).
).
(b). Genetic effects on gastrointestinal parasitism
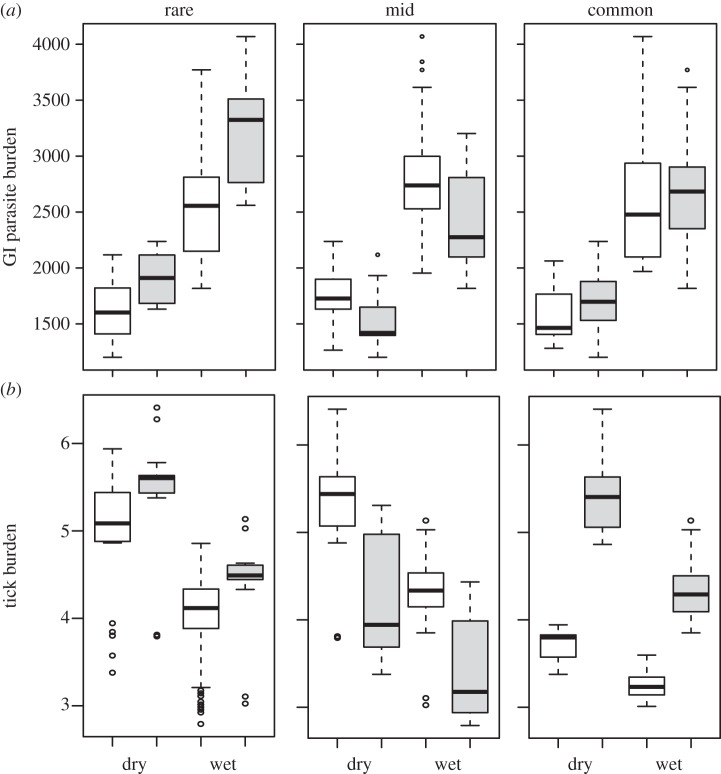
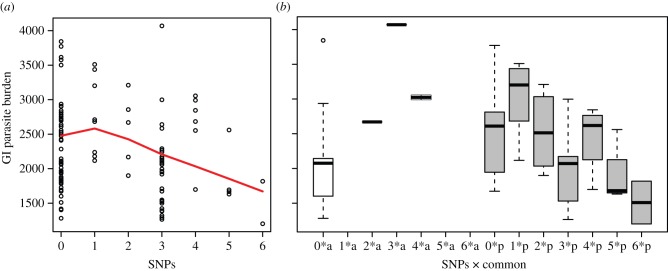
The inclusion of genetic variables improved the GI parasite model fit (ΔQIC = 86.13, per cent improvement = 0.08%; 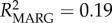 ) with the best-fit model including SNPs, common and rare alleles. Consistent with null model results, the best-fit model indicated a significant effect of season (0.201 ± 0.057, p < 0.001; table 1 and figure 2a; electronic supplementary material, figure S3). The presence of a rare allele in the DRA genotype predicted increased GI parasite burden (0.150 ± 0.042, p < 0.001), while there was no significant effect of common alleles. This model also included a significant interaction between common allele presence and number of SNPs at the DRA locus (−0.078 ± 0.036, p = 0.028), signifying a negative relationship between SNPs and GI parasites in heterozygotes with a common allele (figure 3). Coefficient estimates of explanatory variables from a maximal genetic GI parasitism model were consistent with both null and best-fit models (electronic supplementary material, table S5).
) with the best-fit model including SNPs, common and rare alleles. Consistent with null model results, the best-fit model indicated a significant effect of season (0.201 ± 0.057, p < 0.001; table 1 and figure 2a; electronic supplementary material, figure S3). The presence of a rare allele in the DRA genotype predicted increased GI parasite burden (0.150 ± 0.042, p < 0.001), while there was no significant effect of common alleles. This model also included a significant interaction between common allele presence and number of SNPs at the DRA locus (−0.078 ± 0.036, p = 0.028), signifying a negative relationship between SNPs and GI parasites in heterozygotes with a common allele (figure 3). Coefficient estimates of explanatory variables from a maximal genetic GI parasitism model were consistent with both null and best-fit models (electronic supplementary material, table S5).
Table 1.
Coefficient estimates and significance of parameters in best-fit models predicting GI and ectoparasitism (ECTO) in zebras. Coefficient s.e., Wald Z-test statistics, p-values and effect sizes (with 95% CIs) are reported. Maximum cluster size was 5 for both models.
| response | coefficients | estimate | s.e. | z-value | p(>|z|) | effect size |
|---|---|---|---|---|---|---|
| GIa | (intercept) | 3.718 | 0.119 | 927.09 | <2 × 10−16*** | |
| season (wet) | 0.201 | 0.057 | 12.54 | 4.0 × 10−4*** | 1.22 (1.09–1.37) | |
| sex (male) | 0.126 | 0.102 | 1.52 | 0.218 | 1.13 (0.93–1.39) | |
| age | –0.011 | 0.008 | 1.65 | 0.199 | 0.99 (0.97–1.00) | |
| SNPs | 0.039 | 0.032 | 1.52 | 0.217 | 1.04 (0.98–1.11) | |
| common (presence) | 0.104 | 0.090 | 1.33 | 0.249 | 1.11 (0.93–1.32) | |
| rare (presence) | 0.150 | 0.042 | 12.71 | 3.6 × 10−4*** | 1.16 (1.07–1.26) | |
| SNPs × common | –0.078 | 0.036 | 4.82 | 0.028* | 0.92 (0.86–0.99) | |
| ECTO | (intercept) | 1.179 | 0.271 | 18.95 | 1.3 × 10−5*** | |
| season (wet) | –0.191 | 0.101 | 3.58 | 0.059 | 0.83 (0.68–1.01) | |
| sex (male) | –0.046 | 0.173 | 0.07 | 0.789 | 0.96 (0.68–1.34) | |
| age | 0.021 | 0.026 | 0.65 | 0.422 | 1.02 (0.97–1.07) | |
| common (presence) | 0.344 | 0.172 | 4.02 | 0.045* | 1.41 (1.01–1.98) | |
| mid (presence) | –0.019 | 0.175 | 0.01 | 0.913 | 0.98 (0.70–1.38) | |
| rare (presence) | 0.056 | 0.173 | 0.10 | 0.748 | 1.06 (0.75–1.48) |
aResponse variable for GI parasitism was square-root transformed.
*p < 0.05, ***p < 0.001.
Figure 2.
Model predictions for frequency-dependent ELA-DRA effects on (a) GI parasitism, in terms of EPG of faeces, and (b) ectoparasitism, in terms of number of ticks. Effects of rare (less than 5%) mid-frequency (10–15%) and common alleles (more than 20%) on parasite burden are shown. Factors are defined as the presence (grey) or absence (white) of an allele in an individual's genotype by season (dry and wet). Ninety-five per cent confidence intervals are indicated by the dashed lines.
Figure 3.
Model predictions for SNPs effects on GI parasitism in terms of EPG of faeces. Effects of (a) SNPs and (b) the interaction between SNPs and a common allele on parasite burden are shown. Presence (p, grey) or absence (a, white) of a common allele is indicated. Lines in panel (a) represent a curve-smoothing function of the predicted data (lowess [61]). (Online version in colour.)
(c). Genetic effects on ectoparasitism
Inclusion of genetic variables improved the model fit (ΔQIC = 666.82, per cent improvement = 27.16%; 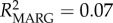 ) and the best-fit model included common, mid-frequency and rare alleles as genetic explanatory variables (table 1). Again, marginal significance for a seasonal effect was found (−0.191 ± 0.101, p = 0.059) and the presence of a common DRA allele predicted increased tick burden (0.344 ± 0.172, p = 0.045; figure 2b; electronic supplementary material, figure S3). The maximal model did not reveal any significant effects (electronic supplementary material, table S5).
) and the best-fit model included common, mid-frequency and rare alleles as genetic explanatory variables (table 1). Again, marginal significance for a seasonal effect was found (−0.191 ± 0.101, p = 0.059) and the presence of a common DRA allele predicted increased tick burden (0.344 ± 0.172, p = 0.045; figure 2b; electronic supplementary material, figure S3). The maximal model did not reveal any significant effects (electronic supplementary material, table S5).
(d). DRA allele effects
The inclusion of allelic variables into the model improved null models for both GI (percentage improvement = 33.5%; 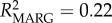 ) and ectoparasitism (percentage improvement = 5%;
) and ectoparasitism (percentage improvement = 5%; 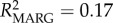 ). Allelic models corroborated seasonal effects on GI parasite load (0.200 ± 0.053, p < 0.001; electronic supplementary material, table S6 and figure S4). In further agreement, the presence of a rare allele in an individual's genotype predicted increased GI parasite intensity (DRA*07: 0.245 ± 0.088, p = 0.005; DRA*10: 0.273 ± 0.126, p = 0.030; DRA*11: 0.474 ± 0.189, p = 0.012), while SNPs had a negative relationship with GI parasitism (−0.085 ± 0.044, p = 0.05). The ectoparasite allelic model revealed that common alleles are significant predictors of increased tick burden (DRA*03: 0.879 ± 0.301, p = 0.004; DRA*04: 1.014 ± 0.362, p = 0.005; electronic supplementary material, table S6 and figure S4). Effects of a mid-frequency (DRA*05; 0.484 ± 0.236, p = 0.041) and rare allele (DRA*10; 0.730 ± 0.359, p = 0.042) were also uncovered but were not significant after applying the FDR critical p-value (α = 0.016).
). Allelic models corroborated seasonal effects on GI parasite load (0.200 ± 0.053, p < 0.001; electronic supplementary material, table S6 and figure S4). In further agreement, the presence of a rare allele in an individual's genotype predicted increased GI parasite intensity (DRA*07: 0.245 ± 0.088, p = 0.005; DRA*10: 0.273 ± 0.126, p = 0.030; DRA*11: 0.474 ± 0.189, p = 0.012), while SNPs had a negative relationship with GI parasitism (−0.085 ± 0.044, p = 0.05). The ectoparasite allelic model revealed that common alleles are significant predictors of increased tick burden (DRA*03: 0.879 ± 0.301, p = 0.004; DRA*04: 1.014 ± 0.362, p = 0.005; electronic supplementary material, table S6 and figure S4). Effects of a mid-frequency (DRA*05; 0.484 ± 0.236, p = 0.041) and rare allele (DRA*10; 0.730 ± 0.359, p = 0.042) were also uncovered but were not significant after applying the FDR critical p-value (α = 0.016).
(e). Relationships between parasite types
Results from statistical models of GI parasitism and ectoparasitism warranted further post hoc investigation into whether a direct inverse relationship (i.e. a co-infection effect) exists between these broadly grouped parasite types. We used the joint parasite data from a subset of our dataset (n = 106) to conduct an independent statistical analysis, analysing tick abundance as an explanatory variable for GI parasitism. We found that the co-infection coefficient was not significant (−0.005 ± 0.008, p = 0.51). Furthermore, when tick abundance was added as a covariate to null, maximal and best-fit GI models, the results were similar.
4. Discussion
In this study, we found significant immunogenetic effects predicting parasite intensity in E. quagga. These data suggest that the MHC locus, ELA-DRA, is centrally involved in a complex interplay between host and parasite, and support the occurrence of parasite-mediated frequency-dependent and/or fluctuating selection acting on this locus. Alleles present at opposing population frequencies conferred increased susceptibility to different parasite types; rare alleles were associated with increased GI parasitism and common alleles with increased ectoparasitism. This highlights a possible host immunogenetic trade-off, with multiple parasite groups competitively shaping patterns of MHC diversity. Although we found little conclusive evidence for heterozygote advantage, an interaction between an allele effect and heterozygosity, in terms of allelic divergence within the DRA genotype, implies that multiple selective mechanisms may act in concert.
(a). Roles of parasite-mediated selective mechanisms
Heterozygote advantage has been supported by individual heterozygosity at MHC loci associated with increased parasite resistance under both natural [15,25,26] and experimental conditions [24]. Others found no association with heterozygosity [21] or even reduced heterozygote fitness [66], probably due to resistance being recessive rather than dominant. Here, we found that DRA heterozygosity was not a significant predictor of parasite intensity. Our analysis, however, contrasted population-averaged parasite loads of heterozygotes versus homozygotes and did not account for overdominance, where the heterozygote is superior to both homozygotes of the respective alleles in its genotype. This is worthy to note given that only overdominance has been shown to maintain balanced allele frequencies [8]. Thus, we emphasize that further examination of the effects of specific genotypes is needed to assess the importance of heterozygote advantage in this system. However, our data suggest heterozygosity defined more explicitly at the molecular level (i.e. genotypic mutational differences) had an inverse relationship with GI parasitism. These results support the divergent allele advantage hypothesis [27], which asserts that more divergent alleles will increase functionality in the peptide-binding repertoire and have higher adaptive value.
Frequency-dependent selection may play a role in maintaining MHC diversity if new or rare alleles confer a fitness advantage to the individual [8]. In lemurs, Schad et al. [13] found evidence for frequency-dependent selection; rare MHC alleles were associated with low and common alleles with high nematode loads. Our study similarly supports the occurrence of frequency-dependent selection, with significant effects of specific DRA alleles on parasitism, and moreover by effects of different allele frequency classes. In particular, rare allele presence predicted a 16% increase in the population-average GI parasite load, whereas common allele presence predicted a 41% increase in ectoparasite load (table 1; electronic supplementary material, figure S3). Specific alleles were found to be behind these effects (electronic supplementary material, table S6 and figure S4). Model effect size (R2), although low, indicated that allele variables help explain a greater proportion of the variation in ectoparasitism (null model: R2 = 0.03; genetic model: R2 = 0.07; allele model: R2 = 0.17), whereas allele variables only slightly improved GI parasite model fit over frequency variables (null model: R2 = 0.13; genetic model: R2 = 0.19; allele model: R2 = 0.22). This suggests a greater role for frequency effects on GI parasite response, while specific MHC alleles may be more significant in driving ectoparasite response dynamics.
Fluctuating selection, driven by spatial and temporal variation in parasite communities, may alternatively or concurrently be a plausible explanation for allele–parasite associations. This mode of selection is theoretically capable of preserving MHC diversity, independent of either heterozygote advantage or frequency-dependent selection, if parasite pressures vary temporally and host resistance alleles are dominant [23]. We previously found that the DRA exhibited a skewed allele frequency distribution in E. quagga of ENP, differing from that in KNP [44]. This difference is consistent with spatially fluctuating selection and indicative of critical differences in the drivers of selection between the host populations. The more arid climate of ENP is thought to be responsible for the relatively lower Strongylinae nematode species richness observed in plains zebra of ENP [34]. Congruent with this, tick species richness is lower in ENP—a total of four or five Ixodidae spp. were found previously [65] and in this study, whereas seven Ixodidae spp. have been identified in zebra of KNP [67]. Our data suggest rare DRA alleles in ENP zebra increase susceptibility to GI parasite infections, and thus nematodes may play a greater selective role on the MHC owing to the combination of low species richness, but high prevalence and abundance [35]. Although GI parasites may be significant drivers of selection at this MHC locus, it is possible that we have only observed a snapshot of the process in time and that this MHC–parasite system exists in a state of flux. Further studies that investigate allele/genotype fitness over time and space are warranted for a comprehensive understanding of the selective processes acting on the MHC.
(b). ‘Susceptibility alleles’ and an immunogenetic trade-off
We found specific DRA alleles associated with increased parasitism, thereby conferring susceptibility rather than resistance to both parasite types, in consonance with other studies reporting associations of genetic variants with susceptibility in wildlife [13,28,29]. A mechanism that would allow maladaptive genetic alleles to be maintained is pleiotropy, the phenomenon that a single gene can affect multiple traits. Pleiotopic effects are believed to be widespread in nature and have been theoretically shown to reduce the ability of beneficial alleles to achieve fixation [68]. Kubinak et al. [69] demonstrated how disease-causing MHC alleles may be maintained through antagonistic pleiotropy between a mouse-specific retrovirus and its host, resulting in trade-offs between MHC genotypes. Experimental evidence for MHC heterozygote superiority against multiple pathogens was also presented as a means for persistence of susceptibility alleles [24]. In a study on house sparrows, Loiseau et al. [28] provided evidence for antagonistic effects of a MHC class I gene on multiple malarial parasite strains, suggesting that these effects allowed for the persistence of deleterious ‘susceptibility alleles’ in the population and may arise due to within-host competition between parasites.
Our results show opposing DRA allele associations with ticks versus GI nematodes and imply that an MHC trade-off alters resistance/susceptibility to multiple parasites in this system. Whereas alleles inferred to be beneficial for reducing GI parasites have been driven up to high frequencies, alleles associated with susceptibility have apparently been selected against, and hence are rare in the population. With this, there is a host immunogenetic trade-off for nematode resistance, in the form of increased susceptibility to ticks. The implications of resistance costs in determining an equilibrium level of resistance have been widely discussed and exemplified in predator/pathogen–prey model systems of Escherichia coli [70] and plants [71]. Hence, we hypothesize that the observed skew in the DRA frequency distribution supports the conclusion that GI parasites play a more significant role in shaping patterns of variation at this locus than do ticks, and pleiotropic effects modulate resistance/susceptibility to multiple parasites in this system. Finally, the lack of significant co-infection effects suggests that a direct relationship between parasite types does not exist, corroborating our hypothesis that parasitism is modulated indirectly, through the host immune system. This is strengthened by the fact that these parasites inhabit different areas of the host's body and do not physically interact.
5. Conclusion
This study elucidates the selective mechanisms acting on an MHC locus in the presence of multiple parasites. Strong support was found for selection by parasites on zebra hosts, modulated by frequency-dependent and/or fluctuating selection. Further, these data suggest that heterozygote advantage, under a divergent allele hypothesis, plays a role in conferring resistance to GI parasites. Most significantly, this study uniquely reports antagonistic effects of a MHC gene modulating susceptibility/resistance to multiple parasites, while excluding the possibility of direct parasite competition. We hypothesize that the presence of disadvantageous ‘susceptibility alleles’ in this population reflects an immunogenetic trade-off. These findings underscore the importance of considering multiple parasites when investigating the selective mechanisms driving host immune gene variation.
Acknowledgements
We thank the Etosha Ecological Institute for assistance during captures, particularly W. Kilian, W. Versfeld, G. Shatumbu, M. Kasaona, J. Kapner, S. Kötting and B. Kötting. We are grateful to the veterinarians (O. Aschenborn, N. Brain, C. Cizauskas and M. Jago), who were critical for conducting captures, and I. Horak for identification of tick species. Members and associates of the Getz Lab helped in sample collection: H. Ganz, S. Bellan, C. Cizauskas, M. Tsalyuk, L. Polansky, R. Zidon, O. Spiegel, Z. Havarua, M. Shikongo, H. Sibanda, C. Cloete. We also thank R. Bowie, J. Benavides, L. Raberg and two anonymous reviewers for insightful comments on the manuscript. Any use of trade, firm or product names is for descriptive purposes only and does not imply endorsement by the US Government.
This project was approved by the Namibian Ministry of Environment and Tourism (permit no. 1220/2007) and the UC Berkeley Animal Care and Use Committee (protocol no. R217-0510B).
Data accessibility
All data used in this study, including genetic, parasite and zebra capture data, have been made publically available at the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.p083b.
Funding statement
This project was funded by a NIH Ecology and Evolution of Infectious Disease grant (GM083863) to W.M.G. and a NSF Doctoral Dissertation Improvement grant (MCINS-20091291) to P.L.K.
References
- 1.Haldane JBS. 1949. Disease and evolution. Ric. Sci. Suppl. A. 19, 68–76 [Google Scholar]
- 2.Anderson RM, May RM. 1982. Coevolution of hosts and parasites. Parasitology 85, 411–426 (doi:10.1017/S0031182000055360) [DOI] [PubMed] [Google Scholar]
- 3.Slev PR, Potts WK. 2002. Disease consequences of pathogen adaptation. Curr. Opin. Immunol. 14, 609–614 (doi:10.1016/S0952-7915(02)00381-3) [DOI] [PubMed] [Google Scholar]
- 4.Frank SA. 2000. Polymorphism of attack and defense. Trends Ecol. Evol. 15, 167–171 (doi:10.1016/S0169-5347(99)01814-5) [DOI] [PubMed] [Google Scholar]
- 5.Acevedo-Whitehouse K, Cunningham AA. 2006. Is MHC enough for understanding wildlife immunogenetics? Trends Ecol. Evol. 21, 433–438 (doi:10.1016/j.tree.2006.05.010) [DOI] [PubMed] [Google Scholar]
- 6.Edwards SV, Hedrick PW. 1998. Evolution and ecology of MHC molecules: from genomics to sexual selection. Trends Ecol. Evol. 13, 305–311 (doi:10.1016/S0169-5347(98)01416-5) [DOI] [PubMed] [Google Scholar]
- 7.Doherty PC, Zinkernagel RM. 1975. Enhanced immunological surveillance in mice heterozygous at H-2 gene complex. Nature 256, 50–52 (doi:10.1038/256050a0) [DOI] [PubMed] [Google Scholar]
- 8.Takahata N, Nei M. 1990. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics 124, 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sommer S. 2005. The importance of immune gene variability (MHC) in evolutionary ecology and conservation. Front. Zool. 2, 1–18 (doi:10.1186/1742-9994-2-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hedrick P, Kim T. 2000. Genetics of complex polymorphisms: parasites and maintenance of MHC variation. In Genetics, evolution, and society (eds Singh RS, Krimbas CB.), pp. 205–233 Cambridge, MA: Harvard University Press [Google Scholar]
- 11.Prugnolle F, Manica A, Charpentier M, Guegan JF, Guernier V, Balloux F. 2005. Pathogen-driven selection and worldwide HLA class I diversity. Curr. Biol. 15, 1022–1027 (doi:10.1016/j.cub.2005.04.050) [DOI] [PubMed] [Google Scholar]
- 12.Hill AVS, et al. 1991. Common West African HLA antigens are associated with protection from severe malaria. Nature 352, 595–600 (doi:10.1038/352595a0) [DOI] [PubMed] [Google Scholar]
- 13.Schad J, Ganzhorn JU, Sommer S. 2005. Parasite burden and constitution of major histocompatibility complex in the malagasy mouse lemur, Microcebus murinus. Evolution 59, 439–450 [PubMed] [Google Scholar]
- 14.Paterson S, Wilson K, Pemberton JM. 1998. Major histocompatibility complex variation associated with juvenile survival and parasite resistance in a large unmanaged ungulate population (Ovis aries L.). Proc. Natl Acad. Sci. USA 95, 3714–3719 (doi:10.1073/pnas.95.7.3714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Froeschke G, Sommer S. 2005. MHC class II DRB variability and parasite load in the striped mouse (Rhabdomys pumilio) in the southern Kalahari. Mol. Biol. Evol. 22, 1254–1259 (doi:10.1093/molbev/msi112) [DOI] [PubMed] [Google Scholar]
- 16.Kloch A, Babik W, Bajer A, Sinski E, Radwan J. 2010. Effects of an MHC-DRB genotype and allele number on the load of gut parasites in the bank vole Myodes glareolus. Mol. Ecol. 19, 255–265 (doi:10.1111/j.1365-294X.2009.04476.x) [DOI] [PubMed] [Google Scholar]
- 17.Schad J, Dechmann DKN, Voigt CC, Sommer S. 2012. Evidence for the ‘Good Genes’ model: association of MHC class II DRB alleles with ectoparasitism and reproductive state in the neotropical lesser bulldog bat, Noctilio albiventris. PLoS ONE 7, e37101 (doi:10.1371/journal.pone.0037101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn PO, Bollmer JL, Freeman-Gallant CR, Whittingham LA. 2013. MHC evolution is related to sexually selected ornament, survival, and parasite resistance in common yellowthroats. Evolution 67, 679–687 (doi:10.1111/j.1558-5646.2012.01799.x) [DOI] [PubMed] [Google Scholar]
- 19.Loiseau C, Zoorob R, Robert A, Chastel O, Julliard R, Sorci G. 2011. Plasmodium relictum infection and MHC diversity in the house sparrow (Passer domesticus). Proc. R. Soc. B 278, 1264–1272 (doi:10.1098/rspb.2010.1968) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Savage AE, Zamudio KR. 2011. MHC genotypes associate with resistance to a frog-killing fungus. Proc. Natl Acad. Sci. USA 108, 16 705–16 710 (doi:10.1073/pnas.1106893108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dionne M, Miller KM, Dodson JJ, Bernatchez L. 2009. MHC standing genetic variation and pathogen resistance in wild Atlantic salmon. Phil. Trans. R. Soc. B 364, 1555–1565 (doi:10.1098/rstb.2009.0011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spurgin LG, Richardson DS. 2010. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proc. R. Soc. B 277, 979–988 (doi:10.1098/rspb.2009.2084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hedrick PW. 2002. Pathogen resistance and genetic variation at MHC loci. Evolution 56, 1902–1908 [DOI] [PubMed] [Google Scholar]
- 24.McClelland EE, Penn DJ, Potts WK. 2003. Major histocompatibility complex heterozygote superiority during coinfection. Infect Immun. 71, 2079–2086 (doi:10.1128/IAI.71.4.2079-2086.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans ML, Neff BD. 2009. Major histocompatibility complex heterozygote advantage and widespread bacterial infections in populations of Chinook salmon (Oncorhynchus tshawytscha). Mol. Ecol. 18, 4716–4729 (doi:10.1111/j.1365-294X.2009.04374.x) [DOI] [PubMed] [Google Scholar]
- 26.Oliver MK, Telfer S, Piertney SB. 2009. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris). Proc. R. Soc. B 276, 1119–1128 (doi:10.1098/rspb.2008.1525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakeland EK, Boehme S, She JX, Lu CC, McIndoe RA, Cheng I, Ye Y, Potts WK. 1990. Ancestral polymorphisms of MHC class-II genes: divergent allele advantage. Immunol. Res. 9, 115–122 (doi:10.1007/BF02918202) [DOI] [PubMed] [Google Scholar]
- 28.Loiseau C, Zoorob R, Garnier S, Birard J, Federici P, Julliard R, Sorci G. 2008. Antagonistic effects of a MHC class I allele on malaria-infected house sparrows. Ecol. Lett. 11, 258–265 (doi:10.1111/j.1461-0248.2007.01141.x) [DOI] [PubMed] [Google Scholar]
- 29.Kloch A, Baran K, Buczek M, Konarzewski M, Radwan J. 2013. MHC influences infection with parasites and winter survival in the root vole Microtus oeconomus. Evol. Ecol. 27, 635–653 (doi:10.1007/s10682-012-9611-1) [Google Scholar]
- 30.Trager W. 1939. Acquired immunity to ticks. J. Parasitol. 25, 57–81 (doi:10.2307/3272160) [Google Scholar]
- 31.Allen JR, Kemp DH. 1982. Observations on the behavior of Dermacentor andersoni larvae infesting normal and tick resistant guinea pigs. Parasitology 84, 195 (doi:10.1017/S0031182000044760) [DOI] [PubMed] [Google Scholar]
- 32.Ditchkoff SS, Hoofer SR, Lochmiller RL, Masters RE, Van Den Bussche RA. 2005. MHC-DRB evolution provides insight into parasite resistance in white-tailed deer. Southw. Nat. 50, 57–64 (doi:10.1894/0038-4909(2005)050<0057:MEPIIP>2.0.CO;2) [Google Scholar]
- 33.Huntley BJ. 1982. Southern African savannas. In Ecology of tropical savannas (eds Huntely BJ, Walker BH.), pp. 101–119 Berlin, Germany: Springer [Google Scholar]
- 34.Matthee S, Krecek RC, McGeoch MA. 2004. A comparison of the intestinal helminth communities of equidae in Southern Africa. J. Parasitol. 90, 1263–1273 (doi:10.1645/GE-3353) [DOI] [PubMed] [Google Scholar]
- 35.Turner WC, Getz WM. 2010. Seasonal and demographic factors influencing gastrointestinal parasitism in ungulates of Etosha National Park. J. Wildl. Dis. 46, 1108–1119 (doi:10.7589/0090-3558-46.4.1108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowman DD. 2003. Georgis’ parasitology for veterinarians, 8th edn Philadelphia, PA: WB Saunders [Google Scholar]
- 37.Dardiri AH, Salama SA. 1988. African horse sickness: an overview. J. Equine Vet. Sci. 8, 46–49 (doi:10.1016/S0737-0806(88)80110-2) [Google Scholar]
- 38.Dwinger RH. 1999. Ticks and tick-borne diseases of equids. In Equine Infectious Diseases VIII: Proc. of the Eighth Int. Conf., Dubai, 23–26 March 1998, pp. 306–310 Newmarket, Canada: R & W Publications [Google Scholar]
- 39.Turner WC, Imologhome P, Havarua Z, Kaaya GP, Mfune JKE, Mpofu IDT, Getz WM. 2013. Soil ingestion, nutrition and the seasonality of anthrax in herbivores of Etosha National Park. Ecosphere 4 (doi:10.1890/ES12-00245.1) [Google Scholar]
- 40.Janova E, Matiasovic J, Vahala J, Vodicka R, Van Dyk E, Horin P. 2009. Polymorphism and selection in the major histocompatibility complex DRA and DQA genes in the family Equidae. Immunogenetics 61, 513–527 (doi:10.1007/s00251-009-0380-0) [DOI] [PubMed] [Google Scholar]
- 41.Kamath PL, Getz WM. 2011. Adaptive molecular evolution of the major histocompatibility complex genes, DRA and DQA, in the genus Equus. BMC Evol. Biol. 11, 128 (doi:10.1186/1471-2148-11-128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner JL, Burnett RC, Storb R. 1999. Organization of the canine major histocompatibility complex: current perspectives. J. Hered. 90, 35–38 (doi:10.1093/jhered/90.1.35) [DOI] [PubMed] [Google Scholar]
- 43.Takada T, Kikkawa Y, Yonekawa H, Amano T. 1998. Analysis of goat MHC class II DRA and DRB genes: identification of the expressed gene and new DRB alleles. Immunogenetics 48, 408–412 (doi:10.1007/s002510050452) [DOI] [PubMed] [Google Scholar]
- 44.Kamath PL, Getz WM. 2012. Unraveling the effects of selection and demography on immune gene variation in free-ranging plains zebra (Equus quagga) populations. PLoS ONE 7, e50971 (doi:10.1371/journal.pone.0050971) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Roux CJG, Grunow JO, Morris JW, Bredenkamp GJ, Scheepers JC. 1988. A classification of the vegetation of the Etosha National Park. South Afr. J. Bot. 54, 1–10 [Google Scholar]
- 46.Smuts GL. 1974. Age determination in Burchells zebra Equus burchelli antiquorum from the Kruger National Park. J. South. Afr. Wildl. Manage. Assoc. 4, 103–115 [Google Scholar]
- 47.Gordon HM, Whitlock HV. 1939. A new technique for counting nematode eggs in sheep faeces. J. Counc. Sci. Ind. Res. Melbourne 12, 50–52 [Google Scholar]
- 48.Gibbons LM, Jacobs DE, Fox MT, Hansen J. 2005. The RVC/FAO guide to veterinary diagnostic parasitology: faecal examination of farm animals for helminth parasites. See http://www.rvc.ac.uk/review/Parasitology/Index/Index.htm. [Google Scholar]
- 49.Coltman DW, Pilkington JG, Smith JA, Pemberton JM. 1999. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution 53, 1259–1267 (doi:10.2307/2640828) [DOI] [PubMed] [Google Scholar]
- 50.Krecek RC, Reinecke RK, Malan FS. 1987. Studies on the parasites of zebras. V. Nematodes of the Burchell and Hartmann mountain zebras from the Etosha National Park, South West Africa/Namibia. Onderstepoort J. Vet. Res. 54, 71–78 [PubMed] [Google Scholar]
- 51.Drummond A, et al. 2010. Geneious v. 5.0 See http://www.geneious.com (accessed 1 November 2012).
- 52.Stephens M, Smith NJ, Donnelly P. 2001. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (doi:10.1086/319501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang KY, Zeger SL. 1986. Longitudinal data analysis using generalized linear models. Biometrika 73, 13–22 (doi:10.1093/biomet/73.1.13) [Google Scholar]
- 54.Zuur AF, Ieno EN, Walker N, Saveliev AA. 2009. Mixed effects models and extensions in ecology in R, 1st edn New York, NY: Springer Science+Business Media, LLC [Google Scholar]
- 55.Hardin JW, Hilbe JM. 2003. Generalized estimating equations, xiii + 222 pp. Boca Raton, FL: Chapman and Hall/CRC [Google Scholar]
- 56.Turner WC. 2009. The ecology of orally ingested parasites in ungulates of Etosha National Park. PhD, University of California at Berkeley, Berkeley, CA [Google Scholar]
- 57.Pan W. 2001. Akaike's information criterion in generalized estimating equations. Biometrics 57, 120–125 (doi:10.1111/j.0006-341X.2001.00120.x) [DOI] [PubMed] [Google Scholar]
- 58.Burnham K, Anderson D. 2002. Model selection and multimodel inference: a practical information-theoretic approach. New York, NY: Springer [Google Scholar]
- 59.Zheng BY. 2000. Summarizing the goodness of fit of generalized linear models for longitudinal data. Stat. Med. 19, 1265–1275 (doi:10.1002/(SICI)1097-0258(20000530)19:10<1265::AID-SIM486>3.0.CO;2-U) [DOI] [PubMed] [Google Scholar]
- 60.Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29, 1165–1188 (doi:10.1214/aos/1013699998) [Google Scholar]
- 61.R Development Core Team. 2011. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 62.Halekoh U, Hojsgaard S, Yan J. 2006. The R package geepack for generalized estimating equations. J. Stat. Softw. 15, 1–11 [Google Scholar]
- 63.Zuur AF, Ieno EN, Walker N, Saveliev AA, Smith GM.2009. Mixed effects models and extensions in ecology with R. New York, NY: Springer.
- 64.Carey V.2004. yags: yet another GEE solver. R package version 4.0-2.2. See http://www.biostat.harvard.edu/∼carey .
- 65.Horak IG, Anthonissen M, Krecek RC, Boomker J. 1992. Arthropod parasites of springbok, gemsbok, kudus, giraffes and Burchell and Hartmann zebras in the Etosha and Hardap Nature-Reserves, Namibia. Onderstepoort J. Vet. Res. 59, 253–257 [PubMed] [Google Scholar]
- 66.Ilmonen P, Penn DJ, Damjanovich K, Morrison L, Ghotbi L, Potts WK. 2007. Major histocompatibility complex heterozygosity reduces fitness in experimentally infected mice. Genetics 176, 2501–2508 (doi:10.1534/genetics.107.074815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horak IG, Devos V, Deklerk BD. 1984. Parasites of domestic and wild animals in South-Africa.17. Arthropod parasites of Burchell zebra, Equus burchelli, in the Eastern Transvaal Lowveld. Onderstepoort J. Vet. Res. 51, 145–154 [PubMed] [Google Scholar]
- 68.Otto SP. 2004. Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc. R. Soc. Lond. B 271, 705–714 (doi:10.1098/rspb.2003.2635) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kubinak JL, Ruff JS, Hyzer CW, Slev PR, Potts WK. 2012. Experimental viral evolution to specific host MHC genotypes reveals fitness and virulence trade-offs in alternative MHC types. Proc. Natl Acad. Sci. USA 109, 3422–3427 (doi:10.1073/pnas.1112633109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lenski RE. 1988. Experimental studies of pleiotropy and epistasis in Escherichia coli 0.1. Variation in competitive fitness among mutants resistant to virus T4. Evolution 42, 425–432 (doi:10.2307/2409028) [DOI] [PubMed] [Google Scholar]
- 71.Simms EL. 1992. Costs of plant resistance to herbivory. In Plant resistance to herbivores and pathogens (eds Fritz RS, Simms EL.), pp. 392–425 Chicago, IL: University of Chicago Press [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data used in this study, including genetic, parasite and zebra capture data, have been made publically available at the Dryad Digital Repository: http://dx.doi.org/10.5061/dryad.p083b.