Abstract
KDM1A/LSD1, a histone H3K4/K9 demethylase and epigenetic regulator with roles in both gene activation and repression, has increased expression in multiple cancer types. Harris et al., in this issue of Cancer Cell, and Schenk et al. show that KDM1A may be a viable therapeutic target in treating AML.
Epigenetic regulation of gene expression, through both histone modification and DNA methylation, provides cells with a heritable mechanism for controlling gene expression without altering the DNA nucleotide sequence. KDM1A/lysine specific demethylase 1 (LSD1) was discovered in 2004 as the first histone demethylase, with specificity for mono- and dimethyl histone H3 lysine 4 and mono- and dimethyl histone H3 lysine 9. Prior to its identification, methylation of histones was thought to be a relatively permanent epigenetic mark. KDM1A is a member of the flavin adenine dinucleotide (FAD)-dependent family of amine oxidases, which require FAD to oxidize the mono- or dimethyl lysine to an imine intermediate that is further hydrolyzed to unmodified lysine and formaldehyde (Shi et al., 2004).
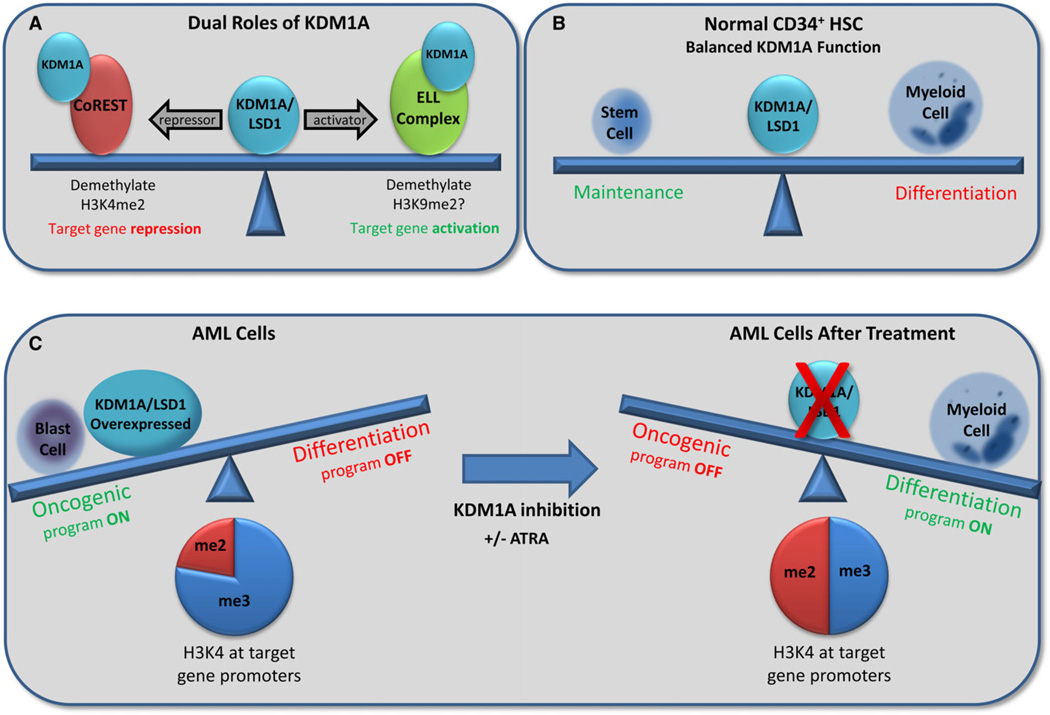
KDM1A was originally identified as a member of the CoREST repressor complex (You et al., 2001) (Figure 1A). When this complex is targeted to lineage-specific genes, KDM1A demethylates the activating H3K4me2 mark to silence their expression. Additional repressive complexes containing KDM1A have since been identified (Wang et al., 2007).
Figure 1. KDM1A/LSD1 Maintains a Balance in Gene Expression through Activating and Repressive Mechanisms that Are Disrupted in Acute Myeloid Leukemia.
(A and B) In normal cells, KDM1A activates or represses genes through its histone demethylase activity (A), maintaining the balance between hematopoietic stem cells and differentiation to mature myeloid cells (B).
(C) In AML, increased KDM1A expression promotes an oncogenic gene expression program, causing a block in differentiation associated with increased H3K4me3 to H3K4me2 ratio at the promoter of target genes. Inhibition of KDM1A restores this balance, promoting blast cell differentiation.
Conversely, KDM1A has been found to interact with multiple proteins/complexes that function in gene activation (Figure 1A). KDM1A is required for transcription of androgen receptor (AR) and estrogen receptor (ER) target genes, where it is recruited via interaction with AR or ER, respectively, and is thought to demethylate the repressive H3K9me2 mark to allow for gene activation (Metzger et al., 2005). KDM1A is also a member of a transcription elongation complex composed of ELL (elongation factor RNA polymerase II), pTEFb, AF4, and AFF4 (Biswas et al., 2011). Additionally, KDM1A is a component of an MLL supercomplex associated with active transcription (Nakamura et al., 2002). MLL itself is an epigenetic modifier as a histone methyltransferase with specificity for H3K4.
In normal hematopoietic development, hematopoietic stem cells undergo a series of changes in gene expression which both promote differentiation to mature blood cell lineages and repress genes necessary for maintaining stem cell identity. These changes are mediated, in part, by epigenetic modifiers such as KDM1A and MLL (Figure 1B). In leukemia, this normal process of cellular maturation goes awry; the leukemic stem cells (LSCs) do not differentiate appropriately, with resultant accumulation of immature blast cells.
The mixed lineage leukemia gene (MLL) is frequently involved in chromosomal translocation with one of a variety of partner genes in acute leukemias of myeloid (AML) or lymphoid lineage. As a result, functional oncogenic fusion proteins are produced that promote the constitutive expression of MLL target genes, thereby blocking differentiation and promoting proliferation of immature blast cells. Recent studies have shown increased KDM1A expression in AML regardless of subtype/cytogenetic status (http://www.proteinatlas.org; Berglund et al., 2008) as well as in a variety of other tumor types (Hayami et al., 2011). This raises the hypothesis that KDM1A may be an attractive target for therapeutic development.
One therapeutic approach that has been successful in treating the acute promyelocytic leukemia (APL) subset of leukemias is forced differentiation of immature leukemic cells into mature myeloid cells. In the majority of APLs, chromosomal translocations involving the retinoic acid receptor alpha gene (RARA) and the promyelocytic leukemia gene (PML) produce the PML-RARα fusion protein. PML-RARα aberrantly interacts with corepressor molecules such as NCOR and HDAC to prevent expression of RAR target genes necessary for differentiation. The use of all-trans retinoic acid (ATRA) can lift the differentiation block by promoting expression of RAR-responsive genes. However, the ability of ATRA to promote differentiation of PML-RARα AML cells is specific to this subset of leukemia; non-APL AML, such as AML harboring a MLL translocation, requires other treatment modalities.
In this issue of Cancer Cell, Harris et al. (2012) used microarray data from murine models of MLL leukemia to determine a correlation between KDM1A expression level and the leukemia colony-forming ability, often used as a surrogate assay to quantify LSC potential. Using both shRNAs and pharmacological inhibitors synthesized to target the enzymatic activity of KDM1A, the authors show that inhibition of KDM1A results in induction of differentiation in both murine and primary human MLL-fusion cells (Figure 1C). Cells without active KDM1A were unable to form colonies (indicative of loss of LSC potential), exhibited differentiated cell morphology and could not cause leukemia when introduced into mice. Gene expression analysis suggested that KDM1A is responsible for promoting the oncogenic gene program associated with MLL-AF9 leukemia. KDM1A colocalized to genes bound by MLL-AF9, and the presence of KDM1A correlated with a decreased ratio of H3K4me2 to H3K4me3 on these target genes. Upon inhibition of KDM1A, no global changes in H3K4me2 were evident, but the genes targeted by MLL-AF9 showed an increase in the H3K4me2 mark at their promoter and 5′ gene regions. This regional increase in H3K4me2 marks increased the ratio of H3K4me2 to H3K4me3 at the locus, making the prevalence of the marks more equal.
Complementary findings were recently reported in Nature Medicine (Schenk et al., 2012). The authors examined the effect of combining ATRA differentiation therapy with KDM1A inhibition in AML cells. They found that KDM1A inhibition, in combination with ATRA therapy, could sensitize otherwise ATRA-insensitive cells toward differentiation. This caused decreased leukemia-initiating activity (i.e., engraftment) as well as decreased tumor burden in human xenograft models. These combination-treated cells demonstrated increased expression of genes associated with myeloid-lineage differentiation and a concomitant increase in H3K4me2 at their promoters.
Together, these two studies illustrate the importance of KDM1A in maintaining expression of oncogenic gene programs and blocking differentiation in multiple subtypes of AML and highlight the potential therapeutic implications of targeting this important epigenetic regulator in leukemia. Despite these exciting findings, some very important questions remain to be addressed. First, what is the mechanism by which KDM1A functions at these target genes in AML? The presence of KDM1A at MLL-AF9 target genes decreases the H3K4me2 to H3K4me3 ratio, but how does KDM1A demethylation of H3K4me2/me1 lead to an increase in H3K4me3? Additionally, what proteins/protein complexes are recruited to these loci when KDM1A is present? Finally, what will be the long-term effect on normal hematopoietic stem cells and hematopoietic cell homeostasis and differentiation programs when treated with KDM1A inhibitors? Harris et al. (2012) start to address this question in their article. They found that while the overall frequency of colony forming cells remains the same upon KDM1A inhibition, erythroid lineage differentiation is decreased. In mice, this manifested as lethal anemia. How will these in vitro and in vivo results translate to patients that may be treated long-term with KDM1A inhibitors? Although the questions are plentiful in this emerging field of targeting epigenetic regulators in cancer, the results presented in these articles highlight the potential of using such therapies, not only in AML, but perhaps in other cancers that are dependent on aberrant epigenetic activity for survival.
REFERENCES
- Berglund L, Björling E, Oksvold P, Fagerberg L, Asplund A, Szigyarto CA, Persson A, Ottosson J, Wernérus H, Nilsson P, et al. Mol. Cell. Proteomics. 2008;7:2019–2027. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- Biswas D, Milne TA, Basrur V, Kim J, Elenitoba-Johnson KS, Allis CD, Roeder RG. Proc. Natl. Acad. Sci. USA. 2011;108:15751–15756. doi: 10.1073/pnas.1111498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris WJ, Huang X, Lynch JT, Spencer GJ, Hitchin JR, Li Y, Cicero F, Blaser JG, Greystroke BF, Jordan AM, et al. Cancer Cell. 2012;21:473–487. doi: 10.1016/j.ccr.2012.03.014. this issue. [DOI] [PubMed] [Google Scholar]
- Hayami S, Kelly JD, Cho HS, Yoshimatsu M, Unoki M, Tsunoda T, Field HI, Neal DE, Yamaue H, Ponder BAJ, et al. Int. J. Cancer. 2011;128:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. Mol. Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Schenk T, Chen WC, Göllner S, Howell L, Jin L, Hebestreit K, Klein HU, Popescu AC, Burnett A, Mills K, et al. Nature Medicine. 2012 doi: 10.1038/nm.2661. Published online March 11, 2012. 10.1038/nm.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- You A, Tong JK, Grozinger CM, Schreiber SL. Proc. Natl. Acad. Sci. USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]