Abstract
Candidatus Magnetoglobus multicellularis (Ca. M. multicellularis) is a member of a group of uncultured magnetotactic prokaryotes that possesses a unique multicellular morphology. To better understand this organism's physiology, we used a genomic approach through pyrosequencing. Genomic data analysis corroborates previous structural studies and reveals the proteins that are likely involved in multicellular morphogenesis of this microorganism. Interestingly, some detected protein sequences that might be involved in cell adhesion are homologues to phylogenetically unrelated filamentous multicellular bacteria proteins, suggesting their contribution in the early development of multicellular organization in Bacteria. Genes related to the behavior of Ca. M. multicellularis (chemo-, photo- and magnetotaxis) and its metabolic capabilities were analyzed. On the basis of the genomic–physiologic information, enrichment media were tested. One medium supported chemoorganoheterotrophic growth of Ca. M. multicellularis and allowed the microorganisms to maintain their multicellular morphology and cell cycle, confirming for the first time that the entire life cycle of the MMP occurs in a multicellular form. Because Ca. M. multicellularis has a unique multicellular life style, its cultivation is an important achievement for further studies regarding the multicellular evolution in prokaryotes.
Keywords: magnetotactic, pyrosequencing, multicellular prokaryotes, culture medium development, Candidatus Magnetoglobus multicellularis
Introduction
The development of multicellularity is one of the most significant evolutionary developments in the history of life on Earth (Sachs, 2008). Multicellularity in organisms evolved independently many times during evolution, but the transition from a unicellular to a multicellular lifestyle has only been studied in detail in the Eukarya. In many natural environments, prokaryotes live essentially as multicellular microorganisms (Shapiro, 1998), for example, in biofilms where different bacterial species have complex traits of interaction that characterize ‘bacterial social engagements' (Henke and Bassler, 2004). Interestingly, multicellular relationships are observed in some bacteria that have cellular speciation or unusual life cycles (Angert, 2005). However, almost all known prokaryotes spend at least some of their time as individual unicellular organisms.
There is one group of prokaryotes that are considered to spend their entire life cycle as multicellular organisms: the magnetotactic multicellular prokaryotes (MMPs) (Abreu et al., 2007; Simmons and Edwards, 2007; Wenter et al., 2009; Shapiro et al., 2011; Zhou et al., 2012). One MMP, known as Candidatus Magnetoglobus multicellularis, consists of an organized assemblage of genetically identical Gram-negative cells that contain greigite (Fe3S4) magnetosomes (Abreu et al., 2007), which are membrane-bound nano-sized magnetic particles used for orientation and navigation along magnetic field lines (Bazylinski and Frankel, 2004). The advantage of responding to the magnetic field is related to the ability of locating and maintaining optimal conditions for survival and reproduction in environments characterized by vertical chemical gradients (Frankel et al., 1997; Bazylinski and Frankel, 2004), a behavior called magnetotaxis (Blakemore, 1975). The biology of Ca. M. multicellularis is intriguing because its morphological and reproductive features strongly support the idea that this microorganism is a truly multicellular organism (Abreu et al., 2007; Keim et al., 2007). Multicellular characteristics observed in this microorganism include a coordinated collective motion and magnetic response, individual cells lacking viability and a completely multicellular life cycle where there is no single-cell stage, a feature that has not been reported in other bacteria.
Although some biological information of Ca. M. multicellularis has been obtained using culture-independent techniques (Keim et al., 2007), its physiology and genetic composition is largely unknown, mostly because of the inability to grow this microorganism in axenic culture. Phylogenetic analyses of MMPs, based on 16S rRNA genes sequences, have shown that this unusual group of prokaryotes is closely related to some dissimilatory sulfate-reducing bacteria in the Deltaproteobacteria (Abreu et al., 2007), suggesting that MMPs are sulfate-reducing bacteria. Despite this information, however, the development of a suitable culture medium for these microorganisms is yet to be achieved. Except for their 16S rRNA genes, mam genes associated with the biomineralization of magnetosomes (Abreu et al., 2011) and genes associated with dissimilatory sulfate reduction including that for dissimilatory sulfite reductase (dsrAB) and dissimilatory adenosine-5′-phosphate reductase (aprA) (Wenter et al., 2009), no other genomic information currently exists for MMPs. Here, we used pyrosequencing to obtain genome data that could contribute to the understanding of the metabolic requirements necessary for the growth of Ca. M. multicellularis in the laboratory. Although our approach was culture-independent, based on results from light microscopy, transmission electron microscopy (TEM) and 16S rRNA gene sequencing, Ca. M. multicellularis was the only microorganism in our samples (Abreu et al., 2011). Our data, along with previously described ultrastructural data, were used to characterize this uncultivated organism and to understand its physiology in turn to predict important physiological features and determine a suitable growth medium for Ca. M. multicellularis. An enrichment culture for this unique multicellular prokaryote was obtained providing the means to confirm the previously proposed multicellular life cycle and to study the genetic determinants in this microorganism responsible for multicellarity, which are pivotal to the understanding of the evolution of multicellularity.
Materials and methods
Sampling
Sediment and water were collected from the Araruama Lagoon (22°50′S, 42°13′W) and maintained in the laboratory at room temperature. Magnetic enrichment of cells, genomic amplification and sequencing were performed according to Abreu et al. (2011). Sampling occurred during 2007 until 2009; the double magnetic enrichment (Abreu et al., 2011) was performed directly after sampling and the purified Ca. M. multicellularis samples were stored at −20 °C. The number of Ca. M. multicellularis recovered from the environmental sample varied between samples. Sampling and magnetic enrichment were performed several times to obtain enough DNA for genome sequencing. The total amount of Ca. M. multicellularis was approximately 107. Significant loss occurred during the double magnetic enrichment and washing steps. After DNA extraction, we obtained approximately 1 μg of DNA, which was used in genomic amplification with REPLI-g mini kit (Qiagen, Hilden, Germany) (performed once) and the total amount of DNA obtained was 3.8 μg. The presence of Ca. M. multicellularis and the purity of samples were determined by observing the sample after the double magnetic enrichment and before storing the sample at −20 °C on a Zeiss Axiostar plus microscope (Carl Zeiss, Oberkochen, Germany), and also by 16S rRNA gene amplification using universal primers 8bF and 1512uR (Eder et al., 1999). Amplification products were cloned using Inst/Aclone polymerase chain reaction product cloning kit (Fermentas, Burlington, ON, Canada) or pGEMT Easy Vector kit (Promega Corporation, Madison, WI, USA). Approximately 50 clones from different magnetically enriched samples were sequenced, indicating that the samples were highly purified. Also observing microscopy samples from magnetically enriched Ca. M. multicellularis assured that no other magnetotactic bacteria was present in the samples.
Sequencing, assembly and annotation
Shotgun libraries were constructed from approximately 3 μg of randomly sheared DNA using a Covaris S2 System (Covaris, Woburn, MA, USA). Shearing conditions were as follows: number of cycles=15, duty cycle=2%, intensity=5, cycles per burst=200 and temperature=5 °C. The remaining steps of the library preparation were performed according to the GS FLX Titanium General Library Preparation Method Manual (Roche Diagnostics GmbH/454 Life Sciences Corporation, Branford, CT, USA). Titration, emulsion polymerase chain reaction and sequencing were carried out according to the manufacturer's protocol. A two-region 454 sequencing run was performed on a 70 × 75 PicoTiterPlate using the Genome Sequencer System (Roche Diagnostics GmbH/454 Life Sciences Corporation); both regions were loaded with the same preparation. Before assembly, artificially replicated sequences generated by 454-based pyrosequencing were identified and removed using the Replicates software program (Gomez-Alvarez et al., 2009). The remaining reads were filtered to remove short sequences (<150 bp) or reads with a phred quality score ⩽20 using the LUCY program (Chou and Holmes, 2001). Assembly was performed using Newbler Assembler software (version 2.5.3; Roche Diagnostics GmbH/454 Life Sciences Corporation) with a ‘-rip' flag that allows for outputting each read in only one contig. Reads identified by the GS De Novo Assembler (Roche Diagnostics GmbH/454 Life Sciences Corporation) as problematic or showing high-quality discrepancies were filtered out. A new assembly and filtering steps were performed. These steps were repeated until no remaining reads were identified as problematic or showed a discrepant base. The coverage of the final assembly and the G+C content are 18 × and 37%, respectively. These values calculated for the contigs that are larger than N50 are 18 × and 39%, both compatible with that calculated for the final assembly.
DNA sequences were annotated and analyzed with the System for Automated Bacterial Integrated Annotation (Almeida et al., 2004). An automatic annotation was performed using the following criteria. Open reading frames (ORFs) that had (1) no BlastP hits against the NCBI-nr, KEGG (Kyoto Encyclopedia of Genes and Genomes), UniProtKB/Swiss-Prot, TCDB and InterPro databases; (2) a Blast best-hit product containing one of the following keywords: ‘hypothetical', ‘putative', ‘unknown', ‘uncharacterized' or ‘DUF' in UniProtKB/Swiss-Prot, NCBI-NR or KEGG; (3) only hits to InterPro; and (4) no reliable BlastP hits against UniProtKB/Swiss-Prot, KEGG or NCBI-nr, subject coverage <50 and query coverage ⩾80 were categorized as ‘hypothetical'. ORFs on borders of contigs or that had amino-acid sequence lengths <60 residues were invalidated. ORFs with reliable BlastP hits against the KEGG, NCBI-nr or UniProtKB/Swiss-Prot database, with subject and query coverage ⩾60%, identities ⩾25% and positives ⩾40%, were categorized as ‘valid'. For these ORFs, the product and the EC (Enzyme Commission) number were imported from the database. A manual annotation was performed for those ORFs that did not fit the above criteria. The whole-genome shotgun sequencing project developed in this study has been deposited at DDBJ/EMBL/GenBank under the accession AFB (PRJNA52963 ID: 52963). The comparative analysis parameters were 1e−05, 22% and 49% for the E-value, coverage value and similarity value, respectively.
Enrichment culture
The enrichment medium developed in this study contained, per liter of distilled water: NaCl 342 mM, MgCl2·6H2O 5 mM, Na2SO4 35 mM, CaCl2 5 mM, KCl 6 mM, C2H3NaO2 6 mM, C4H4Na2O4·6H2O 2 mM, NH4Cl 5.6 mM, cysteine-HCl 2.5 mM, 5 ml modified Wolfe's minerals (Frankel et al., 1997), 1 ml vitamin solution (Frankel et al., 1997), 0.00004% resazurin, NaHCO3 60 mM, Na2S·9H2O 1.6 mM and ferric quinate 0.02 mM. The final pH of the medium was adjusted to 7.4 and the medium was purged with a mix of 93% N2:7% CO2.
Phylogenetic analysis based on biomineralization genes
DNA and amino-acid sequences were aligned using T-Coffee (Notredame et al., 2000) and were manually checked. A DNA or amino-acid substitution model was previously selected for phylogenetic reconstruction using ModelGenerator version 0.85 (Keane et al., 2006) and Bayesian information criterion. A maximum-likelihood phylogeny was then carried out with PHYML version 3.0 (Guindon and Gascuel, 2003). The tree topology space was searched using the best of the Nearest-Neighbor Interchange and Subtree Pruning and Regrafting algorithms, from five random starting trees generated by the BioNJ algorithm (Guindon and Gascuel, 2003; Guindon et al., 2010). Branch support was calculated using the approximate likelihood ratio test with SH-like interpretation (SH-aLRT). Maximum-likelihood reconstructions based on DNA sequences were carried out for 16S rRNA encoding gene, including both magnetotactic and non-magnetotactic representatives of each of the classes Alpha-, Beta-, Gamma-, Epsilon- and Deltaproteobacteria and the phylum Nitrospirae.
Phylogenetic analysis of the putative multicellular morphogenesis-related proteins
Amino-acid sequences of ORFs annotated as hemagglutinin-like proteins, adhesion-like proteins, glycoproteins, integrin-like proteins, fibronectin-like proteins and proteins that were more similar according to BlastP analysis were aligned with the CLUSTALW multiple alignment tool in the BioEdit sequence alignment editor (Hall, 1999). Phylogenetic trees were prepared with MEGA version 5.0 (Tamura et al., 2011), applying the neighbor-joining method (Saitou and Nei, 1987). Bootstrap values were calculated with 1000 replicates.
Fluorescent in situ hybridization
Fluorescent in situ hybridization was performed according to Pernthaler et al. (2001). The formamide concentration was 30% for the Ca. M. multicellularis probe (Abreu et al., 2007); MMP 1991-specific probe (DeLong et al., 1993) and the EUB 338, EUB 338II and EUB 338III bacterial probes (Amann et al., 1990; Daims et al., 1999). Microscopy observations were performed using a Zeiss Axioplan 2 (Carl Zeiss, Oberkochen, Germany), microscope equipped for fluorescence microscopy.
Transmission electron microscopy
Samples were prepared for TEM according to Keim et al. (2004a). Thin sectioning was performed in a Leica ultramicrotome (Leica Microsystems, Vienna, Austria), and the grids were stained with uranyl acetate and lead citrate and observed on a FEI Morgagni (FEI Company, Eindhoven, The Netherlands) TEM at 80 kV.
Results
General genomic features
The general genomic features of Ca. M. multicellularis are summarized in Table 1. On the basis of sequencing data, the genome size is approximately 12.5 Mb. However, this figure might be overestimated because not all contigs could be assembled and superposed regions might not have been identified. However, we consider our value higher than unicellular phylogenetically related bacteria-like Desulfobacterium autotrophicum with a 5.6 Mb genome (Strittmatter et al., 2009). Similar unusual large genome sizes have been reported for Myxococcus xanthus, which has a complex multicellular developmental cycle characterized by fruiting-body formation (Chen et al., 1990). According to Lander and Waterman (1988), we estimate that approximately 90% of the entire genome has been covered by pyrosequencing.
Table 1. General information from pyrosequencing data.
| Genome coverage | 18 × |
| Length (bp) | 12 479 246 |
| G+C content (%) | 37.27 |
| Coding density (%) | 76% |
| Average of ORF length (bp) | 954 |
| Number of contigs | 3706 |
| Total number of ORFs | 9987 |
| Contig average size (bp) | 3367 |
| Single copy genes average coverage | 15 × |
| Number of known protein ORFs | 3616 |
| Number of partial ORFs | 28 |
| Number of truncated ORFs | 140 |
| Number of hypothetical ORFs | 6203 |
| rRNA | 3 |
| rRNA 16s | 1 |
| rRNA 23s | 1 |
| rRNA 5s | 1 |
| tRNA | 46 |
| KEGG matches | 75% |
| InterPro matches | 82% |
Abbreviations: KEGG, Kyoto Encyclopedia of Genes and Genomes; ORF, open reading frames.
The KEGG classification of ORFs, according to function, show that the majority of predicted ORFs are related to carbohydrate and amino-acid metabolism. Other major groups of ORFs are involved in cell motility including chemotaxis, signal transduction and cofactor and vitamin metabolism. Ca. M. multicellularis contains at least one copy of an rRNA operon and 46 genes for tRNAs. Comparison of the partial genome of Ca. M. multicellularis with those of Desulfatibacillum alkenivorans, Desulfococcus oleovorans and Desulfobacterium autotrophicum, bacteria most similar to Ca. M. multicellularis whose genomes have been sequenced, shows that approximately 18% of the ORFs from Ca. M. multicellularis cluster with ORFs from these bacteria. According to BlastP analysis (1e−05 minimum E-value, 60% coverage and 50% similarity), Desulfatibacillum alkenivorans, Desulfococcus oleovorans and Desulfobacterium autotrophicum have 1417, 1245 and 1358 similar genes with Ca. M. multicellularis, respectively. Similar genes shared by these bacteria form 907 clusters, which contain mostly ORFs encoding proteins involved in DNA and RNA metabolism (198 clusters of ORFs), structural proteins (135 clusters of ORFs) and hypothetical proteins (55 clusters of ORFs).
Comparison among magnetotactic bacterial genomes
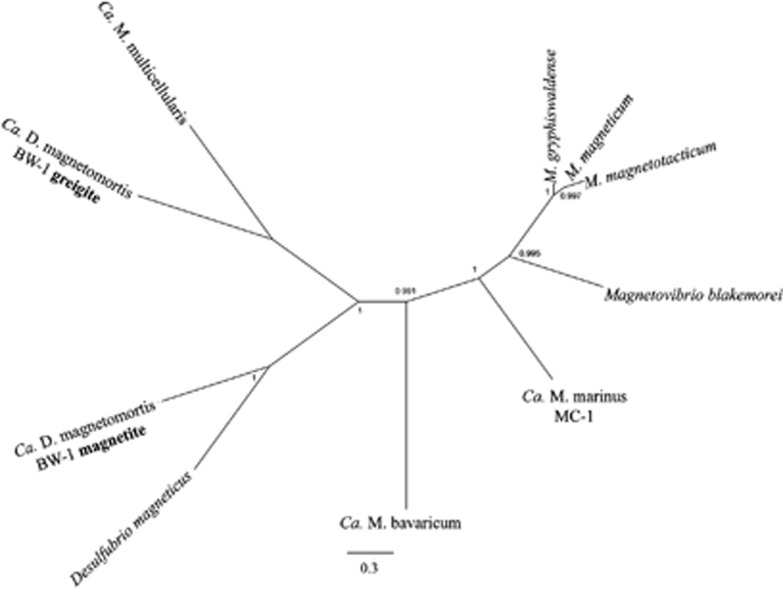
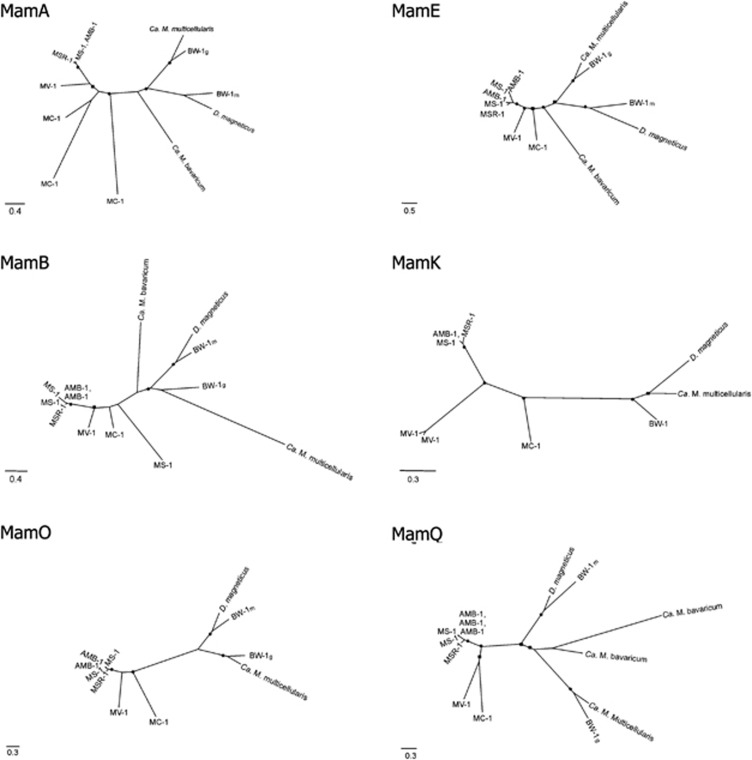
ORFs homologous to those thought to be important in magnetosome biomineralization including mamA, -B, -E, -K, -M, -O, -P, -Q and -T were previously described in Ca. M. multicellularis and phylogenetic analysis indicated that the biomineralization of magnetite and greigite evolved from a common ancestor (Abreu et al., 2011; Lefèvre et al., 2013a). Here, we compared magnetotactic bacterial genomes based on the coverage and similarity value percentages used to analyze biomineralization genes on a previous study (1e−05 E-value, 22% coverage and 50% similarity; Abreu et al., 2011). Ca. M. multicellularis and Desulfovibrio magneticus, both of which belong to the Deltaproteobacteria group, have 938 genes that cluster. Among the Alphaproteobacteria, Magnetospirillum magnetotacticum had the closest relationship with Ca. M. multicellularis, with 865 genes that cluster. Some of the shared ORFs encode hypothetical proteins, chemotactic proteins, integrases, transposases and CRISP-associated protein. Phylogenetic analysis based on concatenated amino-acid sequences of conserved mam genes indicate that the Ca. M. multicellularis mam genes are more similar to those of Candidatus Desulfamplus magnetomortis strain BW-1 (Lefèvre et al., 2011, 2013a), which are believed to be responsible for greigite magnetosome synthesis (Figure 1). When repeated mam genes (mamA, -B, -E, -K, -O and -Q) were analyzed separately (Figure 2), Magnetovibrio blakemorei and Magnetococcus marinus appeared to be most closely related to each other in most cases. On the basis of the phylogenetic analysis of MamQ, Mv. blakemorei and Mc. marinus are clustered, suggesting a more recent divergence between these species, which is not congruent to the phylogenetic analysis based on 16S rRNA gene. Pairwise protein distances matrix using similarity for MamA, -B, -E, -K, -O and -Q are displayed on Supplementary Table 1. The results also showed that the Magnetospirillum species are closely related and major sequence divergence among duplicated sequences might have resulted from gene duplication and mutation. The close relationship among Magnetospirillum species, which can be identified by the formation of a cluster containing these species in any gene tree analysis and the positioning of Mv. blakemorei and Mc. marinus in a clade, suggests that the hypothesis presented by Jogler et al. (2009), which states that magnetosome genes were acquired by recent ancestors of Magnetospirillum that subsequently separated into different species with independent horizontal gene transfer of magnetosome genes from an unknown ancestor into Mc. marinus and Mv. blakemorei, is reasonable. Considering the recent hypothesis that magnetotaxis originated monophyletically in the Proteobacteria phylum that states that the common ancestor of all Proteobacteria was magnetotactic and transferred the magnetosome genes by descendent (Lefèvre et al., 2013a), the sequence similarity between Mc. marinus and Mv. blakemorei might be a result of a lower mutation rate in the mamQ gene.
Figure 1.
Phylogenetic tree based on concatenated amino-acid sequence of conserved magnetosome biomineralization genes. Note that concatenated sequences of Ca. M. multicellularis are most closely related to the sequences likely involved in greigite biomineralization from cultivated Ca. Desulfamplus magnetomortis strain BW-1. Numbers close to branches are SH-aLRT values.
Figure 2.
Maximum-likelihood phylogenetic analyses of available sequences of magnetotactic bacteria based on individual genes including mamA, mamB, mamE, mamK, mamO and mamQ. Phylogenies of mamA and mamQ was constructed using the LG+I+Γ amino-acid substitution model; mamB and mamK using the LG+Γ model; mamE using the LG+I+Γ+F model; mamO using the LG+Γ+F. Black circles are SH-aLRT values between 0.90 and 1.00; and black squares are SH-aLRT values between 0.80 and 0.89. To understand mam gene duplication, repeated ORFs available on magnetotactic bacteria were analyzed. Note that usually sequences from the same species grouped together. Magnetospirillum species are always closely related to each other and Magnetovibrio blakemorei (MV-1) and Magnetococcus marinus (MC-1) MamQ formed a clade, suggesting recent divergence between these proteins in these species. Ca. Desulfamplus magnetomortis (BW-1) magnetite-related sequences are grouped with Desulfovibrio magneticus sequences, whereas its greigite magnetosome sequences are closely related to Ca. M. multicellularis.
Individual Mam protein analysis of Ca. D. magnetomortis, Desulfovibrio magneticus, Ca. M. multicellularis and Ca. M. bavaricum protein sequences, which involve both magnetite and greigite magnetosome production, show that these sequences are more related to each other than to Alphaproteobacteria magnetite-related protein sequences (Figure 2). This result is in agreement with previously studies based on concatenated Mam protein phylogenetic analysis (Abreu et al., 2011; Lefèvre et al., 2013a). Phylogeny based on MamQ shows that both Ca. M. bavaricum protein sequences are more related to the greigite-related protein belonging to Ca. M. multicellularis and Ca. D. magnetomortis, indicating an ancient divergence of greigite biomineralization from the magnetite biomineralization process. Recently, comparative genomic analysis of magnetotactic bacteria from the Deltaproteobacteria showed that these bacteria have a conserved set of genes specific for the group, the mad genes (magnetosome associated deltaproteobacteria genes). Interestingly, among all known magnetotactic bacteria, only Ca. M. bavaricum has genes homologous to the mad genes (Lefèvre et al., 2013b).
In addition to those genes related to magnetosome synthesis, other ORFs of Ca. M. multicellularis are present in other magnetotactic bacteria. According to the bidirectional Blast conducted for the sequences of Ca. M. multicellularis and magnetotactic bacterial species (M. gryphiswaldense, M. magneticum, M. magnetotacticum, Mc. marinus, Mv. blakemorei, Desulfovibrio magneticus), 17% of Ca. M. multicellularis ORFs cluster with other magnetotactic bacterial genes. Interestingly, the ferrous iron transporter-encoding ORF found near mam genes in Ca. M. multicellularis is more closely related to that from Ca. D. magnetomortis (99% coverage and 60% identity) and to D. magneticus (96% coverage and 46% identity) than to the 16S rRNA gene phylogenetically related bacteria. Two other ferrous iron transporters encoded in the partial genome are more similar to different non-magnetotactic Deltaproteobacteria: Desulfatibacillum alkenivorans, Desulfobacterium autotrophicum and Desulfobacter postgatei (coverage higher than 93% and similarity higher than 67%). Comparison of the Ca. M. multicellularis FeoB whose gene is in close proximity to the mam genes and other FeoB encoding gene found in the partial genome showed coverage of approximately 26% and similarity ranging from 29% to 39%. This result indicates that ferrous iron transport, which is directly related to magnetosome production, can be considered specific to this process and might have emerged and evolved in the same way than other magnetosome-related genes (Lefèvre et al., 2013b).
Genomic analysis of Ca. M. multicellularis reveals genes involved in circadian cycle, chemotaxis and phototaxis
The circadian cycle genes kaiC and kaiB were identified in Ca. M. multicellularis. Each of these genes is present in two copies: the kaiC copies are 56% similar and have 76% identity, whereas the kaiB copies are 22% similar and have 51% identity. BlastP analysis showed that both copies of kaiC are approximately 45% identical and have 68% positives compared with D. postgatei. For the kaiB copies, the most similar sequences belong to cyanobacterium Cyanobacterium stanieri PCC 7202, with 42% identity and 65% positives for one copy of the gene, whereas the other copy is more closely related to that of Desulfonatronospira thiodismutans ASO3-1, with 40% identity and 63% positives. Observation of environmental samples containing Ca. M. multicellularis immediately after collection during the day and night showed that the microorganism moves downward at night (see Supplementary Figure 1); the number of dividing microorganisms during this period was higher, suggesting that the multicellular life cycle might be under the control of a circadian rhythm. Cell motility-related genes were classified as structural (40 ORFs) or chemotactic components (120 ORFs). Chemotaxis- and signal-transduction-related genes represented the majority of ORFs assigned according to KEGG functional classification (see Supplementary Table 2).
Regarding photo-sensing mechanisms, two putative genes were identified as red-light sensors in the partial genome of Ca. M. multicellularis; one of them is related to the DSM 4028 phytochrome sensor protein of the sulfate-reducing bacterium Desulfomicrobium baculatum (coverage 95% E-value 1e−176; identity 46%), whereas the other is similar to the Hxd3 putative phytochrome sensor protein of Desulfococcus oleovorans (coverage 95% E-value 7e−46; identity 42%). Importantly, we detected putative genes that shared homology with LOV (light, oxygen, voltage) domain proteins and photoactive yellow protein, both of which are involved in blue-light sensing in bacteria; genes related to protoporphyrin IX, which also senses blue light; and genes that share homology with the amino-acid motif GGDEF (Gly-Gly-Asp-Glu-Phe) and its conversed residues EAL domain proteins, which might be involved in light-dependent gene regulation, particularly blue light. The GGDEF and EAL domain proteins are responsible for increasing or decreasing cellular levels of cyclic di-GMP, the second messenger specific to bacteria that is responsible for the transition from single motile cells to sessile multicellular communities (Gomelsky and Hoff, 2011). Finally, also of note was that no carotenoid synthesis-related genes were detected.
Genes putatively responsible for multicellularity in Ca. M. multicellularis are closely related to genes present in filamentous bacteria.
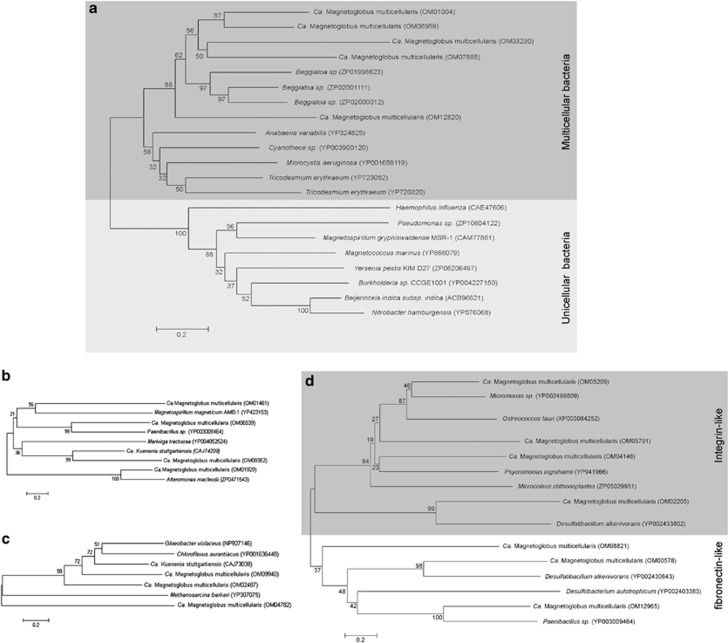
Adhesion molecules and possibly calcium-dependent proteins have important roles in the unique morphology and life cycle of Ca. M. multicellularis (Keim et al., 2004b; Abreu et al., 2013). Thus, we examined the genome of Ca. M. multicellularis thoroughly for potential proteins involved in cell adhesion and the development of multicellular morphogenesis in this organism (Figure 3). Annotated proteins include cell adhesion domain-containing proteins, adhesion-like proteins, fibronectin type III domain-containing proteins, large exoproteins exhibiting hemagglutination activity, filamentous hemagglutinin outer membrane proteins, cell surface glycoproteins, carbohydrate-binding proteins, integrin-like repeat-containing proteins, and α-β-propeller repeat-containing integrins. Type IV pilus-encoding ORFs and extracellular polysaccharide biosynthesis-related genes were also identified that might be involved in cellular adhesion as mentioned previously. The cell adhesion proteins and fibronectin domain-containing proteins found in Ca. M. multicellularis are also associated with those of unicellular bacteria, including the heme utilization/adhesion protein of Magnetospirillum magneticum; these adhesion proteins are involved in the formation of biofilms, a form of multicellular lifestyle (Shank and Kolter, 2011). Interestingly, hemagglutinin protein encoding ORFs detected in Ca. M. multicellularis sequences are mostly associated with those of filamentous multicellular bacteria (Figure 3a). Gene homologues of tyrosine kinases, C-type lectin and cadherins, which are the first known protozoan homologues of animal genes involved in cell signaling and cell adhesion (King et al., 2003), were also identified in the genome of Ca. M. multicellularis.
Figure 3.
Phylogenetic tree based on amino-acid sequences of proteins possibly involved in the multicellular morphogenesis of Ca. Magnetoglobus multicellularis. (a) Hemagglutinin-like protein; (b) adhesion-like protein; (c) glycoprotein; and (d) integrin- and fibronectin-like proteins. Note in (a) that Ca. M. multicellularis hemagglutinin-like proteins cluster with those belonging to multicellular filamentous bacteria.
The partial genome of Ca. M. multicellularis was compared with those of distantly related eukaryotic (Dictyostelium discoideum and Volvox carteri) and prokaryotic (Myxococcus xanthus, Anabaena variabilis, Beggiatoa alba) multicellular microorganisms to detect genes possibly related to the development of a multicellular morphology. B. alba has the highest number of similar ORFs (937), followed by Myxococcus xanthus (932). For A. variabilis, D. discoideum and V. carteri; the numbers of clustered ORFs were 851, 522 and 369, respectively. Among these clusters, a few groups of genes can be considered interesting candidates for the development of a multicellular morphology. The genomes of Ca. M. multicellularis, B. alba, Myxococcus xanthus and A. variabilis contain genes closely related to von Willebrand factor A, which is a glycoprotein with cell adhesion properties involved in blood coagulation in higher eukaryotes. One gene of Ca. M. multicellularis that encodes von Willebrand factor A is similar to von Willebrand factor A genes in Flavobacterium columnare (90% coverage; 3e−78 E-value; 60% identity), Chitinophaga pinensis (100% coverage; 6e−76 E-value; 57% identity) and Microcoleus chthonoplastes (99% coverage; 4e−75 E-value; 57% identity); all these bacteria have a filamentous morphology but are not closely related to Ca. M. multicellularis phylogenetically. Other genes of Ca. M. multicellularis related to this protein are more closely related to a hypothetical protein of Desulfatibacillum alkenivorans, whereas two others are more closely related to von Willebrand factor type A from the same bacterium. Von Willebrand-like genes are also found in unicellular magnetotactic bacteria, but some sequences present in Ca. M. multicellularis are more closely related to nonmagnetic bacteria with filamentous morphology. Clusters of ORFs containing protein–protein interaction motifs, WD40 repeat-containing protein and ankyrin repeat-containing protein included all the multicellular microorganisms analyzed. Different clusters of ORFs from Ca. M. multicellularis, Myxococcus xanthus, B. alba and Deinococcus radiodurans related to the GGDEF family of proteins were also present.
Energy metabolism and physiology of Ca. M. multicellularis based on genomic analysis
Ca. M. multicellularis is an anaerobic microorganism that produces energy from the oxidation of organic compounds and the dissimilatory reduction of sulfate. Carbon metabolism enzymes are related to organic and fatty acid and alcohol utilization. TRAP (tripartite ATP-independent periplasmic) transporters involved in C4-dicarboxylate uptake were also identified. Our data suggest that Ca. M. multicellularis is able to utilize succinate, acetate, formate and malate as substrates as the Ca. M. multicellularis genes for enzymes involved in the utilization of these carbon sources correspond to their respective homologs in other sulfate-reducing bacteria. Ca. M. multicellularis can most likely also completely oxidize acetyl-CoA through the tricarboxylic acid cycle but also has genes homologous to those of the reductive acetyl-CoA pathway indicating that Ca. M. multicellularis is capable of both heterotrophic and autotrophic growth.
The genes for key enzymes for sulfate reduction were identified in the genome of Ca. M. multicellularis. Some of these ORFs encode ATP-sulfurylase (Sat) and sulfate permease probably involved in sulfate transport. Adenyl-sulfate reductase (apr; α and β subunits) and sulfite reductases (dsr; α and β subunits), responsible for the activation of sulfate during sulfate reduction, were also identified. The protein products of AprA and AprB are more similar to those of Desulfatibacillum alkenivorans and have an 86% identity in both cases. The protein products of DsrA and DsrB were more related to Desulfococcus oleovorans, having 83% and 81% identity, respectively. Phylogenetic analysis (not shown) of Aps, Dsr and Sat indicates Desulfatibacillum alkenivorans as the most closely related species to Ca. M. multicellularis.
ORFs involved in protection against reactive oxygen species are present in the partial genome of Ca. M. multicellularis. Several ORFs are related to cytochrome d ubiquinol oxidase subunits I and II, which are involved in oxygen respiration and have a protective purpose (Cypionka, 2000), and to superoxide reductase and rubrerythrin (involved in oxygen detoxification). Several predicted ORFs in Ca. M. multicellularis encode proteins similar to the DcrA protein, which is involved in an oxygen-sensing mechanism in Desulfovibrio vulgaris, indicating that this mechanism might also exist in Ca. M. multicellularis. Thus, this microorganism might not be a strict anaerobe and might have protective mechanisms that guarantee its survival in oxic environments.
Ca. M. multicellularis inhabits hypersaline lagoons, thus it is reasonable to assume that it uses a Na+-dependent energy transduction mechanism similar to those described for many marine bacteria (Oh et al., 1991). The presence of ORFs encoding Na+-transporting NADH:ubiquinone oxidoreductase subunits A–F, an F-type H+-transporting ATPase (which translocates either protons or sodium ions) and other sodium transport-related proteins demonstrates the great adaptation of the strain to life under high salt concentrations. It is also possible that Ca. M. multicellularis utilizes a Na+ pump-dependent respiration.
Enrichment culture
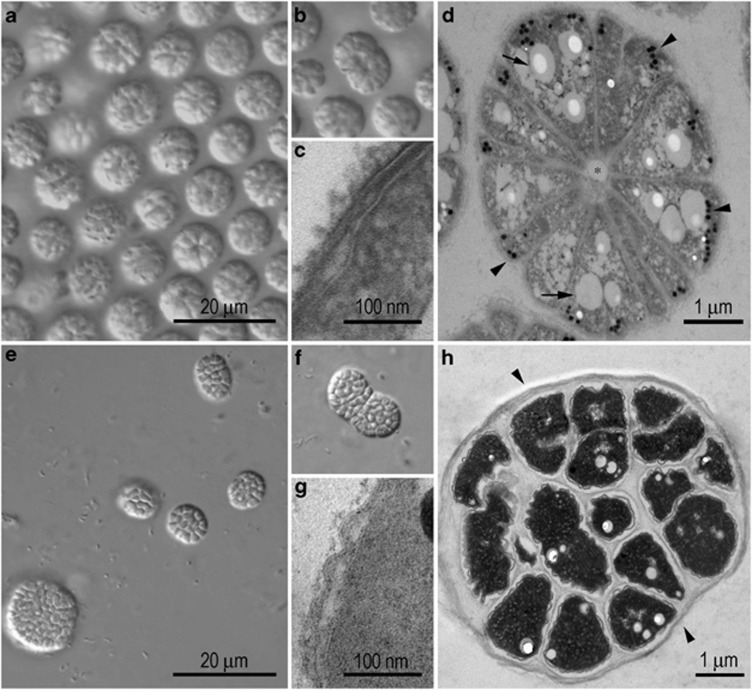
Magnetically enriched Ca. M. multicellularis, approximately 5 × 103 individuals obtained after magnetic enrichment, was inoculated into semisolid medium incubated at 28 °C. Up to 2 weeks after inoculation, the MMPs lost motility and the ability to respond to magnetic fields. Bacterial growth, in the form of turbidity, was observed after a month of incubation, including that of uncharacterized unicellular and multicellular bacteria (Figures 5c and d). Light microscopy and TEM showed that a multicellular microorganism similar in morphology to Ca. M. multicellularis grew in the medium (Figures 4a–g). Occasionally, they appeared to lose the ability to control the division process, resulting in huge aggregates of cells with a diameter of about approximately 40 μm. Despite these differences, fluorescence in situ hybridization with probes specific to MMPs (DeLong et al., 1993; Abreu et al., 2007) confirmed that the microorganisms in culture were Ca. M. multicellularis (Figure 5). Nevertheless, we did not detect independent unicellular cells of Ca. M. multicellularis by fluorescent in situ hybridization labeling, confirming that the previously proposed completely multicellular life cycle of uncultivated Ca. M. multicellularis (Keim et al., 2004a) also occurs in the microorganisms in culture (Figure 4e). Most of the predicted genes for cell envelope and appendages are classified as cell envelope constituents. Notably, those involving capsular polysaccharide and exopolysaccharide biosynthesis were also identified, corroborating our microscopy observations that Ca. M. multicellularis possesses a loosely linked capsule on the cell surface in contact with the external environment. When maintained in enrichment culture, its surface was covered by a thick layer (Figure 4g), and the microorganisms grew in clumps that adhered to the tube, indicating exopolysaccharide production, known to occur in biofilm formation (Figure 4f). Flagellar component-related (for export), several pilus-related and two fimbria-related genes were also identified, including that encoding a mannose-sensitive hemagglutinin pilus. In Ca. M. multicellularis, these pili-related genes might influence adherence among cells possessing a multicellular morphology and/or for substrate adherence under certain conditions as observed in the culture medium. Because Ca. M. multicellularis grew as clumps usually adhering to the glass surface, it was impossible to determine cell density after inoculation and doubling time.
Figure 4.
Morphology of Ca. Magnetoglobus multicellularis. Differential interference microscopy of uncultivated (a) and dividing (b) Ca. M. multicellularis. TEM image of an ultrathin section of uncultivated Ca. M. multicellularis showing a Gram-negative type of cell wall (c). (d) EM image of an ultrathin section of an intact uncultured Ca. M. multicellularis showing magnetosomes (short arrowheads), lipid granules (long arrows) and the central acellular compartment (asterisk). Enrichment cultures show growth of a microorganism with a similar morphology (e) and cell division pattern (f) as Ca. M. multicellularis viewed here with differential interference light microscopy. TEM image of an ultrathin section of cultured Ca. M. multicellularis again showing a Gram-negative cell envelope profile. (g) TEM image of an ultrathin section of a Ca. M. multicellularis from the enrichment culture showing a thick capsule layer (arrowhead).
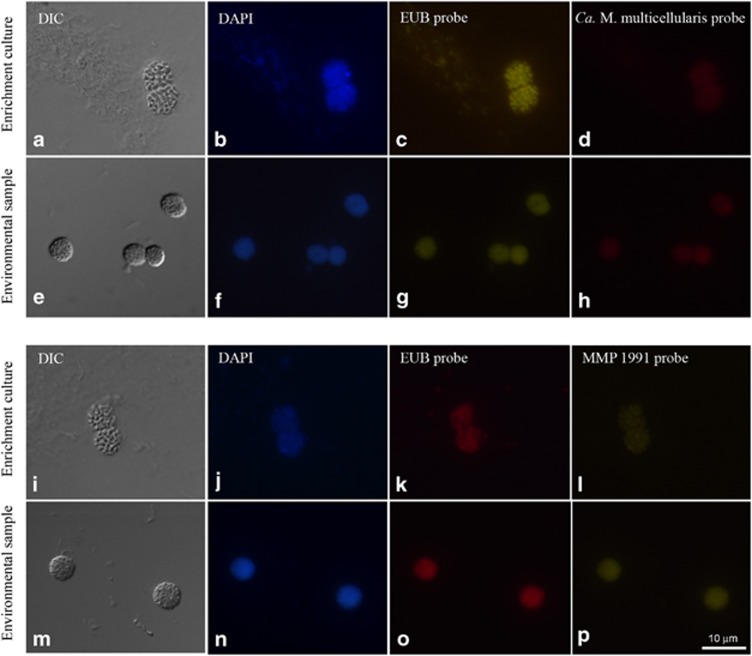
Figure 5.
Fluorescent in situ hybridization (FISH) of environmental and cultivated Ca. Magnetoglobus multicellularis. (a, e, i and m) Differential interference contrast images of Ca. M. multicellularis; (b, f, j and n) Ca. M. multicellularis strained with 4',6-diamidino-2-phenylindole (DAPI); (c and g) Bacteria-specific probe (Amann et al., 1990; Daims et al., 1999) labeled with Alexa 488; (k and o) Bacteria-specific probe labeled with Alexa 594; (d and h) Ca. M. multicellularis- specific probe labeled with rhodamine (Abreu et al., 2007); (l and p) MMP 1991-specific probe labeled with FITC (DeLong et al., 1993). Despite the fact that contaminant bacteria are present in both cultured and uncultured preparations of Ca. M. multicellularis, and stain with DAPI, these cells do not hybridize with specific probes for Ca. M. multicellularis or MMPs.
Discussion
We used a culture-independent, genomic approach to characterize the uncultured multicellular microorganism Ca. M. multicellularis. We have shown that the magnetotactic response of swimming uncultured Ca. M. multicellularis and the use of the double magnetic enrichment performed here is an efficient method to obtain highly purified suspensions of Ca. M. multicellularis from environmental water and sediment samples. Although we obtained an enrichment culture, the microorganisms were not motile or magnetotactic, which makes the purification of the enrichment culture individuals using the magnetic field orientation unrealistic. When we extracted DNA from the enrichment culture, amplified and sequenced 16S rRNA coding genes, we detected contaminant bacterial 16S rRNA gene sequences not detected in the uncultured samples. Thus, we concluded that the magnetically concentrated MMPs were more appropriate for genome sequencing than the cultivated ones because there was a higher possibility of retrieving non-Magnetoglobus bacterial genes from the cultivated than the uncultured individuals that might have made assembly of the genome more arduous. Moreover, the much thicker capsule of the cultivated Ca. M. multicellularis made the extraction of DNA from these MMPs difficult and was thus most likely responsible for the preferential amplification of 16S rRNA genes from the unicellular unrelated bacteria. In addition, our cultures could not be considered completely homogeneous because the population is not clonal (it is an enrichment, not a pure culture). Therefore, we expected that our cultures would exhibit the same degree of cell-to-cell variability and thus there appeared to be no advantage to sequencing MMPs from the enrichment culture over sequencing those motile, magnetically purified individuals collected from environmental samples. In addition, there was also the possibility of gene loss, for example, the loss of mam genes, which has been shown to occur in cultures of some magnetotactic bacteria that also lost the magnetotactic trait (Ullrich et al., 2005; Kolinko et al., 2011). This unique application of genomic sequencing and analysis of microbial environmental DNA contributed to the understanding of many physiological features involved in the behavior and multicellular life of Ca. M. multicellularis. The detection of metabolism-related genes guided our development of an enrichment medium for Ca. M. multicellularis. The medium developed here supported chemoorganoheterotrophic growth of Ca. M. multicellularis, allowing them to maintain their multicellular morphology and cell cycle. Unfortunately, its magnetic properties and motility were lost during cultivation. This might have been related to the absence of a chemical gradient in the culture medium or to certain growth conditions, as occurs in the loss of flagella reported for Escherichia coli when grown under certain favorable or unfavorable conditions (e.g., high temperature, high salt concentration, high carbohydrate concentrations) (Li et al., 1993). Recently, it was shown that MMPs from yellow sea were specifically adapted to variable environments such as intertidal zones (Zhou et al., 2012); it is possible that such adaptation is also present in Ca. M. multicellularis and that specific gradient media may be needed to obtain motile microorganisms.
Figure 6 depicts our interpretation of the biology of Ca. M. multicellularis based on genomic pyrosequencing data and microscopy observations. According to our results, this microorganism is an anaerobic bacterium that swims between the anoxic and suboxic layers of sediment because of the different availabilities of nutrients such as iron and nitrogen (Sobrinho et al., 2011). The presence of mam genes (Abreu et al., 2011) and others that encode chemotaxis- and phototaxis-related proteins indicate that Ca. M. multicellularis uses the interactions between magnetotaxis, chemotaxis and phototaxis to determine their optimal position in environmental gradients and swim according to their nutrient requirements. The presence of genes involved in protective mechanisms against oxygen radicals would help the organism to survive during excursions to the aerobic layers of sediment (or in aerobic preparations of the organism for observation with light microscopy). It is reasonable to assume that these microorganisms continuously swim across chemical gradients in the environment and that the differential spatial availability of nutrients may create metabolic imbalances, which might be reflected in the production of storage granules of polyhydroxyalkanoates (genes encoding several granule synthesis proteins were identified and annotated in the Ca. M. multicellularis genome), polyphosphates and others (Silva et al., 2008). In the anoxic zone, sulfate reduction and H2 energy metabolism might occur, and magnetosome formation should also be favored, whereas migration to the suboxic or oxic zone could be driven by nutrient requirements. In environmental samples, a fraction of Ca. M. multicellularis always displays north-seeking behavior (Keim et al., 2007), which is the opposite of the expected swimming direction in the southern hemisphere. In this case, north-seeking behavior might be regulated by light (Shapiro et al., 2011), stimulating the microorganism to move upwards to find nitrogen. In this sense, the magnetic field orientation is supported by phototaxis (Shapiro et al., 2011) and aerotaxis (Frankel et al., 1997), resulting in a more efficient chemotaxis. Therefore, the presence of light and an excess of oxygen would result in the same type of behavior, reversal of the swimming direction along the axis determined by the magnetic field, possibly because of the generation of a high concentration of reactive oxygen species that could not be reduced by cellular protective mechanisms. This hypothesis is also supported by the fact that, when exposed to high-intensity UV light, multicellular magnetotactic bacteria changed their magnetotactic behavior and began swimming in the opposite direction and did not reverse direction even if illuminated again (Shapiro et al., 2011).
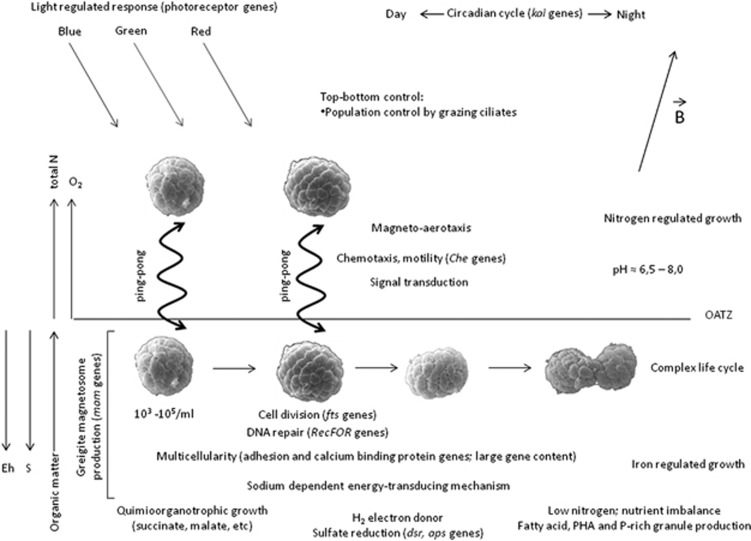
Figure 6.
Schematic representation of the potential ecological roles of Ca. Magnetoglobus multicellularis based on genomic pyrosequencing data and phenotypic characterization. Ca. M. multicellularis is usually found in anoxic and suboxic layers of the sediment; it appears to swim to an optimal position in vertical chemical gradients present at the site depending on its metabolic requirement. Sulfidic-reducing conditions are present in the bottom anoxic layers of the sediment, whereas the concentrations of organic matter and nitrogen sources are higher in the suboxic layers (Sobrinho et al., 2011). Because of the presence of several light receptor-related genes in the partial genome of Ca. M. multicellularis, microscopic observations of environmental samples and previous studies (Lefèvre et al., 2010; Shapiro et al., 2011), Ca. M. multicellularis appears to also use light for orientation in addition to magnetoaerotaxis and chemotaxis. See text for details.
The presence of protoporphyrin IX-related genes in the genome of Ca. M. multicellularis, which encode blue-light-sensing proteins and the absence of carotenoid synthesis genes, which encode proteins responsible for protection from light damage (Yang et al., 1995) support the existence of an intracellular signaling response similar to that under either intense light exposure or high-oxygen conditions. In the absence of carotenoids, the reactive oxygen species generated by intense light would lead to cell damage in a similar way when compared with oxygen exposure. Phototaxis away from blue light is observed in an Escherichia coli mutant that accumulates protoporphyrin IX (Yang et al., 1995). The tumbling response in this case was hypothesized to be induced by reactive oxygen species generated by photosensitization of porphyrin. Protoporphyrin IX is also involved in the lysis of myxobacteria by intense blue light in the absence of carotenoids (Yang et al., 1995); similar lysis occurred during light microscopy observation of Ca. M. multicellularis. Preliminary chemotaxis experiments under low-oxygen conditions showed that the magnetic polarity did not revert even in the presence of undesirable substrates. In this situation, Ca. M. multicellularis started to engage in ping-pong movements but did not change their magnetic polarity. Therefore, this ping-pong movement could be an alternative escape mechanism for rapid egress from a potentially toxic zone using the geomagnetic field (Keim et al., 2007; Martins et al., 2007).
The detection of other genes that encode blue-light-sensing receptors and circadian cycle proteins, the presence of genes encoding GGDEF and EAL domain proteins, previous observations of light-dependent behavior (Almeida et al., 2013) and the fact that during the day Ca. M. multicellularis localized to the upper layers of sediment indicate that a physiological process besides orientation might be governed by light sensing. The multicellular life cycle is considered essential to maintaining the magnetic polarity of newly formed individual cells (Keim et al., 2004a) and might be regulated by light. In many bacteria, blue-light receptors are mainly considered to be involved in the transition between individual planktonic and biofilm lifestyles or between environmental and host-associated lifestyles (Gomelsky and Hoff, 2011). This light-dependent lifestyle is supported by the fact that classes of blue-light receptors are implicated in controlling different aspects of this transition in many bacteria and also by the fact that bioinformatics analysis indicates that these photoreceptors are linked to the GGDEF and EAL protein domains, which are involved in cell signaling pathways (Gomelsky and Hoff, 2011).
Evolution of multicellularity in the Domain Bacteria is not completely understood. Multicellularity evolved independently several times in Eukarya domain (Sachs, 2008) and the same could have occurred in Bacteria. The analysis of multicellular bacterial genomes and of gene expression under different culture conditions might be fundamental for characterizing the metabolic pathways and cellular structure needed for sustaining multicellularity. According to our results, Ca. M. multicellularis has several potential genes dedicated to multicellular organization and lifestyle. Thus, the establishment of a pure culture, the complete sequencing of bacterial genomes and the development of a genetic system and proteomic analysis under different growth conditions are necessary to improve our understanding of multicellular evolution in bacteria. For now, however, the information acquired through the metagenomic approach presented here and the enrichment culture developed based on this information represent a fundamental step toward understanding the requirements for multicellularity.
Acknowledgments
This work was financially supported by the Brazilian agencies CNPq, CAPES and FAPERJ. DAB is supported by US National Science Foundation Grant EAR-0920718. CTL is the recipient of an award from the Fondation pour la Recherche Médicale (FRM: SPF20101220993). FFN is supported by the National Evolutionary Synthesis Center (NESCent) Grant NSF #EF-0423641.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abreu F, Cantão EG, Nicolás MF, Barcellos FG, Morillo V, Almeida LGP, et al. Common ancestry of iron oxide- and iron-sulfide-based biomineralization in magnetotactic bacteria. ISME J. 2011;5:1634–1640. doi: 10.1038/ismej.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu F, Martins JL, Silveira TS, Keim CN, Lins de BarrosHGP, Gueiros-Filho F, et al. ‘Candidatus Magnetoglobus multicellularis', a multicellular magnetotactic prokaryote from a hypersaline environment. Int J Syst Bacteriol. 2007;57:1318–1322. doi: 10.1099/ijs.0.64857-0. [DOI] [PubMed] [Google Scholar]
- Abreu F, Silva KT, Leão P, Guedes IA, Keim CN, Farina M, et al. Cell adhesion, multicellular morphology and magnetosome distribution in multicellular magnetotactic prokaryote Candidatus Magnetoglobus multicellularis. Microsc Microanal. 2013;19:535–543. doi: 10.1017/S1431927613000329. [DOI] [PubMed] [Google Scholar]
- Almeida FP, Viana NB, Lins U, Marcos F, Keim CN. Swimming behavior of the multicellular magnetotactic prokaryote ‘Candidatus Magnetoglobus multicellularis' under applied magnetic fields and ultraviolet light. Ant van Leeuw J Microb. 2013;103:845–857. doi: 10.1007/s10482-012-9866-0. [DOI] [PubMed] [Google Scholar]
- Almeida LG, Paixão R, Souza RC, Costa GC, Barrientos FJ, Santos MT, et al. A system for automated bacterial (genome) integrated annotation—SABIA. Bioinformatics. 2004;20:2832–2833. doi: 10.1093/bioinformatics/bth273. [DOI] [PubMed] [Google Scholar]
- Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angert ER. Alternatives to binary fission in bacteria. Nat Rev Microbiol. 2005;3:214–224. doi: 10.1038/nrmicro1096. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- Blakemore R. Magnetotactic bacteria. Science. 1975;190:377–379. doi: 10.1126/science.170679. [DOI] [PubMed] [Google Scholar]
- Chen H, Keseler IM, Shimkets LJ. Genome size of Myxococcus xanthus determined by pulsed-field gel electrophoresis. J Bacteriol. 1990;172:4206–4213. doi: 10.1128/jb.172.8.4206-4213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H-H, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Cypionka H. Oxygen respiration by Desulfovibrio species. Annu Rev Microbiol. 2000;54:827–848. doi: 10.1146/annurev.micro.54.1.827. [DOI] [PubMed] [Google Scholar]
- Daims H, Brühl A, Amann R, Schleifer K-H, Wagner M. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Frankel RB, Bazylinski DA. Multiple evolutionary origins of magnetotaxis in bacteria magnetotaxis. Science. 1993;259:803–806. doi: 10.1126/science.259.5096.803. [DOI] [PubMed] [Google Scholar]
- Eder W, Ludwig W, Huber R. Novel 16S rRNA gene sequences retrived from highly saline brine sediments of Kebrit Deep, Red Sea. Arch Microbiol. 1999;172:213–218. doi: 10.1007/s002030050762. [DOI] [PubMed] [Google Scholar]
- Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. Magneto-aerotaxis in marine coccoid bacteria. Biophys J. 1997;73:994–1000. doi: 10.1016/S0006-3495(97)78132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomelsky M, Hoff WD. Light helps bacteria make important lifestyle decisions. Trends Microbiol. 2011;9:441–448. doi: 10.1016/j.tim.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NY. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Henke JM, Bassler BL. Bacterial social engagements. Trends Cell Biol. 2004;14:648–656. doi: 10.1016/j.tcb.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Jogler C, Kube M, Schübbe S, Ullrich S, Teeling H, Bazylinski DA, et al. Comparative analysis of magnetosome gene clusters in magnetotactic bacteria provides further evidence for horizontal gene transfer. Environ Microbiol. 2009;15:1267–1277. doi: 10.1111/j.1462-2920.2009.01854.x. [DOI] [PubMed] [Google Scholar]
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29–45. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keim CN, Abreu F, Lins U, Barros HGP, Farina M. Cell organization and ultrastructure of a magnetotactic multicellular organism. J Struct Biol. 2004;145:254–262. doi: 10.1016/j.jsb.2003.10.022. [DOI] [PubMed] [Google Scholar]
- Keim CN, Martins JL, Abreu F, Rosado AS, Lins De Barros HP, Borojevic R, et al. Multicellular life cycle of magnetotactic prokaryotes. FEMS Microbiol Lett. 2004;240:203–208. doi: 10.1016/j.femsle.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Keim CN, Martins JL, Lins de BarrosHP, Lins U, Farina M.2007Structure, behavior, ecology and diversity of multicellular magnetotactic prokaryotesIn: Schüler D (ed.)Magnetoreception and Magnetosomes in Bacteria—Microbiological Monographs Springer: Berlin, Heidelberg, Germany; 103–132. [Google Scholar]
- King N, Hittinger CT, Carroll SB. Evolution of key cell signaling and adhesion protein families predates animal origins. Science. 2003;301:361–363. doi: 10.1126/science.1083853. [DOI] [PubMed] [Google Scholar]
- Kolinko I, Jogler C, Katzmann E, Schüler D. Frequent mutations within the genomic magnetosome island of Magnetospirillum gryphiswaldense are mediated by RecA. J Bacteriol. 2011;193:5328–5334. doi: 10.1128/JB.05491-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. doi: 10.1016/0888-7543(88)90007-9. [DOI] [PubMed] [Google Scholar]
- Lefèvre CT, Abreu F, Lins U, Bazylinski DA. Non-magnetotactic multicellular prokaryotes from low saline, nonmarine aquatic environments and their unusual negative phototactic behavior. Appl Environ Microbiol. 2010;76:3220–3227. doi: 10.1128/AEM.00408-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefèvre CT, Menguy N, Abreu F, Lins U, Pósfai M, Prozorov T, et al. A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science. 2011;334:1720–1723. doi: 10.1126/science.1212596. [DOI] [PubMed] [Google Scholar]
- Lefèvre CT, Trubitsyn D, Abreu F, Kolinko S, Almeida LGP, Vasconcelos ATR, et al. Monophyletic origin of magnetotaxis and the first magnetosomes. Environ Microbiol. 2013;15:2267–2274. doi: 10.1111/1462-2920.12097. [DOI] [PubMed] [Google Scholar]
- Lefèvre CT, Trubitsyn D, Abreu F, Kolinko S, Jogler C, Almeida LGP, et al. Comparative genomic analysis of magnetotactic bacteria from the Deltaproteobacteria provides new insights into magnetite and greigite magnetosome genes required for magnetotaxis. Environ Microbiol. 2013;15:2712–2735. doi: 10.1111/1462-2920.12128. [DOI] [PubMed] [Google Scholar]
- Li C, Louise CJ, Shi W, Adler J. Adverse conditions which cause lack of flagella in Escherichia coli. J Bacteriol. 1993;175:2229–2235. doi: 10.1128/jb.175.8.2229-2235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins JL, Silveira TS, Abreu F, Silva KT, Silva-Neto ID, Lins U. Grazing protozoa and magnetosome dissolution in magnetotactic bacteria. Environ Microbiol. 2007;9:2775–2781. doi: 10.1111/j.1462-2920.2007.01389.x. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Oh S, Kogure K, Ohwada K, Simidu U. Correlation between possession of a respiration-dependent Na+ pump and Na+ requirement for growth of marine bacteria. Appl Environ Microbiol. 1991;57:1844–1846. doi: 10.1128/aem.57.6.1844-1846.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J, Glöckner FO, Schönhuber W, Amann R.2001Fluorescence in situ hybridization with rRNA-targeted oligonucleotide probesIn: Paul J (ed.)Methods in Microbiology: Marine Microbiology Academic Press: London, UK; 207–226. [Google Scholar]
- Sachs JL. Resolving the first steps to multicellularity. Trends Ecol Evol. 2008;23:245–248. doi: 10.1016/j.tree.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Shank EA, Kolter R. Extracellular signaling and multicellularity in Bacillus subtilis. Curr Opin Microbiol. 2011;14:741–747. doi: 10.1016/j.mib.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro JA. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–104. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- Shapiro OH, Hatzenpichler R, Buckley DH, Zinder SH, Orphan VJ. Multicellular photo-magnetotactic bacteria. Environ Microbiol Rep. 2011;3:233–238. doi: 10.1111/j.1758-2229.2010.00215.x. [DOI] [PubMed] [Google Scholar]
- Simmons SL, Edwards KJ. Unexpected diversity in populations of the many-celled magnetotactic prokaryote. Environ Microbiol. 2007;9:206–215. doi: 10.1111/j.1462-2920.2006.01129.x. [DOI] [PubMed] [Google Scholar]
- Silva KT, Abreu F, Keim CN, Farina M, Lins U. Ultrastructure and cytochemistry of lipid granules in the many-celled magnetotactic prokaryote, ‘Candidatus Magnetoglobus multicellularis'. Micron. 2008;39:1387–1392. doi: 10.1016/j.micron.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Sobrinho R, Lins U, Bernardes M. Geochemical characteristics related to the greigite-producing multicellular magnetotactic prokaryote ‘Candidatus Magnetoglobus multicellularis' in a hypersaline lagoon. Geomicrobiol J. 2011;28:705–713. [Google Scholar]
- Strittmatter AW, Liesegang H, Rabus R, Decker I, Amann J, Andres S, et al. Genome sequence of Desulfobacterium autotrophicum HRM2, a marine sulfate reducer oxidizing organic carbon completely to carbon dioxide. Environ Microbiol. 2009;11:1038–1055. doi: 10.1111/j.1462-2920.2008.01825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum mikelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S, Kube M, Schübbe S, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphyswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenter R, Wanner G, Schüler D, Overmann J. Ultrastructure, tactic behaviour and potential for sulfate reduction of a novel multicellular magnetotactic prokaryote from North Sea sediments. Environ Microbiol. 2009;11:1493–1505. doi: 10.1111/j.1462-2920.2009.01877.x. [DOI] [PubMed] [Google Scholar]
- Yang H, Inokuchi H, Adler J. Phototaxis away from blue light by an Escherichia coli mutant accumulating protoporphyrin IX. Proc Natl Acad Sci USA. 1995;92:7332–7336. doi: 10.1073/pnas.92.16.7332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Zhang WY, Yu-Zhang K, Pan HM, Zhang SD, Zhang WJ, et al. A novel genus of multicellular magnetotactic prokaryotes from the Yellow Sea. Environ Microbiol. 2012;14:405–413. doi: 10.1111/j.1462-2920.2011.02590.x. [DOI] [PubMed] [Google Scholar]
- Zhou K, Zhang WY, Pan HM, Li JH, Yue HD, Xiao T, et al. Adaptation of spherical multicellular magnetotactic prokaryotes to the geochemically variable habitat of an intertidal zone. Environ Microbiol. 2012;15:1595–1605. doi: 10.1111/1462-2920.12057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.