Abstract
Metagenomics has opened new windows on investigating viral diversity and functions. Viromic studies typically require large sample volumes and filtration through 0.2 μm pore-size filters, consequently excluding or under-sampling tailed and very large viruses. We have optimized a targeted viromic approach that employs fluorescence-activated sorting and whole genome amplification to produce dsDNA-enriched libraries from discrete viral populations from a 1-ml water sample. Using this approach on an environmental sample from the Patagonian Shelf, we produced three distinct libraries. One of the virus libraries was dominated (79.65% of sequences with known viral homology) by giant viruses from the Mimiviridae and Phycodnaviridae families, while the two other viromes were dominated by smaller phycodnaviruses, cyanophages and other bacteriophages. The estimated genotypic richness and diversity in our sorted viromes, with 52–163 estimated genotypes, was much lower than in previous virome reports. Fragment recruitment of metagenome reads to selected reference viral genomes yields high genome coverage, suggesting little amplification and sequencing bias against some genomic regions. These results underscore the value of our approach as an effective way to target and investigate specific virus groups. In particular, it will help reveal the diversity and abundance of giant viruses in marine ecosystems.
Keywords: giant viruses, metagenome, NCLDV, viral diversity
Introduction
With as many as 107 viruses per milliliter of seawater (Bergh et al., 1989) and 109 viruses per gram of marine sediments (Danovaro et al., 2001) and soil in terrestrial environments (Williamson et al., 2003), viruses are the most abundant and genetically diverse entities in the earth's biosphere. In the ocean, viruses affect the biogeochemistry and genetic variability that sustain plankton communities through lysis of their unicellular hosts and through DNA or RNA transduction (Brüssow et al., 2004; Suttle, 2007). Yet, knowledge of the exact extent of viral diversity has so far been hindered by limitations in sampling methods and in isolation and maintenance of host-virus systems in the laboratory. An added difficulty in the study of viruses is the lack of universal genetic markers (Rohwer and Edwards, 2002) that would allow deriving viral diversity, phylogeny and taxonomic relationships based on a monophyletic origin of all viruses in the same way that, for example, rRNA markers allow for prokaryotes and eukaryotes (Amann et al., 1995). However, conserved genes exist for comparative phylogenetic analysis within certain virus groups. Yutin et al. (2009) identified clusters of orthologous genes for functional and evolutionary analysis of Nucleo-Cytoplasmic Large DNA Viruses (NCLDVs). The NCLDVs group consists of at least six families of eukaryotic viruses with large dsDNA genomes that infect animals as well as protists (Wilson and Allen, 2009). Alternatively, culture-independent sequence analysis of viral assemblages (viromes) has been applied since 2002 (Breitbart et al., 2002) to provide insights into viral functions, community composition and structure in the environment. A clear advantage of this approach is that it does not rely on the presence of any particular gene in every single virus particle. The success of virus metagenomics has increased rapidly with the advancement of sequencing technology and the development of bioinformatics tools. Environmental viromic studies suggest that less than 1% of the extant viral diversity has been explored so far (Mokili et al., 2012). Furthermore, the majority (usually, 60–99%) of sequences in viromes from any environment have no significant sequence similarity to other sequences in databases or have higher homology to prokaryotic or eukaryotic genes (Breitbart et al., 2002, 2003, 2004; Angly et al., 2006; Schoenfeld et al., 2008; Blomström et al., 2010).
One of the main barriers that limit our knowledge of viral diversity in marine environments is the fact that no single method exists that allows targeting the entire viral assemblage within a discrete water sample at once. Frequently, studies focus on either RNA or DNA viruses, single-stranded or double-stranded. Frequently, viruses are operationally defined as nucleic acid-containing particles that pass through 0.2 μm pore-size filters, which is a necessary step to eliminate the cellular fraction (Thurber et al., 2009; Wommack et al., 2010; Steward and Preston, 2011). This standard filtration procedure leads to removal and under-sampling of giant viruses, that is, NCLDV with genomes larger than 300 Kb and capsids close to or larger than 200 nm in diameter (Claverie et al., 2006; Wilson and Allen, 2009; Van Etten, 2011). Additionally, tailed viruses have also been shown to be preferentially lost during filtration (Cochlan et al., 1993). New approaches to target under-represented virus groups, such as giant viruses, are necessary to supplement technical advances in metagenomic sequencing and bioinformatics analysis to help unveil the true degree of viral diversity and ecological functions. We have developed a novel targeted viromic approach employing fluorescence-activated sorting and whole genome amplification to produce dsDNA-enriched viromes from distinct viral populations, determined by flow cytometry (FCM). Here we present data obtained with this approach from a single one-milliliter water sample collected from the Patagonian Shelf.
Materials and methods
Sampling
Sampling was carried out during the COPAS'08 cruise onboard of the RV Roger Revelle (Cruise Knox22RR, 4 December 2008—2 January 2009) along the Patagonian Shelf. Seawater samples were collected at discrete depths at 152 conductivity–temperature–depth stations using a Seabird SBE 911 conductivity–temperature–depth equipped with 24 × 20 l Go-Flow Niskin bottles. One-milliliter water subsamples from each depth and station were fixed with 0.1% or 0.5% glutaraldehyde (final concentrations, for molecular or transmission electron microscopy (TEM) analysis, respectively) for 30 min at 4 °C and snap frozen in liquid nitrogen. The samples were stored at −80 °C until further processing. For the present study, we selected a single sample collected on 30 December 2008 at station 142 (52° 36.382 S, 60° 16.869 W) at 15.5 m depth, collection time GMT 03:43.
Fluorescence-activated sorting
On 7 August 2009, approximately after 7 months in storage at −80 °C, the 0.1% and 0.5% glutaraldehyde-fixed samples were thawed on ice, diluted 100-fold with 0.2 μm-filtered 10:1 TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and stained with SYBR Green I (Molecular Probes Inc., Eugene, OR, USA) as described by Brussaard (2004). Sorting of virus was done at the JJ MacIsaac Facility for Aquatic Cytometry, at Bigelow Laboratory for Ocean Sciences, with an Influx (BD Biosciences, San Jose, CA, USA) flow cytometer using a 488 nm argon laser for excitation. The ‘single 1 drop' mode was used to ensure the absence of non-target particles within the target particle drop and the surrounding drops. Sorting instruments and reagents were decontaminated as previously described (Stepanauskas and Sieracki, 2007). The cytometer was triggered on side scatter and the sort gates were based on SYBR Green I green fluorescence and side scatter signals. Approximately 5000 virus-like particles (VLPs) from each of the distinct groups were sorted for genomic analysis (0.1% glutaraldehyde-fixed sample), and between 5000 and 15 000 VLPs from each of the groups were sorted for microscopy analysis (0.5% glutaraldehyde-fixed sample) into separate 1.5 ml Eppendorf tubes and stored at −80 °C until further processing.
Transmission electron microscopy
The sorted VLPs were spotted onto carbon-coated copper microscopy grids and stained with 2% uranyl acetate following general recommendations by Ackermann and Heldal (2010). Imaging analysis was performed at the Applied Medical Sciences department at the University of Southern Maine, Portland, ME, USA.
Whole genome amplification of DNA from sorted particles and sequencing
The sorted particles were lysed and their genetic material amplified employing GenomePlex Single Cell Whole Genome Amplification (WGA4) Kit (Sigma-Aldrich, St Louis, MO, USA) following the manufacturer's recommendations. WGA-amplified genomic material was purified using GenElute PCR Clean-up kit (Sigma-Aldrich) and quantified using a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Library construction and sequencing were performed at the Broad Institute (Cambridge, MA, USA). VLP DNA samples were sequenced using 1/4 of a 454 Titanium picotitre plate. Sequences were deposited to CAMERA (http://camera.calit2.net) under the following project accessions: CAM_SMPL_000988, CAM_SMPL_001013 and CAM_SMPL_000959.
Bioinformatics analyses
The sequences were initially processed to trim linkers, allowing a maximum of 10 mismatches at both ends, using TAG Cleaner tool (http://edwards.sdsu.edu/cgi-bin/tagcleaner/tc.cgi). Sequences shorter than 65 bp and/or that contained any ‘Ns' were removed and a low-complexity threshold of 70 (using entropy) was applied using PRINSEQ v0.20.3 (http://edwards.sdsu.edu/cgi-bin/prinseq/prinseq.cgi). Natural and artificial duplicates were removed from the pyrosequencing runs using the program cdhit-454 (http://weizhong-lab.ucsd.edu/cdhit_454/cgi-bin/index.cgi?cmd=cdhit_454) (Niu et al., 2010).
Taxonomic analysis
Quality-screened, curated sequences were submitted to the VIROME pipeline for analysis, including functional and taxonomic Open reading frame (ORF) characterization and environmental characterization derived by BLASTp comparison to the UniRef 100 and MetaGenomes online databases (as of September 2012). Details about the VIROME pipeline can be found in Wommack et al. (2012). The analysis was supplemented by comparing the virome libraries to a custom protein database comprised of 77 taxonomically diverse viral genomes (see GenBank genome accession numbers and additional information in Supplementary Table S1) using the NCBI's BLAST × algorithm (version 2.2.26+) (Altschul et al., 1990) with an e-value cut-off of ⩽10−5. Additionally, the virome libraries were compared by BLASTn (e-value cut-off of ⩽10−10) to the non-redundant nucleotide database in NCBI to identify rRNA fragment sequences. Sequences in the libraries with only significant homology to bacterial members for which rRNA sequences were detected were labeled as contaminants and removed from further analysis. Reads with no significant homology match in the databases were assigned as ORFans and sequences homologous to ORFs in databases were classified as viral, bacterial, archaeal, eukaryotic or ‘other' (unclassified or unknown origin). When a particular metagenomic read had more than one homology match, only top viral hits or hits with the highest bitscore value were selected. Viral homologs were further classified into viral families. BLAST results were used to calculate viral community richness and evenness, phylogenetic analysis of NCLDV genes (Yutin et al., 2009, 2013) identified within the libraries, and fragment recruitment against selected viral genomes.
Viral community structure and richness
Viral assemblage structure and richness of each sorted metagenome was compared using the PHACCS tool ((Angly et al., 2005); http://sourceforge.net/projects/phaccs). A random sub-sample of 50 000 sequences from each metagenome was used in the calculation of each contig spectrum using Circonspect (Angly et al., 2006), with 98% sequence similarity of 35 bp or greater. Average genome size was determined as the weighted average genome size of the viruses to which the sequences in each library had significant homology.
Phylogenetic analysis of NCLDVs
NCLDV gene homologs in the viromic libraries were identified by BLASTx comparison (e-value cut-off of ⩽10−5) to reference databases for D5-like helicase-primase, A2L-like transcription factor, A32-like packaging ATPase, superfamily II (SF II) helicase, small subunit ribonucleotide reductase, and DNA polymerase family B genes available at ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/NCVOG/cl2fa (Yutin et al., 2009). The databases were supplemented with the respective genes (where available) from Organic Lake Phycodnavirus 2, Bathycoccus prasinos-virus 1, Ostreococcus lucimarinus-virus 3, Micromonas pusilla-virus SP1, Phaeocystis globosa-virus 12 T, Cyanophage strain STIM5 and Synechococchus-phage S-SSM7. Metagenomic reads from the BLAST analysis larger than 165 bp were translated into amino acids and identical sequences were eliminated. The metagenomic reads were aligned to the reference sequences using the MAFFT multiple sequence alignment program (http://www.ebi.ac.uk/Tools/msa/mafft/). All the alignments were manually checked for the conservation of domain architecture. Preliminary maximum-likelihood trees of the reference sequences, in Newick format, and tree statistics files were produced using the PhyML v3.0 tool (Guindon et al., 2010), with LG model of amino acids substitution, allowing for estimated proportion of invariable sites and four substitution rate categories. Reference Newick trees, their statistics files and the alignments of the virome reads to the reference sequences were then used as the input for the phylogenetic placement and visualization software package placer (Matsen et al., 2010) to produce final maximum-likelihood trees.
Viral fragment recruitment analysis
Metagenome sequences were compared to 16 viral genomes (Supplementary Table S2) representing a range of genome sizes using tBLASTx, with an e-value cut-off of ⩽10−5. A random subsample of 70 065 sequences from each metagenome was used in the recruitment analysis. Average coverage and percentage coverage of each genome ⩾1 × was determined from tBLASTx results by converting protein-based alignments to nucleotide positions along each reference genome using a custom Python script.
Results
Sorting, DNA amplification and sequencing
We successfully sorted VLPs from three discrete populations, G4000, G4001 and G4002, observed by FCM (Figures 1 and 2 insert). TEM analysis revealed the presence of VLPs and the absence of bacterial or other cellular organisms. However, the VLPs were lumped together instead of evenly distributed on the microscopy grids, and their capsids appeared damaged, possibly because of the sorting procedure, hindering proper morphological analysis (Supplementary Figure S1). Whole genome amplification on sorted particles yielded at least 2 μg of total DNA from each sample. A total of 184 906, 212 996 and 202 525 reads were obtained for the G4000, G4001 and G4002 libraries, respectively. Curation and removal of low quality and duplicate reads left 70 065, 94 900 and 72 217 reads (G4000, G4001 and G4002, respectively) for further analysis (Table 1).
Figure 1.
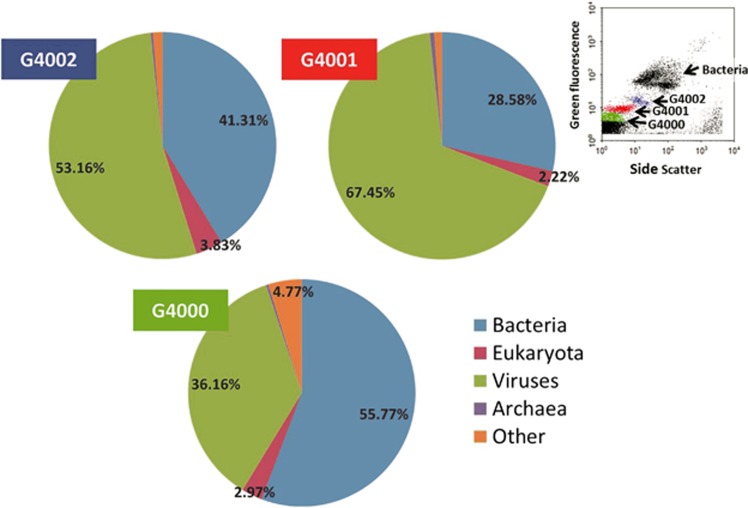
Viromes domain taxonomy. Bacteria, Eukaryota, Viruses and Archaea percentage of library reads with homology to known sequences in each of the three target-sorted viromes. Reads were taxonomically assigned based on BLAST comparison to UniRef 100 and MetaGenomes online databases, and to our own database, including 77 taxonomically diverse complete viral genomes (see Supplementary Table 1). ‘Other' refers to unclassified ORFs and mobile elements such as plasmids. Top right inset: representative flow cytometry plot showing bacteria and virus populations in the water sample. Discrimination of groups was based on green fluorescence and side scatter signatures. Sorted populations G4000, G4001 and G4002 are indicated in green, red and blue, respectively.
Figure 2.
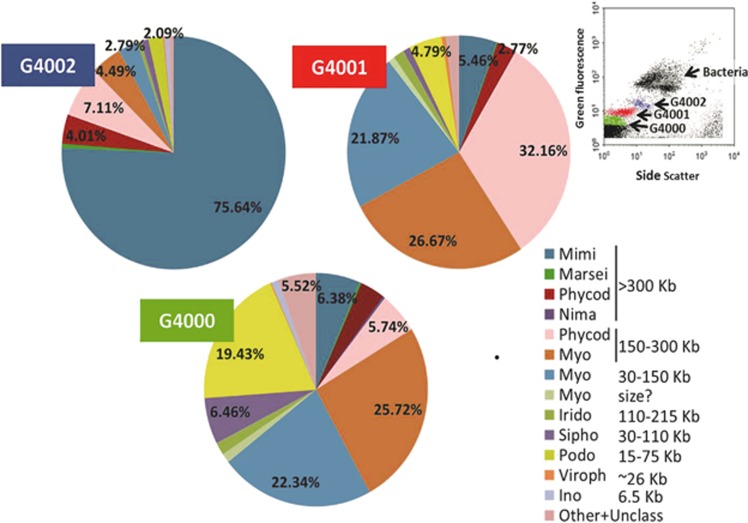
Viral families taxonomy. Summary classification of hits from the three target-sorted libraries with homology to known viruses into viral families. Reads were taxonomically assigned based on BLAST comparison to UniRef 100 and MetaGenomes online databases, and to our own database, including 77 taxonomically diverse complete viral genomes (see Supplementary Table 1). Unclass=hit to unclassified virus sequence. Top right inset: representative flow cytometry plot showing bacteria and virus populations in the water sample. Discrimination of groups was based on green fluorescence and side scatter signatures. Sorted populations G4000, G4001 and G4002 are indicated in green, red and blue, respectively.
Table 1. Summary of total number of reads per library before and after bioinformatics curation.
| G4000 | G4001 | G4002 | |
|---|---|---|---|
| No. of total 454-reads | 184 906 | 212 996 | 202 525 |
| No. of rRNA reads | 531 | 163 | 473 |
| No. of putative contaminant reads removed | 13 199 | 3053 | 9010 |
| No. of 454-reads (curated, no duplicates) | 70 065 | 94 900 | 72 217 |
| No. of ORFan reads | 55 455 | 72 279 | 49 953 |
| No. of reads with homology in databases | 14 601 | 22 621 | 22 264 |
Taxonomic composition of targeted viromes
Although we did not observe any whole unicellular organisms by TEM analysis, we found a small number of partial rRNA sequences in all three metagenomic libraries: 531 in G4000, 163 in G4001 and 473 in G4002 (Table 1). Of those, 1–2% were identified as 18S rRNA from fungi or insect (additionally, two of the sequences in G4001 had the highest similarity to human 18S rRNA). The remaining 98–99% rRNA reads were identified as prokaryotic 16S rRNA, 23S rRNA or 16-23S intergenic spacer regions (90–100% sequence identitiy, e-values ⩽1E-66). In all three libraries, rRNA sequences were dominated by members of the order Burkholderiales, mainly of the genus Ralstonia. Propionibacterium spp. (Actinomycetales) rRNA fragments were also found in all three libraries (Supplementary Table S3). Any sequences that only had significant homology to genes from Propionibacterium spp. or from Burkholderiales for which rRNA sequences were detected were considered external contamination and removed from further analysis. Additionally, we removed approximately 200 reads from each library that were identified as mammalian genes (partial sequences, most similar to human). To be conservative in our analyses, we did not remove sequences with hits to other Burkholderiales members for which rRNA homologues were not found. We also kept all sequences in the libraries that were most similar to genes from bacterial species, but for which 16S rRNA partial sequences were not found in all three metagenomic libraries (Supplementary Table S3).
As commonly found in other marine viral metagenomic studies (Breitbart et al., 2002, 2003, 2004; Angly et al., 2006; Schoenfeld et al., 2008; Blomström et al., 2010; Hurwitz and Sullivan, 2013), the majority (79.16% G4000, 76.16% G4001, 69.17% G4002) of the reads in our libraries were ORFans, that is, ORFs that have no similarity to sequences in public databases (Supplementary Figure S2). Of the reads with homology to database sequences, viruses accounted for approximately 36% in G4000, 67% in G4001 and 53% in G4002; hits to members of the domain Bacteria accounted for about 56%, 29% and 41%, respectively; and hits to Eukarya accounted for only 3%, 2% and 4%, respectively. The very few remaining reads included Archaea, unclassified ORFs and mobile elements such as plasmids (referred to as ‘other') (Figure 1). A closer look at the virus fraction provided further insights. Top viral hits for the G4000 library, which originated from the VLPs group with the lowest green fluorescence and side scatter signals in FCM (green cluster, Figures 1 and 2 insert), were mostly to myoviruses and podoviruses of the abundant SAR11 (Pelagibacter) and SAR116 (e.g., Puniceispirillum marinum) bacterial clades, cyanophage myoviruses and other bacteriophages with genome sizes smaller than 300 Kb. In particular, 25.72% of the reads were most similar to cyanophage myoviruses with genomes between 150 Kb and 300 Kb, and 52.44% of the reads had top hits to other myoviruses, iridoviruses, siphoviruses, podoviruses and inoviruses with genome sizes in the 6.5–200 Kb range. The G4001 library, from VLPs with intermediate green fluorescence and side scatter values (red cluster, Figures 1 and 2 insert), was dominated by hits to phytoplankton viruses of the families Phycodnaviridae (32.16%, prasinoviruses) and Myoviridae (26.67%, mainly cyanophages) with genomes between 150 Kb and 300 Kb. An additional 21.87% of the reads were assigned to Myoviridae (mainly to Pelagibacter-myovirus HTVC008M) with 30–150 Kb genome-sizes. The sequences in the G4002 library from the VLPs with the highest green fluorescence and side scatter (blue cluster, Figures 1 and 2 insert) were dominated (∼80%) by members of the Mimiviridae family and giant algal viruses of the Phycodnaviridae family, all with genome sizes in excess of 300 Kb (Figure 2).
Viral diversity
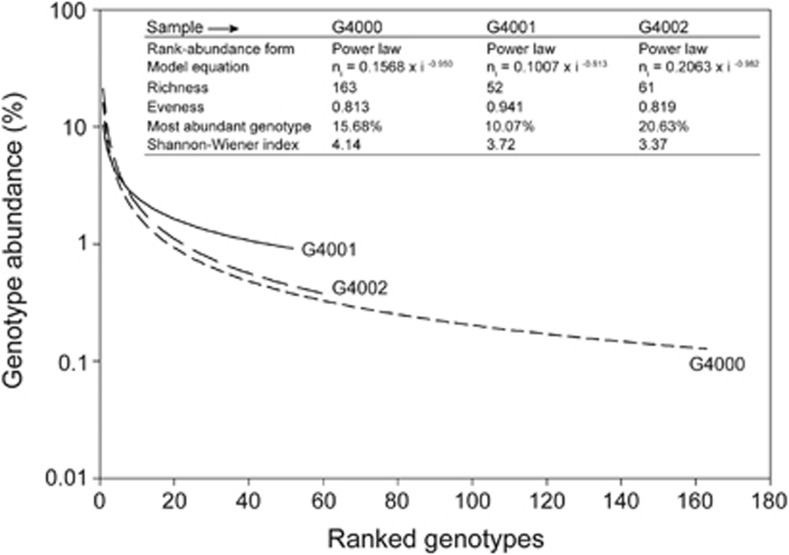
We estimated genotype richness, evenness and diversity within each virus sorted group using the PHACCS analysis system (Angly et al., 2005) (Figure 3). The estimated average genome sizes for the viruses to which the virome library sequences had significant homology were 420 057 bp, 209 540 bp and 186 395 bp for the G4002, G4001 and G4000, respectively. The power law rank-abundance form was the best fit for describing the viral assemblages' structure. The G4000 viral metagenome was the most genotype-rich, with 163 predicted genotypes, and diverse (H′=4.14). Whereas the G4002 (61 predicted genotypes) was the least diverse (H′=3.37), the G4001 metagenome gave the lowest number of predicted genotypes, only 52. The number of predicted virus genotypes in our metagenomic libraries is markedly lower than the several thousand genotypes reported in previous marine virome studies (Breitbart et al., 2002; Angly et al., 2006).
Figure 3.
Viral community diversity. Comparison of viral community structure and richness between sorted viromes using the PHACCS tool. Rank abundance curves were obtained by plotting the abundance of each genotype versus its rank-abundance.
Phylogeny
BLAST comparison of G4000 reads to the core NCLDV genes returned relatively few well-supported hits. Therefore, we did not proceed with any further phylogenetic analysis of reads from this fraction. Phylogenetic analysis of NCLDV gene homologs unequivocally placed the majority of the reads in the G4001 library with members of the Phycodnaviridae family, closely related to prasinoviruses, which infect picoeukaryote hosts (Supplementary Figures S3–S8). However, one of the reads with highest similarity to the A2L-like transcription factor gene fell among members of the Mimiviridae family (Supplementary Figure S4), and two and four reads similar to the ribonucleotide reductase and the DNA polymerase family B genes, respectively, were phylogenetically closer to cyanophages (Supplementary Figures S7 and S8). Most reads in library G4002 identified as core NCLDV genes clustered with Mimiviridae family members (Supplementary Figures S9–S14), except for one A2L-like transcription factor read and three SF II helicase reads that were most closely related to prasinoviruses (Supplementary Figures S10 and S12). Additionally, phylogenetic analysis of ribonucleotide reductase and DNA polymerase family B homologs was inconclusive and resulted in several sequence reads falling with a range of diverse virus groups, that is, poxviruses, asfaviruses and large phycodnaviruses (Supplementary Figures S13 and S14). These phylogenetic results are in agreement with the results from our BLAST analysis.
Viral fragment recruitment to reference viral genomes
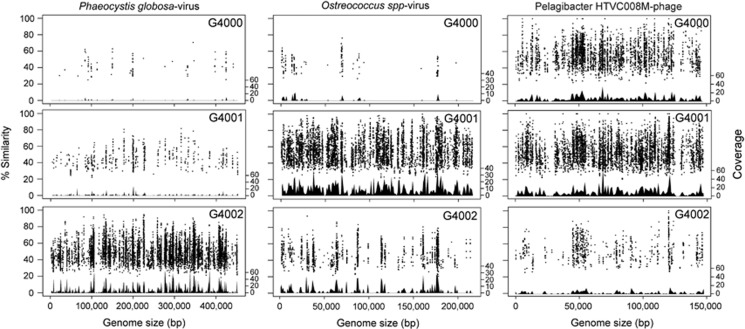
Phylogenetic analysis results and the number of BLAST hits to specific virus genomes aided in selecting reference virus genome candidates for fragment recruitment analysis of our targeted virome libraries (Supplementary Table S2). The G4002 virome yielded the highest genome coverage at ⩾1 × for known algal virus members of the recently expanded Mimiviridae family (Yutin et al., 2013), for example, Phaeocystis globosa-virus PgV-14T. Pelagibacter-myovirus HTVC008M and prasinophyte viruses, such as Micromonas pusilla-virus PL1 and Ostreococcus spp-viruses, had the highest genome coverage within the G4001 virome. The best genome coverage in the G4000 library was for pelagibacter-myovirus HTVC008M and SAR116 Puniceispirillum-podovirus HMO-2011 (Supplementary Table S2). Additionally, the recruitment plots show high percentage of similarity between the metagenomic reads and reference genomes at the amino acid level, and even sequence coverage of the PgV-14T, Ostreococcus spp-virus (strain OlV-4) and pelagiphage HTVC008M genomes in G4002, G4001 and G4000, respectively (Figure 4).
Figure 4.
Fragment recruitment of sorted metagenomes to selected viral genomes. Similarity values are based on tBLASTx alignments, and coverage across each genome was plotted using a sliding window of 4000 bp.
Discussion
To our knowledge, this is first time that distinct virus particles within a natural assemblage have successfully been sorted, and their genetic material amplified and sequenced. The results obtained with our targeted viromic approach represent a significant advantage over a recently published study in which, using a similar approach based on FCM sorting and whole genome amplification, Allen et al. (2011) were able to sort individual viruses from an assemblage comprised of two Escherichia coli bacteriophages in culture.
Homogenization, filtration through 0.2 μm pore-size filters, ultracentrifugation and density gradients are frequently employed to separate and concentrate the viral particles present in a seawater sample (Thurber et al., 2009; Wommack et al., 2010). However, these procedures bias against very large, tailed and/or buoyant viruses. Additionally, due to the typically small fraction of viral DNA and RNA in environmental samples, and the relatively short length of viral genomes, prokaryotic and eukaryotic nucleic acids within the samples can reduce the efficiency of isolation and detection of viral nucleic acids. Samples are commonly treated with chloroform in order to disrupt mitochondrial, bacterial and eukaryotic membranes and release their DNA, which is subsequently removed by DNase digestion (Thurber et al., 2009). Unfortunately, chloroform treatment also renders DNA from the many enveloped viruses with lipid membranes (Ono, 2010; Roine and Bamford, 2012) subject to DNase digestion. Despite the precautions above, virus metagenomes typically contain many sequences with homology to only prokaryotic or eukaryotic genes (e.g. Breitbart et al., 2002; Angly et al., 2006; Schoenfeld et al., 2008). Partly, this may be due to transduction events, but it is also a direct consequence of the still relatively low number of viral reference sequences in databases compared to prokaryotic and eukaryotic sequences. Availability of novel reference viral genomes greatly improves binning of marine viromes; in particular genomes of viruses that infect major microbial groups such as SAR116 and SAR11 bacterial clades (Kang et al., 2013; Zhao et al., 2013).
Our approach for generating viral metagenomic libraries from environmental samples potentially avoids cellular contamination by targeting only VLPs within a seawater sample, independent of their capsid sizes or lipid content, using FCM, a well-established tool for discriminating and enumerating viral populations in water (Marie et al., 1999; Brussaard et al., 2000; Brussaard, 2004), and fluorescence-activated sorting (Shapiro, 2005; Sieracki et al., 2005). However, our method is biased towards viruses with dsDNA genomes because SYBR Green I (Molecular Probes Inc), the fluorescent dye we employed for FCM detection, preferentially binds to dsDNA and has lower affinity for ssDNA and RNA. Other virus groups can be more specifically targeted using different fluorescence dyes (e.g. SYTO RNASelect, Life Technologies, Carlsbad, CA, USA) and/or modified FCM protocols (Tomaru and Nagasaki, 2007; Robertson et al., 2010). Additionally, we did not include in this study very small viruses that may appear in the region with the lowest green fluorescence values measured by FCM (black cluster under G4000 population in Figures 1 and 2 insert). We also sorted VLPs from that region but the resulting sequences were mostly homologs to human and bacteria (mainly Burkholderiales) genes and were consequently excluded from further analysis. Contamination was probably introduced during the two rounds of DNA amplification needed to produce sufficient material for sequencing from this VLPs group, while only one round of DNA amplification was required for the G4000, G4001 and G4002 populations (data not shown).
Seawater samples were not treated with DNase to remove extracellular DNA prior to sorting VLPs. However, only a few picoliters of water are sorted along with each VLP, minimizing the risk of free DNA contamination (Sieracki et al., 2005). Unfortunately, our viromic libraries were not entirely free of non-viral contamination, as revealed by the presence of partial (mostly) bacterial rRNA sequences (Supplementary Table S3). Extensive single cell genomic studies using fluorescence-activated sorting for physical separation of cells have proven the efficiency of this technology to avoid cross-sorting contamination (Martínez Martínez et al., 2011; Swan et al., 2011; Yoon et al., 2011; Martinez-Garcia et al., 2012). However, we found NCLDV genes phylogenetically closer to small algal viruses in G4002, and genes more similar to mimiviruses in G4001, indicating the possibility for some cross-sorting of viral populations, and therefore the possibility of sorting bacteria cells along with the targeted VLPs also exists. In the future, this may be prevented by using more restrictive sort gates. Yet, the lack of bacterial cells in our TEM analysis suggests that very few, if any, bacteria were sorted, and the source of contamination might have resulted from steps other than the sorting process. It is more probable that contamination was introduced during the whole genome amplification step. Notably, only a few members of the order Burkholderiales dominated the rRNA sequences in the three libraries. In a study of bacterial populations inhabiting ultrapure water systems, Kulakov et al. (2002) found that the gram-negative bacteria Ralstonia pickettii (order Burkholderiales), Bradyrhizobium sp., Pseudomonas saccharophilia and Strenotrophomonas spp. were indigenous to all water systems investigated. Additionally, colleagues at the Bigelow Laboratory's Single Cell Genomic Center have detected contamination from Propionibacterium spp. in multiple displacement amplification reagents and/or ultrapure water systems (personal communication). In the future, these contamination issues may be circumvented by applying a stringent decontamination procedure of water and DNA amplification reagents as reported by Woyke et al. (2011).
A significant added benefit of our method is the fact that we were able to produce high-quality virome libraries from glutaraldehyde-fixed samples. Aldehyde cross-linking of proteins preserves the viral capsids integrity and shape, facilitating discrimination of distinct virus groups by FCM. Additionally, glutaraldehyde-preservation allows long-term storage of samples (∼seven months in our study), making it possible to collect, screen and process a larger number of samples than otherwise would be possible if immediate VLP sorting is required. Using as little as 0.1% glutaraldehyde (final concentration) facilitated optimum FCM resolution of viral populations and prevented inhibition of the DNA amplification reaction. High percentage and even coverage (Figure 4) of the reference genomes for viral fragment recruitment indicate that sample preservation, genomic amplification and sequencing did not significantly bias against certain regions of the viral genomes.
An inherent aspect of viromes is the high percentage of generated sequences that have no homology to any sequences in public databases, rendering characterization of new viruses not yet available in culture an arduous task. Another problem associated with viromics is the difficulty in assembling complete genomes or even long contigs. Luo et al. (2012) estimated that to retrieve a genome from a metagenome the genome must have at least 20 × coverage, while at lower coverage the produced assemblies contained numerous chimeras. Additionally, longer, less chimeric assembled contigs were derived from less diverse environments. Therefore, our targeted viromic approach may be employed to ease assembly of individual genomes within the relatively low-diversity metagenomic libraries generated. With only 52–163 estimated genotypes and H′ between 3.37 and 4.14, our three viromes were much less diverse than other marine virome studies, for example, Breitbart et al., 2002; Angly et al., 2006, who reported diverse viral assemblages containing up to several thousand genotypes. This result is not surprising but rather expected, and adds support for the effectiveness of our method targeting specific, distinct components within the total viral assemblage. The G4000 virome contains ∼2.5–3 times more genotypes than the other two viromes analyzed, which is possibly a direct consequence of the different level of diversity within the specific host communities and host ranges for each of the sorted virus groups. Based on our BLAST analysis, G4002 and G4001 likely contain mostly eukaryotic algal viruses, while G4000 is mainly comprised of bacteriophages within the Myoviridae and Podoviridae families (Figure 2) that infect members of the abundant SAR11 and SAR116 bacterial clades. In general, in any given environment, bacterial communities are more abundant and diverse than co-occurring protists. Our results confirm the abundance and ubiquity of SAR11 and SAR116 phages in the marine surface waters as shown in previous studies (Kang et al., 2013; Zhao et al., 2013).
It is unquestionable that we are missing some viral diversity by focusing on only a few groups, within a one-milliliter water sample (though sub-sampled from a well-mixed 20 l water sample collected with a Niskin bottle) from a single depth in the water column. Currently, we lack standards based on real criteria as to what constitutes a useful sample size to study natural viral assemblages. Instead, sampling decisions should be guided by the research questions. For example, our motivation for this study was to investigate large dsDNA viruses, which are commonly under-sampled by other methodologies. Sequencing a handful of giant virus isolates has revealed genomes packed with novelty (e.g. Raoult et al., 2004; Wilson et al., 2005; Fischer et al., 2010; Arslan et al., 2011). Evidence for DNA viruses closely related evolutionarily to Mimivirus (∼1.2 Mbp genome, capsid size ∼750 nm) (La Scola et al., 2003; Raoult et al., 2004) was found in environmental metagenomic sequence data corresponding to the ‘bacterial-sized' fraction of the microbial community from the Sargasso Sea (Ghedin and Claverie, 2005) and from Organic Lake, Antarctica, (Yau et al., 2011). Also, the giant Megavirus chilensis (Arslan et al., 2011) was isolated off the coast of Chile. With our novel targeted viromic approach we have found that giant viruses phylogenetically close to the Mimiviridae family (Supplementary Figures S9–S14) are also present in Patagonian Shelf waters, adding further support for the idea that giant viruses are neither rare nor marginal players in a wide range of marine ecosystems.
Finally, we recommend the following considerations as a means to further improve the potential of our targeted viromic approach for virus discovery: (1) DNase treatment of the water sample prior to sorting to eliminate non-target free DNA carryover into the whole genome amplification reaction; (2) decontamination of molecular reagents as described by Woyke et al. (2011); (3) multiple displacement amplification of DNA to produce overlapping reads that can potentially be assembled more easily than reads from randomly fragmented genomes, as was the case in this study; and (4) increased sequencing effort using different sequencing platforms, for example, MiSeq Illumina.
Viruses constitute one of the largest reservoirs of unexplored genetic diversity, making them an important source for discoveries in viral and microbial ecology, with possible applications for biotechnology companies seeking novel enzymes and compounds from the ocean. Targeted viromics is a powerful, fast and sensitive technique with the potential to become an important high-throughput, cost-efficient bioprospecting tool to help unveil and understand the concealed wealth in the global virosphere, given its potential to generate unequivocal viral genomic information. Here, we have shown the effectiveness of our approach to investigate important low-abundance virus groups that are typically excluded using standard current methodology. Targeted viromics will also prove useful for investigating viral assemblages from environments where the ability to obtain large-volume samples is limited or impractical. In addition, genomic information obtained using the approach described here may ease the investigation of horizontal gene transfer and co-evolution of host-virus systems. Virus sorting can be paired with cell sorting in future studies to cross-correlate viruses with their hosts. Finally, targeted viromics may enable isolation of not yet cultivated viruses by providing clues about potential host candidates. Isolation and maintenance of host-virus systems in the laboratory is still essential to allow experimentation and full exploitation of viral capabilities, such as host population control, and synthesis of enzymes with research and therapeutic and diagnostic uses.
Acknowledgments
We thank Dr WM Balch (chief scientist) and the rest of the crew and scientists aboard the R/V Roger Revelle, in particular L Lubelczyk for her help in collecting and pre-processing water samples. Dr NJ Poulton for operating the Influx flow cytometer. I Gilg and Dr J Labonté for advice about bioinformatics tools. Special thanks to the VIROME pipeline team, in particular to DJ Nasko and KE Wommack. Funds for participation on the COPAS'08 cruise and sample analysis were provided by NSF grants # OCE0849363 and EF0949162. Virome sequencing was supported by the Gordon and Betty Moore Foundation MMI Marine Phage, Virus, and Virome Sequencing Initiative.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ackermann HW, Heldal M.2010Basic electron microscopy of aquatic virusesIn: Wilhelm SW, Weinbauer MG, Suttle CA (eds)Manual of Aquatic Viral Ecology ASLO: Waco, TX, USA; 182–192. [Google Scholar]
- Allen LZ, Ishoey T, Novotny MA, McLean JS, Lasken RS, Williamson SJ. Single virus genomics: a new tool for virus discovery. PLoS One. 2011;6:e17722. doi: 10.1371/journal.pone.0017722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microb Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly F, Rodriguez-Brito B, Bangor D, McNairnie P, Breitbart M, Salamon P, et al. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics. 2005;6:41. doi: 10.1186/1471-2105-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan D, Legendre M, Seltzer V, Abergel C, Claverie J-M. Distant mimivirus relative with a larger genome highlights the fundamental features of megaviridae. Proc Natl Acad Sci USA. 2011;108:17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- Blomström A-L, Widén F, Hammer A-S, Belák S, Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol. 2010;48:4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Felts B, Kelley S, Mahaffy JM, Nulton J, Salamon P, et al. Diversity and population structure of a near-shore marine-sediment viral community. Proc R Soc B. 2004;271:565–574. doi: 10.1098/rspb.2003.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard CPD. Optimization of procedures for counting viruses by flow cytometry. Appl Environ Microbiol. 2004;70:1506–1513. doi: 10.1128/AEM.70.3.1506-1513.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard CPD, Marie D, Bratbak G. Flow cytometric detection of viruses. J Virol Met. 2000;85:175–182. doi: 10.1016/s0166-0934(99)00167-6. [DOI] [PubMed] [Google Scholar]
- Brüssow H, Canchaya C, Hardt W-D. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol Mol Biol Rev. 2004;68:560–602. doi: 10.1128/MMBR.68.3.560-602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverie J, Ogata H, Audic S, Abergel C, Suhre K, Fournier P. Mimivirus and the emerging concept of ‘giant' virus. Virus Res. 2006;117:133–144. doi: 10.1016/j.virusres.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Cochlan WP, Wikner J, Steward GF, Smith DC, Azam F. Spatial-distribution of viruses, bacteria and chlorophyll-a in neritic, oceanic and estuarine environments. Mar Ecol Prog Ser. 1993;92:77–87. [Google Scholar]
- Danovaro R, Dell'anno A, Trucco A, Serresi M, Vanucci S. Determination of virus abundance in marine sediments. Appl Environ Microbiol. 2001;67:1384–1387. doi: 10.1128/AEM.67.3.1384-1387.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer MG, Allen MJ, Wilson WH, Suttle CA. Giant virus with a remarkable complement of genes infects marine zooplankton. Proc Natl Acad Sci USA. 2010;107:19508–19513. doi: 10.1073/pnas.1007615107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghedin E, Claverie J-M. Mimivirus relatives in the Sargasso sea. Virology J. 2005;2:62. doi: 10.1186/1743-422X-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon Sp, Dufayard J-Fo, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hurwitz BL, Sullivan MB. The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS One. 2013;8:e57355. doi: 10.1371/journal.pone.0057355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang I, Oh H-M, Kang D, Cho J-C. Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc Natl Acad Sci USA. 2013;110:12343–12348. doi: 10.1073/pnas.1219930110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulakov LA, McAlister MB, Ogden KL, Larkin MJ, O'Hanlon JF. Analysis of bacteria contaminating ultrapure water in industrial systems. Appl Environ Microbiol. 2002;68:1548–1555. doi: 10.1128/AEM.68.4.1548-1555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Scola B, Audic S, Robert C, Jungang L, de Lamballerie X, Drancourt M, et al. A giant virus in amoebae. Science. 2003;299:2033–2033. doi: 10.1126/science.1081867. [DOI] [PubMed] [Google Scholar]
- Luo C, Tsementzi D, Kyrpides NC, Konstantinidis KT. Individual genome assembly from complex community short-read metagenomic datasets. ISME J. 2012;6:898–901. doi: 10.1038/ismej.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D, Brussaard CPD, Thyrhaug R, Bratbak G, Vaulot D. Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl Environ Microbiol. 1999;65:45–52. doi: 10.1128/aem.65.1.45-52.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia M, Swan BK, Poulton NJ, Gomez ML, Masland D, Sieracki ME, et al. High-throughput single-cell sequencing identifies photoheterotrophs and chemoautotrophs in freshwater bacterioplankton. ISME J. 2012;6:113–123. doi: 10.1038/ismej.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez Martínez J, Poulton NJ, Stepanauskas R, Sieracki ME, Wilson WH. Targeted sorting of single virus-infected cells of the coccolithophore Emiliania huxleyi. PLoS One. 2011;6:e22520. doi: 10.1371/journal.pone.0022520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsen F, Kodner R, Armbrust EV. pplacer: linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinformatics. 2010;11:538. doi: 10.1186/1471-2105-11-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokili JL, Rohwer F, Dutilh BE. Metagenomics and future perspectives in virus discovery. Curr Opin Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Fu L, Sun S, Li W. Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinformatics. 2010;11:187. doi: 10.1186/1471-2105-11-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono A. Viruses and lipids. Viruses. 2010;2:1236–1238. doi: 10.3390/v2051236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, et al. The 1.2-megabase genome sequence of mimivirus. Science. 2004;306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- Robertson KL, Verhoeven AB, Thach DC, Chang EL. Monitoring viral RNA in infected cells with LNA flow-FISH. RNA. 2010;16:1679–1685. doi: 10.1261/rna.2016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwer F, Edwards R. The phage proteomic tree: a genome-based taxonomy for phage. J Bacteriol. 2002;184:4529–4535. doi: 10.1128/JB.184.16.4529-4535.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roine E, Bamford DH. Lipids of archaeal viruses. Archaea. 2012;2012:8. doi: 10.1155/2012/384919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld T, Patterson M, Richardson PM, Wommack KE, Young M, Mead D. Assembly of viral metagenomes from yellowstone hot springs. Appl Environ Microbiol. 2008;74:4164–4174. doi: 10.1128/AEM.02598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro HM. Practical Flow Cytometry. John Wiley & Sons, Inc.: Hoboken, NJ, USA; 2005. Flow Sorting; pp. 257–271. [Google Scholar]
- Sieracki M, Poulton N, Crosbie N.2005Automated isolationtechniques for microalgaeIn: Andersen R (ed)Algal culturing techniques Elsevier: San Diego [Google Scholar]
- Stepanauskas R, Sieracki ME. Matching phylogeny and metabolism in the uncultured marine bacteria, one cell at a time. Proc Natl Acad Sci USA. 2007;104:9052–9057. doi: 10.1073/pnas.0700496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward G, Preston C. Analysis of a viral metagenomic library from 200 m depth in Monterey Bay, California constructed by direct shotgun cloning. Virology J. 2011;8:287. doi: 10.1186/1743-422X-8-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses—major players in the global ecosystem. Nature Rev. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the Dark Ocean. Science. 2011;333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protocols. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- Tomaru Y, Nagasaki K. Flow cytometric detection and enumeration of DNA and RNA viruses infecting marine eukaryotic microalgae. J Oceanogr. 2007;63:215–221. [Google Scholar]
- Van Etten JL. Giant Viruses. American Scientist. 2011;99:304–311. [Google Scholar]
- Williamson KE, Wommack KE, Radosevich M. Sampling natural viral communities from soil for culture-independent analyses. Appl Environ Microbiol. 2003;69:6628–6633. doi: 10.1128/AEM.69.11.6628-6633.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WH, Allen MJ.2009Giant viruses and their genomesIn: Feng Z, Long M (eds)Viral genomes: Diversity, Properties and Parameters Nova Science Publishers, Inc: Hauppage, NY, USA; 145–157. [Google Scholar]
- Wilson WH, Schroeder DC, Allen MJ, Holden M, Parkhill J, Barrell BG, et al. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309:1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- Wommack KE, Bhavsar J, Polson SW, Chen J, Dumas M, Srinivasiah S, et al. VIROME: a standard operating procedure for analysis of viral metagenome sequences. Stand Genomic Sci. 2012;6:13. doi: 10.4056/sigs.2945050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Sime-Ngando T, Winget DM, Jamindar S, Helton RR.2010Filtration-based methods for the collection of viral concentrates from large water samplesIn: Wilhelm SW, Weinbauer MG, Suttle CA (eds)Manual of Aquatic Viral Ecology ASLO: Wako, TX, USA; 110–117. [Google Scholar]
- Woyke T, Sczyrba A, Lee J, Rinke C, Tighe D, Clingenpeel S, et al. Decontamination of MDA reagents for single cell whole genome amplification. PLoS One. 2011;6:e26161. doi: 10.1371/journal.pone.0026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau S, Lauro FM, DeMaere MZ, Brown MV, Thomas T, Raftery MJ, et al. Virophage control of antarctic algal host-virus dynamics. Proc Natl Acad Sci USA. 2011;108:6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Price DC, Stepanauskas R, Rajah VD, Sieracki ME, Wilson WH, et al. Single-cell genomics reveals organismal interactions in uncultivated marine protists. Science. 2011;332:714–717. doi: 10.1126/science.1203163. [DOI] [PubMed] [Google Scholar]
- Yutin N, Colson P, Raoult D, Koonin E. Mimiviridae: clusters of orthologous genes, reconstruction of gene repertoire evolution and proposed expansion of the giant virus family. Virology J. 2013;10:106. doi: 10.1186/1743-422X-10-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutin N, Wolf Y, Raoult D, Koonin E. Eukaryotic large nucleo-cytoplasmic DNA viruses: clusters of orthologous genes and reconstruction of viral genome evolution. Virology J. 2009;6:223. doi: 10.1186/1743-422X-6-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC, et al. Nature. 2013. pp. 357–360. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.