Abstract
Viruses contribute to the mortality of marine microbes, consequentially altering biological species composition and system biogeochemistry. Although it is well established that host cells provide metabolic resources for virus replication, the extent to which infection reshapes host metabolism at a global level and the effect of this alteration on the cellular material released following viral lysis is less understood. To address this knowledge gap, the growth dynamics, metabolism and extracellular lysate of roseophage-infected Sulfitobacter sp. 2047 was studied using a variety of techniques, including liquid chromatography–tandem mass spectrometry (LC-MS/MS)-based metabolomics. Quantitative estimates of the total amount of carbon and nitrogen sequestered into particulate biomass indicate that phage infection redirects ∼75% of nutrients into virions. Intracellular concentrations for 82 metabolites were measured at seven time points over the infection cycle. By the end of this period, 71% of the detected metabolites were significantly elevated in infected populations, and stable isotope-based flux measurements showed that these cells had elevated metabolic activity. In contrast to simple hypothetical models that assume that extracellular compounds increase because of lysis, a profile of metabolites from infected cultures showed that >70% of the 56 quantified compounds had decreased concentrations in the lysate relative to uninfected controls, suggesting that these small, labile nutrients were being utilized by surviving cells. These results indicate that virus-infected cells are physiologically distinct from their uninfected counterparts, which has implications for microbial community ecology and biogeochemistry.
Keywords: marine biogeochemistry, virus infection, bacterial physiology, metabolomics
Introduction
It is estimated that 20–30% of the marine microbial community is infected by viruses at any given time. This contributes to as much as 3 Gt of carbon that are released in the world's oceans each year as a result of virus-mediated lysis (Suttle, 2007). The released dissolved organic matter (DOM) is a major contributor to marine microbial activity (Weinbauer et al., 2011), stimulating both primary and secondary productivity (see, for example, Middelboe and Lyck, 2002; Riemann and Middelboe, 2002; Middelboe and Jorgensen, 2006). Through their lytic activities, viruses influence the flow of carbon, nitrogen and other nutrients in the marine environment (Brussaard et al., 2008).
To better define the role of viruses as global drivers of nutrient flow, we need an improved understanding of the impact of infection on host metabolic processes. Presently, little is known of the influences of bacteriophage infection on global host metabolism beyond isolated model systems. A few marine phage–host systems appear to sequester sufficient resources for nucleic acid synthesis entirely from degradation of host DNA (Wikner et al., 1993), although the sources of other macromolecular building blocks are unknown. Much of the knowledge concerning phage–host interactions comes from studies in Escherichia coli. Both lysogenic and lytic coliphages initially promote similar alterations in host metabolism (Calendar, 2006) that halt host cell DNA synthesis (Kazmierczak and Rothman-Denes, 2006), degrade host DNA (Miller et al., 2003) and assemble the machinery for viral production (Taylor, 1995). These activities drastically alter aspects of host metabolism in a specific manner that favors virus replication (see, for example, Poranen et al., 2006). In fact, a number of viruses contain auxiliary metabolic genes to overcome rate-limiting steps in host biosynthesis (Breitbart, 2012). A dramatic example of host manipulation has been demonstrated in marine cyanophage that encode and express photosynthesis proteins homologous to those found in their hosts (Thompson et al., 2011). However, the extent to which pathway-specific alterations of host metabolism is a universal strategy among phages remains unknown. Furthermore, the hijacking of host metabolic processes may not be fully sufficient to supply the demand for rapid virus production, necessitating the acquisition of resources from the environment. For example, the breakdown of host DNA does not fully supply precursors needed for coliphage DNA synthesis. Thus, an initial lag in E. coli metabolism is followed by rapid uptake of nutrients to be used for de novo synthesis of macromolecular building blocks (Friedman and Gots, 1953). Similarly, it has been found that thymidine uptake by infected Pseudoalteromonas sp. SKA18 occurred at rates similar to uninfected cells during the early stages of infection, yet became markedly enhanced just before onset of cell lysis (Middelboe, 2000). These observations suggest an interplay between the viral life cycle and environmental nutrient availability.
From the standpoint of characterizing the influence of virus activity on ocean biogeochemistry, most studies have focused on characterizing the bulk material properties of lysate DOM (for example, total C, N, Fe, Se; see Gobler et al., 1997; Bratbak et al., 1998; Poorvin et al., 2004; Lønborg et al., 2013) or monitoring a few select molecules (for example, dimethyl sulfide, acrylate) or compound classes, such as amino acids and carbohydrates (see, for example, Weinbauer and Peduzzi, 1995; Middelboe and Jorgensen, 2006; Shelford et al., 2012). In general, these studies reported increases in these molecules in lysates (see, for example, Poorvin et al., 2004; Lønborg et al., 2013) and have hypothesized that viral lysates are rich in free and combined amino acids (Middelboe and Jorgensen, 2006) and may be an important source of labile organic nitrogen (Shelford et al., 2012). Only recently has the technology necessary to monitor metabolism at the systems level become available via nuclear magnetic resonance (Fiehn, 2002) or liquid chromatography–mass spectrometry (LC-MS/MS)-based metabolomics (Siuzdak, 1994; Coon et al., 2005; Bajad et al., 2006), and these same tools can be used to help characterize the chemical composition of DOM.
Bacteria belonging to the Roseobacter lineage, which includes Sulfitobacter species, are abundant in marine systems and carry out critical biogeochemical transformations (Buchan et al., 2005). Herein we report the study of a Sulfitobacter sp. 2047 roseophage infection system in which LC-MS/MS-based metabolomics techniques were employed to both compare the metabolism of an infected population with an uninfected control and to gain insight into the interaction between active infection and small metabolites within the DOM pool. As nonlipid-containing phages comprise primarily nucleic acid and protein, typically in an equal mass ratio (see, for example, Bancroft and Freifelder, 1970), we hypothesized that any influences of viral infection on host metabolism would be manifested in pathways that contribute to the synthesis of the biochemical building blocks for these macromolecules and that this would be reflected in the suite of metabolites released from lysed cells.
Materials and methods
Bacterium and phage isolation
Sulfitobacter sp. 2047 was isolated from a mesocosm study in Raunefjorden, Norway, in 2008 by enrichment with dimethylsufoniopropionate (Pagarete et al., 2011; Budinoff, 2012). Agar plates (0.8% w/v Noble Agar; Difco, Sparks, MD, USA) were made using 0.22 μm filtered fjord water and 10 mM dimethylsufoniopropionate (kindly provided by J Henrikson and W Whitman, UGA, Athens, GA, USA). The strain was subsequently maintained in artificial sea water: 1.0% agar plates (230 mM NaCl, 5.3 mM KCl, 3.9 mM CaCl2, 0.1 mM H3BO3, 11.8 mM MgSO4, 11.2 mM MgCl2, 0.8 mM NaHCO3, 5 mM NH4Cl, 75 μM K2HPO4 and 10 mM Tris-HCl (pH 7.5)) supplemented with 0.25% yeast extract and 0.4% tryptone at 20 °C. Filter-sterilized (0.22 μm) stock solutions were added to the autoclaved basal salt solution along with vitamins, iron and trace metals prepared as previously described (Budinoff and Hollibaugh, 2007). A lytic phage (Φ2047B) (Budinoff, 2012; Ankrah et al., 2014) was isolated from viral concentrates of Raunefjorden sea water using standard bacteriophage enrichment (Van Twest and Kropinski, 2009; Wommack et al., 2009). Plaque purification and preparation of phage stocks were based on previously described methods (Kropinski et al., 2009). Concentrated lysates were made by gently washing soft agar from plaque assay plates with MSB buffer. The final purified phage concentrate was 0.22 μm filtered and stored at 4 °C in the dark. All growth experiments with the bacterium, including infections, were performed in artificial sea water at 20 °C. All chemicals were obtained from Fisher Scientific (Fair Lawn, NJ, USA). Genome analysis of Sulfitobacter sp. 2047 revealed it is lysogenized by a prophage (designate as Φ2047A) that is genetically distinct from Φ2047B (HQ317387).
Bacterial culture conditions and metabolite measurements
To determine whether phage infection shifts the metabolite repertoire of Sulfitobacter sp. 2047, the bacterium was grown in artificial sea water supplemented with 10 mM sodium acetate at 25 °C in the dark with 200 r.p.m. agitation. Once cultures reached an optical density (OD540) of ca. 0.17, phages were added at a multiplicity of infection of 4 (4 phage cell−1). Under these conditions, cell lysis is evident at ∼240 min post infection (p.i.). To monitor host metabolites during all phases of infection, samples were collected before phage addition (t=0) and then 15, 30, 60, 120, 240, 360 and 480 min post phage addition. Extracellular metabolites were measured in cell-free filtrates of the populations at 480 min. Control cultures (without added phages) were maintained and sampled in parallel. Samples taken for microscopic analysis were fixed with gluteraldehyde (0.5%), stored at −80 °C and processed using standard techniques. Samples for quantitative polymerase chain reaction (qPCR) were flash frozen and stored at −80 °C until processing, whereas samples for metabolite analysis were immediately processed.
Following the initial experiment, a separate analysis using stable isotope-labeled nutrients was performed to examine turnover rates and cellular fluxes for a subset of metabolites (Supplementary Table 2). For these studies, 20 mM 13C-labeled sodium acetate (1,2-13C2, 99%, Cambridge Isotope Laboratories, Andover, MA, USA) was added to a set of control and viral infected cultures at either 0 min or 240 min p.i. These two time points were selected to represent each of the two phases of infection that were evident from the metabolite profiles identified in the initial experiment described above. Samples for the flux analysis were taken from each culture at 0, 5, 15, 30, 60 and 120 min post addition of 13C-acetate. However, isotope incorporation was complete at 60 min, and the data presented herein reflect this.
The targeted metabolomic methods employed to measure relative intracellular metabolite concentrations and turnover rates used slight modifications of a known metabolite extraction procedure (Rabinowitz and Kimball, 2007; Bennett et al., 2008; Yuan et al., 2008). Briefly, 10 ml of cells were rapidly collected on Magna nylon filters (Millipore, Billerica, MA, USA) via vacuum filtration and then extracted by placing the filter directly in a petri dish containing extraction solvent at 4 °C as previously described. Extracellular metabolites from both infected and control cultures were collected at 480 min by filtering 10 ml of culture as previously described and collecting the filtrate. The resulting liquids were lyophilized and then resuspended in 300 μl 40:40:20 acetonitrile/methanol/water with 0.1 M formic acid for MS analysis. Two LC-MS/MS analyses, one in each of positive and negative ion modes, were performed for each sample, and relative metabolite levels and fluxes were analyzed as previously described (Rabinowitz and Kimball, 2007; Bennett et al., 2008; Yuan et al., 2008).
Metabolite data analysis
Signal intensity for all metabolites was analyzed as peak area, and each measurement was manually curated using the Xcalibur 2.0.7 Quan Browser analysis package (Thermo Fisher Scientific, Waltham, MA, USA). Metabolite area counts were normalized to cell density to obtain per cell metabolite abundances, and a ratio of these values from each condition was used to compare relative concentrations of metabolites. All heat maps were generated using Gene Cluster 3.0 (de Hoon et al., 2004) and viewed using Java TreeView 1.1.5 (Saldanha, 2004). Interactions among the metabolites were visualized using resources available through the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (www.genome.jp/kegg/).
Data analysis for stable isotope incorporation into metabolites
Peak areas for all compounds detected in the 13C incorporation experiments were calculated as described for the pool size data above. Once the peak areas were determined, the ratios of unlabeled, partially labeled and fully labeled metabolites were determined for each time point. These values were then used to calculate the rate of disappearance (turnover) for the unlabeled material. The data were fit using the following equation: y=Ae(-kx)+c, where k is the rate constant. This equation is most correct for populations that have reached steady state, which may not be the case in the experiments reported herein because of the addition of a pulse of labeled nutrients. Therefore, the ratios of rate constants from the phage-infected and control cultures were calculated and multiplied by the fold difference in metabolite concentration to determine relative fluxes.
Phage enumeration
Phages were enumerated using qPCR assays specific for each of the two phages (Φ2047A and Φ2047B). Genome sequences were obtained for the host and Φ2047B, facilitating the design and optimization of phage-specific primer sets (see Supplementary Materials). qPCR was carried out by using a DNA Engine Opticon 2 real-time PCR detector with the Opticon Monitor 3.1.32 software package (Bio-Rad Laboratories, Inc., Hercules, CA, USA). qPCR reactions were performed in a 25 μl volume with 12.5 μl SYBR Premix Ex Taq cocktail RR041 (Perfect Real Time; Takara Bio, Inc., Shiga, Japan), 500 nM forward and reverse primers and 10.0 μl of template dilutions. The amplification programs for each primer set are as follows: 95 °C for 3 min, 40 cycles of: 95 °C for 20 s, 56 °C or 55 °C (Φ2047A or Φ2047B, respectively) for 20 s, 72 °C for 20 s, followed by 5 min at 72 °C. Fluorescence measurements were conducted at the end of each cycle at 72 °C. Melt curves were generated after each assay to verify the specificity of the amplification by heating from 50 °C to 100 °C, read every 1 °C. Standards were developed from plasmids containing cloned sequences and 10-fold serial dilutions of these samples in 10 mM Tris-HCl were used in the reactions. Standard curves were determined as the correlation between the log of gene copy numbers and the Ct. In all cases, correlation coefficients for standard curves assays were ⩾0.99.
Full instrumentation and additional experimental details are available in the supporting documents.
Results
The Sulfitobacter-phage infection cycle
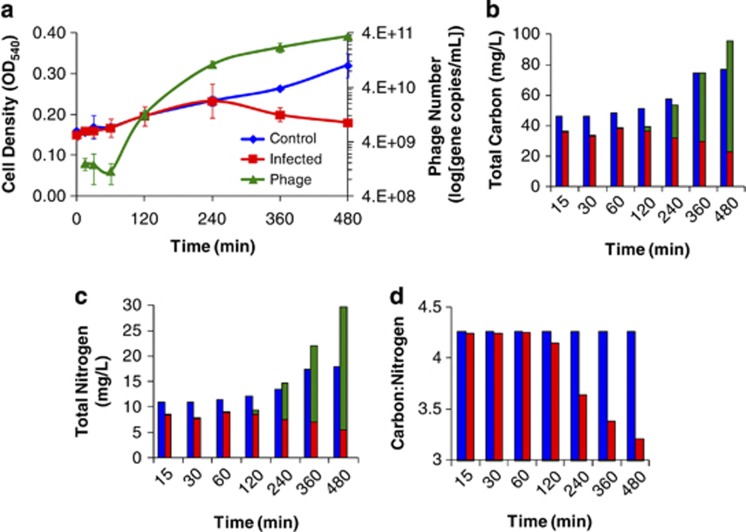
The growth of the infected Sulfitobacter sp. 2047 cultures was nearly identical to that of the uninfected controls until the onset of cell lysis at 240 min p.i. (Figure 1a). Rapid production of phages occurred between 60 and 240 min p.i., at which point it plateaued. By the end of the 480 min experiment, 3.53 × 1011 new virus particles had been produced, the density of bacteria in the flasks receiving phages was ∼30% of that present in the no-virus control cultures, and phages outnumbered bacteria by ∼2500 to 1 (Figure 1). Under the growth conditions employed here, infection of the host with the lytic phage results in induction of a prophage for at least a portion of the host population, leading to the propagation of both phages in batch culture (Supplementary Table S1).
Figure 1.
(a) Sulfitobacter sp. 2047 cell density (OD540) and phage concentration at each metabolite sampling time point reported in Figure 2. Line graphs are color coded as follows: phage-infected culture ( ), control culture (
), control culture ( ) and phage numbers (
) and phage numbers ( ). Turbidity declines are indicative of phage-induced lysis. Phage numbers were derived from qPCR assays. Averages and ranges of biological duplicates are reported. Estimates of (b) carbon and (c) nitrogen content and their (d) ratios for Sulfitobacter sp. 2047 cells and phage during an infection cycle. Bar graphs are color coded as follows: phage (
). Turbidity declines are indicative of phage-induced lysis. Phage numbers were derived from qPCR assays. Averages and ranges of biological duplicates are reported. Estimates of (b) carbon and (c) nitrogen content and their (d) ratios for Sulfitobacter sp. 2047 cells and phage during an infection cycle. Bar graphs are color coded as follows: phage ( ), infected culture (
), infected culture ( ) and control culture (
) and control culture ( ). Bacterial cell densities were determined by microscopy. Phage numbers were determined using qPCR. Bacterial carbon (149 fg C per cell) and nitrogen (35 fg N per cell) were derived from literature values of marine bacteria as reported in text. Phage carbon (0.2 fg C per phage) and nitrogen (0.076 fg N per phage) were derived from theoretical calculations (Jover et al., 2014). Values reflect the average of duplicate biological replicates.
). Bacterial cell densities were determined by microscopy. Phage numbers were determined using qPCR. Bacterial carbon (149 fg C per cell) and nitrogen (35 fg N per cell) were derived from literature values of marine bacteria as reported in text. Phage carbon (0.2 fg C per phage) and nitrogen (0.076 fg N per phage) were derived from theoretical calculations (Jover et al., 2014). Values reflect the average of duplicate biological replicates.
Stoichiometric shifts in carbon and nitrogen associated with phage infection
The distributions of carbon (C) and nitrogen (N) in biological particles for the control culture and the Sulfitobacter-roseophage infection system were calculated based on reported estimates of the C and N content in both marine viruses and bacteria. The total masses of bacteria (cellular) and phage C and N were calculated using previously reported elemental estimates of 0.2 fg C and 0.076 fg N per phage and 149 fg C and 35 fg N per cell (Heldal et al., 1985; Vrede et al., 2002; Chan et al., 2012; Jover et al., 2014). At the final time point, cells in the control culture contained 76.74 (±10.43) and 18.0 (±2.45) mg l−1 of C and N, respectively (Figures 1b and c). During the experiment, this culture was estimated to incorporate 31.33 and 7.33 mg l−1 of C and N, respectively, into new cells. In comparison, phage-infected cultures were found to have 95 mg l−1 of C in both intact bacterial cells (22.35±2.98 mg l−1) and viral particles (72.66±2.05 mg l−1) by the final time point. As for N, 29.59 mg l−1 had been incorporated in total with 5.25 (±0.7) and 24.34 (±0.7) mg l−1 being found in bacteria and phages, respectively (Figures 1b and c). These data were used to calculate the C/N ratio for infected and control populations throughout the course of the experiment. This ratio remained constant for the control populations, but decreased by 32% for the infected cultures by 480 min (Figure 1d).
Intracellular metabolite concentrations throughout the infection cycle
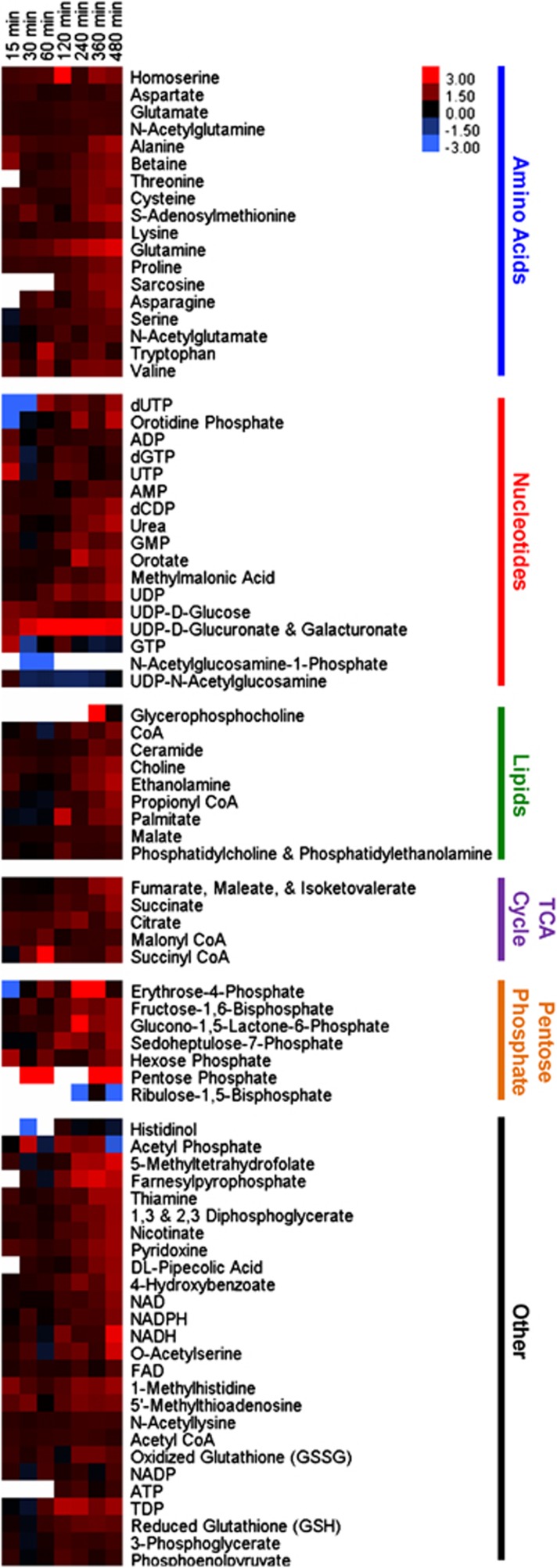
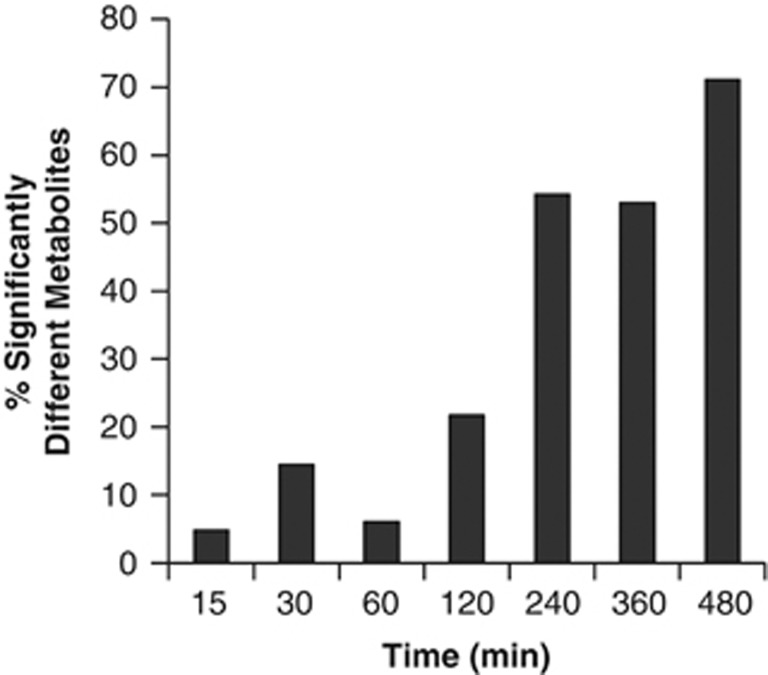
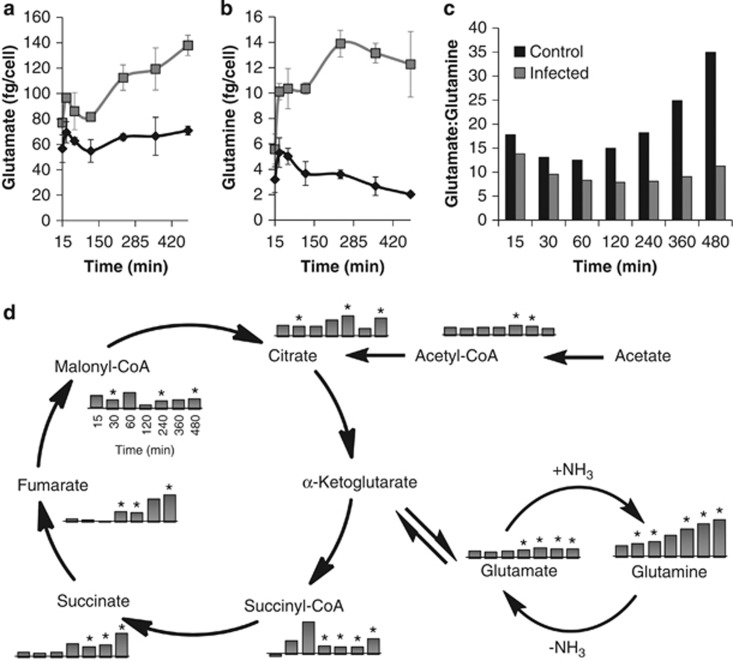
The relative concentrations of 82 central pathway metabolites (the ‘core metabolome') were measured via LC-MS/MS for both infected and control populations at seven discrete time points throughout the infection cycle (Figure 2). During the early infection period (ending at 120 min p.i.), <25% of the measured metabolites were significantly different between the infected and control populations (18 at P⩽0.05 and ⩾1.5-fold change). At the end of the infection cycle (480 min p.i.), ∼75% of metabolite concentrations were significantly elevated (59 at P⩽0.05 and ⩾1.5-fold change) in phage-infected populations (Figure 3). The intracellular metabolite concentration progressively increased in the phage-infected populations for nearly every metabolite over the course of the infection cycle. In fact, almost no metabolites showed a significant decrease in concentration (Supplementary Table S3). For example, dramatic differences were evident in uridine diphosphate (UDP)-activated sugars. The relative concentration of UDP-glucose in the infected population was elevated 1.7-fold 15 min after phage addition and remained fairly constant throughout the experiment. UDP-glucuronate/galacturonate became elevated over time with a 13-fold spike in concentration at 120 min p.i., the time point preceding widespread cell lysis. Similarly, several amino acids were elevated in concentration in the late-phase infected populations relative to controls by 1.6- to 6.0-fold, with an average of 2.9-fold, at 480 min p.i. (that is, alanine, aspargine, aspartate, cysteine, glutamate, glutamine, lysine, proline, serine, threonine and valine). To verify that Sulfitobacter sp. 2047 contained high concentrations of glutamate and glutamine, their absolute concentrations were determined for this experiment and found to be 7.1 × 10−14 g per cell and 2 × 10−15 g per cell, respectively, in the infected culture at the final time point (Figures 4a and b).
Figure 2.
Heat map of intracellular metabolites of phage-infected and control Sulfitobacter sp. 2047 populations. Metabolite concentrations are normalized to bacterial cell number and expressed relative to levels measured in the uninfected host cells at the corresponding time point. Ratios are log2 transformed. Increases in intracellular metabolite concentrations are shown in red and decrease in blue. Columns correspond to min post infection, and rows represent specific metabolites. Values are averages of duplicate biological and technical replicates and are reported in Supplementary Table S3.
Figure 3.
Variation in intracellular metabolite concentrations between phage-infected and control populations during the infection cycle shown in Figure 2. Columns indicate fold changes ⩾1.5 and P⩽0.05, and all data are shown in Supplementary Tables S3 and S4.
Figure 4.
Absolute concentrations of (a) glutamate and (b) glutamine in control (♦) and phage-infected (▪) Sulfitobacter sp. 2047 populations. Values represent duplicate biological and duplicate technical replicates. Error bars show the s.e.m. (c) Glutamate to glutamine ratios for phage-infected and control populations throughout the experimental time course. Bar graphs are coded as indicated by the key in each figure. (d) Selected metabolites are shown to illustrate the relationship between the tricarboxylic acid (TCA) cycle and glutamate and glutamine metabolism in Sulfitobacter sp. 2047. The bar graphs represent concentrations for metabolites in the phage-infected culture and are expressed as fold change relative to the control at the corresponding time point as shown for malonyl-Coenzyme A. Fold changes are log2 transformed and represent averages of duplicate biological and technical replicates. Asterisks designate significant fold changes (⩾1.5 and P⩽0.05).
Metabolic turnover and flux during phage infection
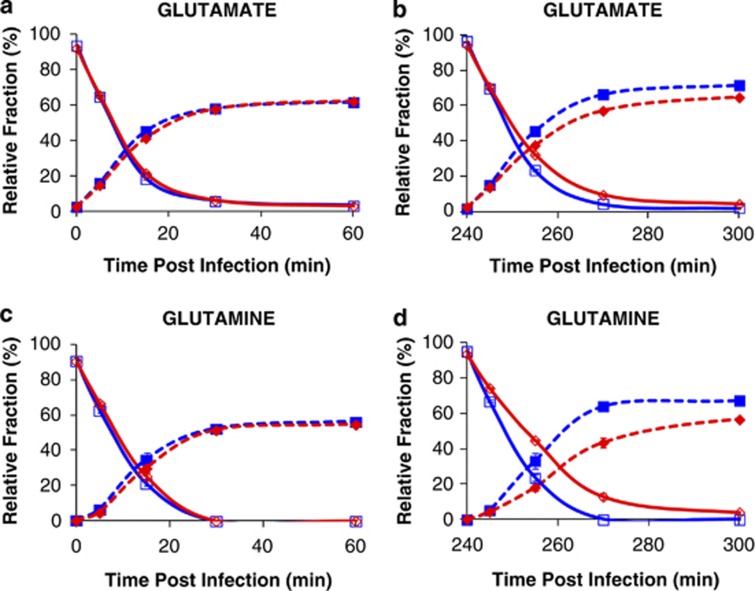
Complementary experiments to analyze metabolite turnover and metabolic flux were also performed using a stable isotope-labeled carbon source. 13C-acetate was added to separate sets of cultures at times that matched both the early (immediately following virus addition) and late (240 min p.i.) phases that were identified from the time course used to determine growth dynamics and overall metabolite concentration (see Figures 1a and 2). Incorporation of the label into 14 selected metabolites representing tricarboxylic acid cycle components, N assimilation intermediates and amino acids was monitored via LC-MS/MS during the first 120 min following 13C acetate addition in both cases. These measurements provide two useful pieces of data: (1) the rate at which the metabolite pool is converted to entirely new metabolites because of biosynthesis and (2) the total amount of nutrients that pass through a specific compound, that is, the flux through them, which is proportional to the turnover rate and metabolite pool size. With the exception of glutamate and glutamine (Table 1 and Figure 5), the turnover rates for all measured metabolites were indistinguishable between the control and viral-treated cultures throughout the infection cycle (Supplementary Tables S5–12). Consequently, the fluxes for these metabolites were increased in infected populations as the larger pools were being replenished at equivalent, or only slightly reduced, rates. The measured turnover rates for glutamate and glutamine are indistinguishable between the control and viral-treated cultures during the early infection period (Figures 5a and c), but they are decreased in the infected cultures during the later stages of infection (Figures 5b and d). The calculated flux through glutamate in the infected cultures is equivalent to controls throughout both phases of the infection cycle. However, flux through glutamine is 70–100% greater and 200–400% greater during the early and late infection periods, respectively (Table 1).
Table 1. Relative flux measurements of glutamate and glutamine.
| Time (min) |
Glutamate |
Glutamine |
||||
|---|---|---|---|---|---|---|
| Relative poola,b | Relative turnovera,c | Relative fluxa | Relative poola,b | Relative turnovera,c | Relative fluxa | |
| 60 (early) | 1.40±0.33 | 0.910±0.001 | 1.28±0.30 | 1.86±0.40 | 0.914±0.004 | 1.71±0.36 |
| 120 (early) | 1.52±0.30 | 0.910±0.001 | 1.38±0.27 | 2.30±0.26 | 0.914±0.004 | 2.10±0.23 |
| 240 (late) | 1.67±0.17 | 0.7961±0.0002 | 1.33±0.14 | 3.52±0.76 | 0.720±0.002 | 2.53±0.55 |
| 360 (late) | 1.60±0.42 | 0.7961±0.0002 | 1.27±0.33 | 5.68±1.68 | 0.720±0.002 | 4.08±1.21 |
Relative pool and turnover were calculated by dividing the measured values from the infected by that of the control. Relative flux was calculated as the product of the relative pool and turnover. In each case, s.d. values were determined for measured values and then propagated through the calculations as appropriate for the mathematical operation being performed.
Data were obtained from the metabolite measurement experiment.
Data were obtained from the stable isotope incorporation experiment.
Figure 5.
Incorporation of acetate-derived 13C into (a, b) glutamate and (c, d) glutamine in phage-infected and control populations during two distinct phases of infection: (a, c) 0–60 min post infection (early) and (b, d) 240–300 min post infection (late). The graphs show the disappearance of unlabeled metabolites for phage-infected ( ) and control (
) and control ( ) populations as well as the appearance of fully 13C-labeled metabolites in phage-infected (
) populations as well as the appearance of fully 13C-labeled metabolites in phage-infected ( ) and control (
) and control ( ) populations. Error bars represent range of the data and are obscured by the data markers, in some cases.
) populations. Error bars represent range of the data and are obscured by the data markers, in some cases.
Composition of extracellular small-molecule components following phage-induced cell lysis
Cell-free filtrates of phage-infected and control cultures were collected at 480 min p.i., and 56 metabolites were detected in filtrates from either infected or control cultures (Table 2). The metabolite concentrations in the infected cultures were not uniformly higher for every molecule. Instead, there was an increase in 29%, no change in 45% and a decrease in 27% of the metabolite concentrations detected in the virus-infected cultures (Table 2). For example, several components of C and N metabolism (that is, α-ketoglutarate, phosphoenolpyruvate, lactate and erythrose-4-phosphate) as well as small cofactors (flavin adenine dinucleotide and nicotinamide adenine dinucleotide) are either decreased or not detected in the filtrates of virus-infected cultures relative to controls. In contrast, cell wall constituents, such as phosphatidylcholine and phosphatidylethanolamine, were present only in the filtrates of virus-infected cultures. In addition, several amino acids (asparagine, methionine, cysteine and homoserine) and urea were elevated by an average of 2- and 2.8-fold, respectively. Finally, all acyl-Coenzyme A metabolites were either elevated or only present in infected culture filtrates (Table 2).
Table 2. Metabolite content of cell-free filtrates from phage-infected Sulfitobacter sp. 2047 relative to control cultures at 480 min post infection.
| Metabolite | Fold changea | Metabolite | Fold changea |
|---|---|---|---|
| Amino Acids | Nucleic acids, nucleosides and nucleotides | ||
| Asparagine | 2.37(+) | Deoxyadenosineb | 4.69(+) |
| Methionineb | 1.83(+) | Thymineb | 2.62(+) |
| Cysteine | 1.74(+) | Cytosineb | 1.55(+) |
| Tryptophan | 0.73 | dCDP | 1.24 |
| Threonine | 0.10(−) | UDP | 1.12 |
| Homoserine | 1.87(+) | 5′-Methylthioadenosine | 1.05 |
| GABAb | 1.45 | Orotate | 0.95 |
| Betaine | 1.29 | Dihydrooroate | 0.87 |
| 1-Methylhistidine | 1.28 | N-acetylglucosamine-1-phosphate | 0.8 |
| O-Acetylserine | 0.91 | TDP | 0.57(−) |
| S-Adenosylmethionine | 0.88 | GMP | 0.42(−) |
| N-Acetylornithineb | 0.56(−) | 5-Methyldeoxycytidineb | 0.35(−) |
| TCA cycle | Pentose phosphate | ||
| Malonyl CoAc | 1000.00(+) | Fructose-1,6-bisphosphate | 0.99 |
| Succinyl CoA | 1.34 | Sedoheptulose-7-phosphate | 0.71 |
| Succinate | 0.78 | Erythrose-4-phosphate | 0.00(−) |
| (Fumarate, maleate and isoketovalerate)d | 0.72 | ||
| Citrate | 0.7 | Cofactors, vitamins and electron carriers | |
| 2-Oxoglutarateb | 0.62(−) | Acetyl CoA | 35.17(+) |
| 3-Phosphoglycerate | 1.89(+) | Pyridoxine | 4.63(+) |
| (1,3 and 2,3 bisphosphoglycerate) | 1.1 | Nicotinate | 0.95 |
| Phosphoenolpyruvate | 0.61(−) | 5-Methyltetrahydrofolate | 0.85 |
| Methylmalonic acid | 0.79 | ||
| Lipids | Thiamine | 0.29(−) | |
| (Phosphatidylcholine, phosphatidylethanolamine)c,d | 1000.00(+) | FAD | 0.00(−) |
| Propionyl CoA | 79.81(+) | NAD | 0.00(−) |
| Ethanolamine | 2.93(+) | ||
| Palmitate | 0.97 | Other | |
| Farnesylpyrophosphate | 0.57(−) | Urea | 2.78(+) |
| pipecolic acid | 1.89(+) | ||
| Acetyl phosphate | 0.92 | ||
| 4-Hydroxybenzoate | 0.81 | ||
| Citraconate | 0.42(−) | ||
| Phenylpyruvateb | 0.37(−) | ||
| Lactate | 0.32(−) |
Abbreviations: Co A, Coenzyme A; dCDP, deoxycytidine diphosphate; FAD, flavin adenine dinucleotide; GMP, guanosine monophosphate; NAD, nicotinamide adenine dinucleotide; TCA, tricarboxylic acid; TDP, thymidine diphosphate; UDP, uridine diphosphate.
Metabolite levels in infected cultures are expressed relative to levels in control cultures at 480 min. Elevated metabolites ((+); fold change ⩾1.5) and depressed metabolites ((−); fold change ⩽0.67).
Not detected in intact cells, detected in filtrates only.
Metabolite was not detected in control cultures, and therefore 1000 was used to denote a large increase in concentration in the infected cells.
Metabolites are indistinguishable with the applied method.
Discussion
A way to view the potential metabolic burden of virion production on a host cell is to consider the elemental resources required by the virus. The C/N ratio of phages (3.00) is predicted to be lower than that of bacteria (4.25) (Vrede et al., 2002; Chan et al., 2012; Jover et al., 2014). Quantitative estimates indicated that phage particles represented a significant portion of redistributed C and N in our experiment, and as a result of phage production these elements were incorporated into biological particles (cells and phages) at greater, but disproportionate, amounts compared with control populations. In fact, the infected cultures had incorporated 49% more C and 148% more N than the controls by the conclusion of the experiment. Thus, partitioning of host N into phage particles represented a potential for stoichiometric disruption in the C/N of infected cells.
To assess the effect of phage production on the metabolism of the host population, we measured intracellular metabolites from both the control and infected cultures. These data highlight the distinct metabolic trajectories of these populations, as it was observed that viral infection led to a progressive and general increase in concentration for nearly all measured compounds. Dynamics in phage production and host population survival can be used to divide the time course of the experiment into two phases: an early infection period (0–120 min) and a later infection period (240–480 min). The per-cell metabolite concentrations also support this division. During the second phase of the time course, phage production had leveled off, the host population had dropped to half of its peak cell density and the variation in the core metabolome became more dramatic relative to the uninfected controls.
The 66 metabolites with significantly elevated concentrations in infected cultures clearly point to the population of infected cells being physiologically distinct from those that are uninfected. Differences in UDP-activated sugars (UDP-glucose, UDP-glucuronate/galacturonate) may reflect alterations in cell wall integrity in phage-infected bacteria. Similarly, several amino acids were elevated in concentration in infected populations relative to controls. Of particular interest were glutamate and glutamine, which together serve as the entry points for 99% of inorganic N into biomolecules (Yan, 2007), and are among the most abundant metabolites in E. coli (Bennett et al., 2009). We confirmed that the absolute concentrations for glutamate and glutamine in Sulfitobacter sp. 2047 are similar to those reported for E. coli (1.5 × 10−14 g cell−1 and 1.1 × 10−15 g cell−1, respectively) (Bennett et al., 2009) and thus are expected to be major components of the metabolome. The ratio of intracellular glutamate to glutamine is an indicator of N availability in many bacterial species, with an increased ratio indicating growth under N limitation (Flynn et al., 1989). An analysis of the trends in this ratio for both cultures indicates that the control population, which shows a progressive increase in this ratio through the growth cycle, is likely experiencing decreased N availability as the cultures reach the end of exponential phase. As sufficient ammonium was provided to supply the N required in both cultures, these data suggest that the infected cultures had access to preferred N-containing nutrient sources not available in the control cultures.
Following the measurement of metabolite concentrations during the infection cycle, stable isotope labeling studies using 13C-acetate were performed to determine whether the general increases in metabolite concentrations were because of increased biosynthesis or decreased utilization of each molecule. The rates of label incorporation for 12 of the 14 metabolites monitored indicated that both populations were metabolically active. As metabolite concentrations and turnover were similar between infected and control populations during the early time points, these data further support the observation that host metabolism was not dramatically altered within the first hour following phage addition. However, the flux through all metabolites at the later time points was altered as the higher cellular concentrations of these metabolites in the infected cultures results in an increased flux through these molecules as the larger pools are being replaced in the same unit time. Although the glutamate and glutamine pools turned over more slowly during the later phase of the infection, the elevated concentrations of these compounds resulted in measured fluxes for these metabolites that were equal for glutamate and 200–400% greater for glutamine than those of the control population. We suggest that the apparent decrease in turnover rates for these two molecules is the result of uptake and incorporation of unlabeled nutrients from lysed kin.
The metabolites with decreased concentrations in the filtrates of the infected cultures were primarily those that are expected to be readily recycled, such as components of central C and N metabolism and small cofactors. This suggests that intact cells in the virus-infected cultures remained active and were able to rapidly consume material from lysed cells to support their own metabolism, consequently depleting selected metabolites in the extracellular milieu. Metabolites with increased relative concentrations in the filtrates were those typically related to cellular stress or those that are potentially too large to be transported into the cell (that is, >600 Da; Benz, 1985; Williams, 2000). The presence of bacterial cell wall constituents (that is, phosphatidylcholine and phosphatidylethanolamine) in the phage-infected culture filtrates is also consistent with the degradation of cell wall material from lysed cells and with prior studies demonstrating the prevalence and recalcitrance of bacterial cell wall components (that is, peptidoglycan) in marine (McCarthy et al., 1998; Pedersen et al., 2001; Jørgensen et al., 2003) and virus-derived DOM (Middelboe and Jorgensen, 2006). Dissolved and free amino acids have also been previously identified in viral lysates (Middelboe and Jorgensen, 2006), but the contribution of individual amino acids was not reported. Of the extracellular amino acids detected here, several of those with the largest increases in concentration for the phage-infected cultures are also part of the normal cellular stress response (for example, asparagine, cysteine, homoserine and methionine; Jozefczuk et al., 2010). Acyl-CoAs may not be efficiently transported and assimilated because of their large size and/or low concentrations in nature, potentially explaining their relative increase in infected culture filtrates. The repertoire of metabolites remaining in the lysate may also be a reflection of the host's substrate preferences. For example, urea, a nitrogen-rich byproduct of de novo nucleotide biosynthesis (Berg et al., 2012), was also elevated in infected cultures.
The small molecule composition of the filtrates as well as the intracellular metabolite concentrations and turnover rates suggest that the phage-infected cultures were incorporating nutrients from recycled sources, that is, from small molecules that were liberated during viral lysis or from breakdown of unlabeled macromolecules. The strongest evidence for this phenomenon comes from the data for glutamate and glutamine that, despite showing high intracellular concentrations in infected populations, were undetectable in the culture filtrates. Given that these compounds are typically preferred sources of N (over ammonium) in bacteria (Kirchman et al., 1989; Middelboe et al., 1996; Riemann and Middelboe, 2002), it is likely that these compounds were released from lysed cells and then rapidly assimilated by metabolically active cells.
Ecological implications and conclusions
This work provides a biochemical basis from which a mechanistic understanding of the influence of phage infection on host metabolism can be developed for extension to a biogeochemical framework. In terms of how virus activity shapes host metabolism, our results show that phage infection in Sulfitobacter sp. 2047 does not lead to any pathway-specific alterations in host metabolism. Instead, a general increase in net host metabolic activity is used to meet the requirements of virus propagation that is facilitated by recycling of metabolites from lysed siblings. Additional studies are necessary to determine whether this observed response is generalizable to other host–phage systems. It is relevant to highlight that in the model system used here simultaneous production of two distinct phages (one lytic and one lysogenic) occurs. Furthermore, by the final sampling time point, some unknown and undistinguishable fraction of the population is resistant to infection, or at least lysis. As lysogeny is hypothesized to be prevalent in marine bacteria in general (Stopar et al., 2004; Leitet et al., 2006; Paul, 2008) and in roseobacters in particular (Chen et al., 2006; Zhao et al., 2010), the complexity of this infection process may, in fact, be reflective of events that are common in nature.
An increased demand for N results in a stoichiometric imbalance of C/N in infected cells that has implications for trophic transfer and stimulation of secondary production. In fact, our observations suggest that infected cells are a functionally different entity than their noninfected counterparts. In pelagic environments, the progression of the lytic cycle may thus be constrained by the availability of host-limiting nutrients because of increased cellular demands. Conversely, removal of constraints on nutrient availability may stimulate host growth, thereby facilitating viral production (Higgins et al., 2009; Weinbauer et al., 2009). Indeed, the interplay between nutrient demand and availability may influence the lytic–lysogeny decision (Hong et al., 1971; Herskowitz and Hagen, 1980; Jiang and Paul, 1998) that phages make when infecting a cell. This could also result in the establishment of a state that may be perceived as pseudolysogeny (Łoś and Węgrzyn, 2012) where the cell is infected but there are insufficient cellular resources to propagate new virions. Alternatively, infected cells with access to sufficient nutrients may themselves be N enriched and more susceptible to grazing. To this end, the idea of the infected cell as a unique biochemical cell type continues to gain traction, while the role of viruses as vectors of biogeochemical change continues to expand.
Acknowledgments
We are grateful to Dr Charles Budinoff for isolation of Sulfitobacter sp. 2047 and its infecting phage and Dr Willie Wilson for encouraging and facilitating our participation in the mesocosm experiment that yielded this bacterium and phage. We thank Thomas Lane for the analysis of the Sulfitobacter sp. 2047 genome. We are indebted to Dr Andrew Lang for insightful comments on an earlier version of this manuscript. This work was supported by NSF Grants OCE-1061352 awarded to AB, SRC and SWW. JLM was supported by OCE-1208784 awarded to SRC.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ankrah NYD, Budinoff CR, Wilson WH, Wilhelm SW, Buchan A.2014. Genome sequence of the Sulfitobacter sp. 2047 infecting lytic phage pCB2047-B. Genome Announc in press. [DOI] [PMC free article] [PubMed]
- Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. Separation and quantitation of water soluble cellular metabolites by hydrophilic interaction chromatography-tandem mass spectrometry. J Chromatogr. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Bancroft FC, Freifelder D. Molecular weights of coliphages and coliphage DNA. J Mol Biol. 1970;54:537–546. doi: 10.1016/0022-2836(70)90124-5. [DOI] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc. 2008;3:1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz R. Porin from bacterial and mitochondrial outer membrane. Crit Rev Biochem Mol Biol. 1985;19:145–190. doi: 10.3109/10409238509082542. [DOI] [PubMed] [Google Scholar]
- Berg JM, Tymoczko JL, Stryer L. Biochemistry. W.H. Freeman: Basingstoke; 2012. [Google Scholar]
- Bratbak G, Jacobsen A, Heldal M. Viral lysis of Phaeocystis pouchetii and bacterial secondary production. Aquat Microb Ecol. 1998;16:11–16. [Google Scholar]
- Breitbart M. Marine viruses: truth or dare. Ann Rev Mar Sci. 2012;4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- Brussaard CPD, Wilhelm SW, Thingstad F, Weinbauer MG, Bratbak G, Heldal M, et al. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2008;2:575–578. doi: 10.1038/ismej.2008.31. [DOI] [PubMed] [Google Scholar]
- Buchan A, Gonzalez JM, Moran MA. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–5677. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budinoff CR. Diversity and activity of roseobacters and roseophage. PhD dissertation, University of Tennessee: Knoxville, TN, USA; 2012. [Google Scholar]
- Budinoff CR, Hollibaugh JT. Ecophysiology of a Mono Lake picocyanobacterium. Limnol Oceanogr. 2007;52:2484–2495. [Google Scholar]
- Calendar R. The Bacteriophage. Oxford Uninversity Press: New York; 2006. [Google Scholar]
- Chan LK, Newton RJ, Sharma S, Smith CB, Rayapati P, Limardo AJ, et al. Transcriptional changes underlying elemental stoichiometry shifts in a marine heterotrophic bacterium. Front Microbiol. 2012;3:159. doi: 10.3389/fmicb.2012.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wang K, Stewart J, Belas R. Induction of multiple prophages from a marine bacterium: a genomic approach. Appl Environ Microbiol. 2006;72:4995–5001. doi: 10.1128/AEM.00056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon JJ, Ueberheide B, Syka JE, Dryhurst DD, Ausio J, Shabanowitz J, et al. Protein identification using sequential ion/ion reactions and tandem mass spectrometry. Proc Natl Acad Sci USA. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- Fiehn O. Metabolomics–the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- Flynn KJ, Dickson DMJ, Al-Amoudi OA. The ratio of glutamine: glutamate in microalgae: a biomarker for N-status suitable for use at natural cell densities. J Plankton Res. 1989;11:165–170. [Google Scholar]
- Friedman S, Gots JS. The purine and pyrimidine metabolism of normal and phage infected Escherichia coli. J Biol Chem. 1953;201:125–135. [PubMed] [Google Scholar]
- Gobler CJ, Hutchins DA, Fisher NS, Cosper EM, Sanudo-Wilhelmy SA. Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnol Oceanogr. 1997;42:1492–1504. [Google Scholar]
- Heldal M, Norland S, Tumyr O. X-Ray microanalytic methods for measurement of dry matter and elemental content of individual bacteria. Appl Environ Microbiol. 1985;50:1251–1257. doi: 10.1128/aem.50.5.1251-1257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I, Hagen D. The lysis-lysogeny decision of phage lambda: explicit programming and responsiveness. Annu Rev Genet. 1980;14:399–445. doi: 10.1146/annurev.ge.14.120180.002151. [DOI] [PubMed] [Google Scholar]
- Higgins JL, Kudo I, Nishioka J, Tsuda A, Wilhelm SW. The response of the virus community to the SEEDS II mesoscale iron fertilization. Deep Sea Res Part (II Top Stud Oceanogr) 2009;56:2788–2795. [Google Scholar]
- Hong JS, Smith GR, Ames BN. Adenosine 3′: 5′-cyclic monophosphate concentration in the bacterial host regulates the viral decision between lysogeny and lysis. Proc Natl Acad Sci USA. 1971;68:2258–2262. doi: 10.1073/pnas.68.9.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Paul J. Significance of lysogeny in the marine environment: studies with isolates and a model of lysogenic phage production. Microb Ecol. 1998;35:235–243. doi: 10.1007/s002489900079. [DOI] [PubMed] [Google Scholar]
- Jørgensen NO, Stepanaukas R, Pedersen AGU, Hansen M, Nybroe O. Occurrence and degradation of peptidoglycan in aquatic environments. FEMS Microbiol Ecol. 2003;46:269–280. doi: 10.1016/S0168-6496(03)00194-6. [DOI] [PubMed] [Google Scholar]
- Jover LH, Effler TC, Buchan A, Wilhelm S, Weitz JS.2014. The elemental composition of virus particles: implications for marine biogeochemical cycles. Nat Rev Micro in press. [DOI] [PubMed]
- Jozefczuk S, Klie S, Catchpole G, Szymanski J, Cuadros-Inostroza A, Steinhauser D, et al. Metabolomic and transcriptomic stress response of Escherichia coli. Mol Syst Biol. 2010;6:364. doi: 10.1038/msb.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak KM, Rothman-Denes LB.2006Bacteriophage N4In: Calendar R (eds). The Bacteriophages Oxford Uninversity Press: New York; 302–314. [Google Scholar]
- Kirchman DL, Keil RG, Wheeler PA. The effect of amino acid on ammonium utilization and regeneration by heterotrophic bacteria in the sub-arctic pacific. Deep Sea Res (I Oceanogr Res Pap) 1989;36:1763–1776. [Google Scholar]
- Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol. 2009;501:69–76. doi: 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- Leitet C, Riemann L, Hagström Å. Plasmids and prophages in Baltic Sea bacterioplankton isolates. J Mar Biol Assoc UK. 2006;86:567–575. [Google Scholar]
- Lønborg C, Middelboe M, Brussaard CD.2013Viral lysis of Micromonas pusilla: impacts on dissolved organic matter production and composition Biogeochemistrydoi: 10.1007/s10533-013-9853-1 [DOI]
- Łoś M, Węgrzyn G. Pseudolysogeny. Adv Virus Res. 2012;82:339–349. doi: 10.1016/B978-0-12-394621-8.00019-4. [DOI] [PubMed] [Google Scholar]
- McCarthy MD, Hedges JI, Benner R. Major bacterial contribution to marine dissolved organic nitrogen. Science. 1998;281:231–234. doi: 10.1126/science.281.5374.231. [DOI] [PubMed] [Google Scholar]
- Middelboe M. Bacterial growth rate and marine virus–host dynamics. Microb Ecol. 2000;40:114–124. doi: 10.1007/s002480000050. [DOI] [PubMed] [Google Scholar]
- Middelboe M, Jorgensen NOG. Viral lysis of bacteria: an important source of dissolved amino acids and cell wall compounds. J Mar Biol Assoc UK. 2006;86:605–612. [Google Scholar]
- Middelboe M, Jorgensen N, Kroer N. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl Environ Microbiol. 1996;62:1991–1997. doi: 10.1128/aem.62.6.1991-1997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelboe M, Lyck PG. Regeneration of dissolved organic matter by viral lysis in marine microbial communities. Aquat Microb Ecol. 2002;27:187–194. [Google Scholar]
- Miller ES, Kutter E, Mosig K, Arisaka F, Kunisawa T, Ruger W. Bacteriophage T4 genome. Microbiol Mol Biol Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagarete A, Corguillé G, Tiwari B, Ogata H, Vargas C, Wilson WH, et al. Unveiling the transcriptional features associated with coccolithovirus infection of natural Emiliania huxleyi blooms. FEMS Microbiol Ecol. 2011;78:555–564. doi: 10.1111/j.1574-6941.2011.01191.x. [DOI] [PubMed] [Google Scholar]
- Paul JH. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas. ISME J. 2008;2:579–589. doi: 10.1038/ismej.2008.35. [DOI] [PubMed] [Google Scholar]
- Pedersen A-GU, Thomsen TR, Lomstein BA, Jørgensen NO. Bacterial influence on amino acid enantiomerization in a coastal marine sediment. Limnol Oceanogr. 2001;46:1358–1369. [Google Scholar]
- Poorvin L, Rinta-Kanto JM, Hutchins DA, Wilhelm SW. Viral release of iron and its bioavailability to marine plankton. Limnol Oceanogr. 2004;49:1734–1741. [Google Scholar]
- Poranen MM, Ravantti JJ, Grahn AM, Gupta R, Auvinen P, Bamford DH. Global changes in cellular gene expression during bacteriophage PRD1 infection. J Virol. 2006;80:8081–8088. doi: 10.1128/JVI.00065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JD, Kimball E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal Chem. 2007;79:6167–6173. doi: 10.1021/ac070470c. [DOI] [PubMed] [Google Scholar]
- Riemann L, Middelboe M. Viral lysis of marine bacterioplankton: implications for organic matter cycling and bacterial clonal composition. Ophelia. 2002;56:57–68. [Google Scholar]
- Saldanha AJ. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Shelford EJ, Middelboe M, Møller EF, Suttle CA. Virus-driven nitrogen cycling enhances phytoplankton growth. Aquat Microb Ecol. 2012;66:41–46. [Google Scholar]
- Siuzdak G. The emergence of mass spectrometry in biochemical research. Proc Natl Acad Sci USA. 1994;91:11290–11297. doi: 10.1073/pnas.91.24.11290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopar D, Černe A, Žigman M, Poljšak-Prijatelj M, Turk V. Viral abundance and a high proportion of lysogens suggest that viruses are important members of the microbial community in the Gulf of Trieste. Microb Ecol. 2004;47:1–8. doi: 10.1007/s00248-002-3009-5. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses: major players in the global ecosystem. Nat Rev Micro. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Taylor K. Replication of coliphage lambda DNA. FEMS Microbiol Rev. 1995;17:109–119. doi: 10.1111/j.1574-6976.1995.tb00192.x. [DOI] [PubMed] [Google Scholar]
- Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci USA. 2011;108:E757–E764. doi: 10.1073/pnas.1102164108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Twest R, Kropinski AM. Bacteriophage enrichment from water and soil. Methods Mol Biol. 2009;501:15–21. doi: 10.1007/978-1-60327-164-6_2. [DOI] [PubMed] [Google Scholar]
- Vrede K, Heldal M, Norland S, Bratbak G. Elemental composition (C, N, P) and cell volume of exponentially growing and nutrient-limited bacterioplankton. Appl Environ Microbiol. 2002;68:2965–2971. doi: 10.1128/AEM.68.6.2965-2971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer M, Chen F, Wilhelm S.2011Microbial Carbon Pump in the OceanIn: Jiao N, Azam F, Sanders S (eds)Science/AAAS Business Office [Google Scholar]
- Weinbauer MG, Arrieta J-M, Griebler C, Herndl GJ. Enhanced viral production and infection of bacterioplankton during an iron-induced phytoplankton bloom in the Southern Ocean. Limnol Oceanogr. 2009;54:774–784. [Google Scholar]
- Weinbauer MG, Peduzzi P. Effect of virus-rich high molecular weight concentrates of seawater on the dynamics of dissolved amino acids and carbohydrates. Mar Ecol Prog Ser. 1995;127:245–253. [Google Scholar]
- Wikner J, Vallino JJ, Steward GF, Smith DC, Azam F. Nucleic acids from the host bacterium as a major source of nucleotides for three marine bacteriophages. FEMS Microbiol Ecol. 1993;12:237–248. [Google Scholar]
- Williams PLB.2000Heterotrophic bacteria and the dynamics of dissolved organic materialIn: Kirchman DL (ed)Microbial Ecology of the Oceans Wiley-Liss, Inc.: New York, NY; 153–200. [Google Scholar]
- Wommack KE, Williamson KE, Helton RR, Bench SR, Winget DM. Methods for the isolation of viruses from environmental samples. Methods Mol Biol. 2009;501:3–14. doi: 10.1007/978-1-60327-164-6_1. [DOI] [PubMed] [Google Scholar]
- Yan D. Protection of the glutamate pool concentration in enteric bacteria. Proc Natl Acad Sci USA. 2007;104:9475–9480. doi: 10.1073/pnas.0703360104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Bennett BD, Rabinowitz JD. Kinetic flux profiling for quantitation of cellular metabolic fluxes. Nat protoc. 2008;3:1328–1340. doi: 10.1038/nprot.2008.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang K, Ackermann H-W, Halden RU, Jiao N, Chen F. Searching for a “hidden” prophage in a marine bacterium. Appl Environ Microbiol. 2010;76:589–595. doi: 10.1128/AEM.01450-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.