Abstract
The influence of specific serum-borne biomolecules (e.g. heparin) on growth factor-dependent cell behavior is often difficult to elucidate in traditional cell culture due to the random, non-specific nature of biomolecule adsorption from serum. We hypothesized that chemically well-defined cell culture substrates could be used to study the influence of sequestered heparin on human mesenchymal stem cell (hMSC) behavior. Specifically, we used bio-inert self-assembled monolayers (SAMs) chemically modified with a bioinspired heparin-binding peptide (termed “HEPpep”) and an integrin-binding peptide (RGDSP) as stem cell culture substrates. Our results demonstrate that purified heparin binds to HEPpep SAMs in a dose-dependent manner, and serum-borne heparin binds specifically and in a dose-dependent manner to HEPpep SAMs. These heparin-sequestering SAMs enhance hMSC proliferation by amplifying endogenous fibroblast growth factor (FGF) signaling, and enhance hMSC osteogenic differentiation by amplifying endogenous bone morphogenetic protein (BMP) signaling. The effects of heparin-sequestering are similar to the effects of supraphysiologic concentrations of recombinant FGF-2. hMSC phenotype is maintained over multiple population doublings on heparin-sequestering substrates in growth medium, while hMSC osteogenic differentiation is enhanced in a bone morphogenetic protein-dependent manner on the same substrates during culture in osteogenic induction medium. Together, these observations demonstrate that the influence of the substrate on stem cell phenotype is sensitive to the culture medium formulation. Our results also demonstrate that enhanced hMSC proliferation can be spatially localized by patterning the location of HEPpep on the substrate. Importantly, the use of chemically well-defined SAMs in this study eliminated the confounding factor of random, non-specific biomolecule adsorption, and identified serum-borne heparin as a key mediator of hMSC response to endogenous growth factors.
A Introduction
Serum is commonly used as a cell culture supplement, as it provides a relatively inexpensive source of biomolecules that mediate cell adhesion and support cell survival. To enhance specific stem cell behaviors, such as proliferation or differentiation, cell culture media are often further supplemented with biomolecules (e.g. growth factors) that activate the behavior of interest. For example, addition of fibroblast growth factor (FGF)-2 to human mesenchymal stem cell (hMSC) cultures up-regulates proliferation and maintains the multipotent phenotype of these cells,1 while addition of bone morphogenetic protein (BMP)-2 enhances hMSC osteogenic differentiation.2 However, eliciting these changes in stem cell behavior typically requires a supraphysiologic concentration of growth factor, which likely provides limited insight into growth factor function within the in vivo context. Therefore, culture systems that can harness the activity of endogenous growth factors may provide better in vitro models to study their importance within physiologically relevant settings.
One approach to harness endogenous growth factor activity could involve mimicking regulatory mechanisms prevalent in the natural extracellular matrix (ECM). For example, heparin proteoglycans (PGs) and glycosaminoglycans (GAGs) integrated within the ECM can bind to soluble growth factors, thereby concentrating them and locally amplifying their activity within distinct extracellular microenvironments.3 This natural mechanism has previously inspired the development of biomaterials decorated with heparin GAGs to augment growth factor release.4 Additionally, we and others have developed biomaterials modified with a heparin-binding peptide as models to probe the role of interactions between cell-surface heparin and the ECM on cell functions, such as adhesion5 or expansion of pluripotent stem cells.6 During in vitro culture, however, soluble, serum-borne heparin is likely localized to the cell-material interface either through non-specific electrostatic mechanisms or through specific interactions with proteins that have adsorbed to the culture substrate, such as fibronectin7 or laminin.8 Yet, to date, the influence of soluble heparin sequestered at the cell-material interface remains poorly characterized due to the lack of model culture systems that can isolate the influence of soluble heparin from other serum-borne biomolecules. Recently, we demonstrated that self-assembled monolayers (SAMs) presenting a heparin-binding peptide (termed “HEPpep”) sequester serum-borne heparin, either as a PG or GAG, and enhance human umbilical vein endothelial cell (HUVEC) proliferation by amplifying the activity of recombinant fibroblast growth factor (FGF)-2.9 Our results suggested that soluble heparin sequestered at the cell-material interface is a key mediator of cell response to the growth factor, as enhanced FGF-mediated proliferation was not observed when HUVECs were cultured in medium lacking heparin or on substrates resistant to heparin binding. However, similar to existing methods to modulate cell behavior, our initial demonstration relied on supraphysiologic, recombinant FGF-2 concentrations to enhance HUVEC proliferation. Here, we hypothesized that soluble, serum-borne heparin sequestered at the cell-material interface can locally amplify the activity of serum-borne or cell-secreted (i.e. endogenous) growth factors to modulate stem cell behavior.
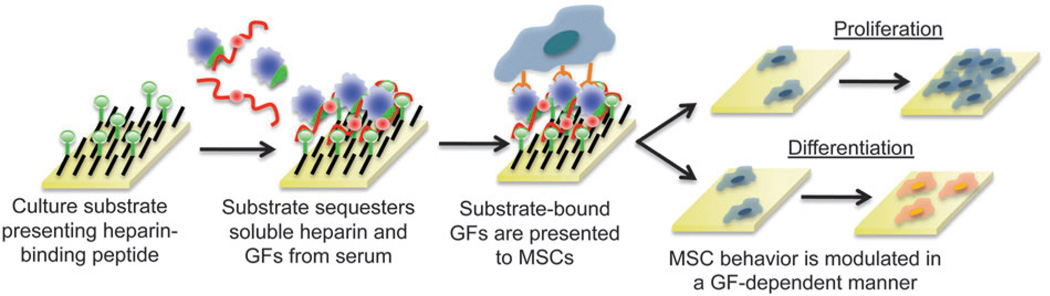
In this report, model SAM substrates that selectively sequester soluble heparin and growth factors via sequential, non-covalent interactions (HEPpep SAMs) were used to study the influence of endogenous heparin and growth factors on stem cell behavior. By sequestering soluble, endogenous heparin and growth factors at the cell-material interface, these substrates enhanced hMSC proliferation in an FGF-dependent manner and enhanced hMSC osteogenic differentiation in a BMP-dependent manner (Fig. 1). In addition, enhanced hMSC proliferation could be spatially controlled by patterning heparin sequestering on the substrate, which provides a level of local regulation that is unattainable by simply adding supraphysiologic concentrations of biomolecules to the cell culture medium. Importantly, growth factor-dependent enhancement of hMSC behaviors was observed in media supplemented only with serum, which suggests that mimicking natural mechanisms to localize and amplify endogenous growth factor activity may circumvent the need for supraphysiologic concentrations of growth factors. In total, this work establishes non-covalent sequestering as a promising strategy to develop in vitro culture models that more closely mimic native microenvironments. This approach may also enable novel approaches to understand the role of endogenous factors in modulating stem cell behavior.
Fig. 1.
Schematic representation of SAMs that non-covalently sequester endogenous heparin PGs and growth factors at the cell-material interface to influence stem cell behaviors, such as proliferation or differentiation.
B Experimental methods
Materials
Gold substrates (5nm Cr, 100nm Au and 2nm Ti, 10 nm Au) were from Evaporated Metal Films (Ithaca, NY). 11-tri-(ethylene glycol)-undecane-1-thiol (HS⋯EG3) was synthesized using a previously described protocol.5 11-carboxyhexa-(ethylene glycol)-undecane-1-thiol (HS⋯EG6⋯COOH) and 11-azido-hexa(ethylene glycol)-undecane-1-thiol (HS⋯EG6⋯N3) were purchased from Prochimia (Sopot, Poland). The peptides GGGKRTGQYKL (HEPpep), GGGTYRKKGLQ (Scramble), GGLWLGGRGDSP, and 3-amino-5-hexynoic acid- GGRGDSP (alkyne-terminated RGDSP) were synthesized using a standard solid phase peptide synthesis protocol for Fmoc-protected amino acids activated by HOBt and DIC. Piperidine, dimethylformamide (DMF), triisoproylsilane (TIPS), and PD 173074 were from Sigma-Aldrich (St. Louis, MO). Fmoc-protected amino acids and Rink amide MBHA peptide synthesis resin were from NovaBiochem (San Diego, CA). Hydroxybenzotriazole (HOBt) was from Advanced Chemtech (Louisville, KY). Diisopropylcarbodiimide (DIC) was from Anaspec (San Jose, CA). Trifluoroacetic acid (TFA) and diethyl ether were from Fisher Scientific (Fairlawn, NJ). Absolute ethanol was from AAPER Alcohol and Chemical Co. (Shelbyville, KY). Human mesenchymal stem cells (hMSCs) were from Cambrex (North Brunswick, NJ). 1× minimum essential medium, alpha (αMEM) and Medium 199 (M199) were from CellGro (Mannassas, VA). MSC-qualified fetal bovine serum was from Invitrogen (Carlsbad, CA). 0.05% Trypsin and penicillin/streptomycin were from Hyclone (Logan, UT). Goat anti-human CD105 antibody was from BD Transductions (San Jose, CA), rabbit anti-human CD90 antibody was from Abgent (San Diego, CA), and alexafluor-488-tagged donkey anti-goat and alexafluor-647-tagged goat anti-rabbit secondary antibodies were from Invitrogen (Carlsbad, CA).
SAM formation and peptide immobilization
Gold substrates were cut, sonicated in ethanol for 3 min, washed with ethanol, and dried under a stream of nitrogen prior to monolayer formation. Monolayers were formed by immersing gold substrates in an ethanolic solution containing 97.5% HS⋯EG3 : 2.5% HS⋯EG6⋯COOH for typical SPR binding experiments, or in an ethanolic solutions containing 96% HS⋯EG3 : 4% HS⋯EG6⋯COOH or 96% HS⋯EG3 : 2% HS⋯EG6⋯COOH: 2% HS⋯EG6⋯N3 for typical cell culture experiments. After monolayer formation, gold substrates were removed from the ethanolic solution, washed with ethanol, and dried under a stream of nitrogen.
Peptide immobilization onto monolayers was achieved using one of two strategies. The first strategy involved a two-step process: (1) Carboxylate groups were “activated” by incubating SAMs in an aqueous solution containing 100 mM NHS and 250 mM EDC for 10 min, followed by washing with DI H2O and ethanol, and drying under a stream of nitrogen, and (2) NHS-activated SAMs were incubated in a 1× phosphate-buffered saline (PBS) solution containing 250 µM total peptide (250 µM HEPpep or scramble for typical SPR experiments; 125 µM GGLWLGGRGDSP and 125 µM HEPpep or scramble for typical cell culture experiments) for 60 min, followed by washing with DI H2O, 0.1% SDS, DI H2O and ethanol, and drying under a stream of nitrogen. In the second strategy, (1) CuBr, sodium ascorbate (Na-Asc), and tris-(Benzyltriazolylmethyl)-amine (TBTA) were dissolved in DMSO at a concentration of 2 mM, (2) acetylene-terminated RGDSP was dissolved in HEPES (0.1 M, pH 8.5) at a concentration of 2mM, (3) the DMSO solution containing CuBr, Na-Asc, and TBTA and the HEPES solution containing peptide were then mixed at a 1 : 1 ratio by vortexing, followed by sonication for 10 min, (4) gold substrates bearing azide-termini were immersed in this solution and allowed to incubate at room temperature for 60 min, and (5) at the reaction endpoint, gold substrates were washed sequentially with DI water, 0.1% sodium dodecyl sulfate in water, DI water, and ethanol, followed by drying under a stream of nitrogen.
Surface plasmon resonance
Heparin GAG and serum-borne biomolecule binding onto mixed SAMs consisting of HS–EG3 and HEPpep or scramble were analyzed using a BIAcore 2000 system (Piscataway, NJ). Typical binding experiments were performed as follows: (1) phosphate buffered saline (pH 7.4) (PBS) was passed over the SAM to establish the plasmon resonance baseline of the surface, (2) 100 µL of 1× PBS solutions containing various concentrations of heparin or FBS were passed over the SAM, and 3) PBS was passed over the SAM to establish the change in surface plasmon resonance signal due to the presence of adsorbed protein. koff and kobs were calculated using non-linear regression analysis in Prism software using the one-phase decay and one-phase association models, respectively.
hMSC proliferation
Passage 6 hMSCs were seeded on SAM substrates at a density of 2000 cells/cm2 in αMEM supplemented with 10% FBS. After overnight attachment, SAMs were washed with 1× PBS to remove any loosely bound cells. For proliferation experiments, SAMs were maintained in one of the following culture conditions for 72 h: αMEM supplemented with 0.01, 0.1, 1, or 10% FBS; αMEM supplemented with 10% FBS and 200 nM PD 173054; serum-free αMEM supplemented with 10 ng/mL FGF-2; αMEM supplemented with 10% FBS and 0, 1 or 5 ng/mL FGF-2. T = 0 h for each experiment was designated as the point immediately after placing the SAMs in the appropriate culture medium. Brightfield photomicrographs (40× mag) of each substrate were collected on an Olympus IX51 inverted epifluorescent microscope at t = 0, 24, 48, and 72 h and the total number of cells per viewing area was counted manually.
Immunocytochemistry of hMSC CD90 and CD105 expression
hMSCs were fixed to the substrates using 4% paraformaldehyde in 1× PBS, samples were blocked with 0.1 wt% bovine serum albumin, substrates were incubated in 1× PBS containing a CD105 antibody (BD Transductions, San Jose, CA) or a CD90 antibody (Abgent, San Diego, CA) at room temperature for 60 min, substrates were incubated in a 1× PBS solution containing an alexfluor-tagged secondary antibody (Invitrogen, Carlsbad, CA) at room temperature for 60 min, and brightfield and fluorescent photomicrographs of antibody stained hMSCs were then collected using an Olympus IX51 inverted epifluorescent microscope equipped with a FITC filter cube set.
Patterned SAMs
Microfluidic devices with arrays of microchannels were created using soft lithography and designed to facilitate “passive pumping” in which differences in surface tension across the microchannel were used to drive fluid transport. Briefly, master molds were fabricated from SU-8 (Microchem, Newton, MA) spin-coated silicon wafers using conventional photolithography techniques. Multiple layers of photoresist were used to create channels with geometries of 4500 × 750 × 250 µm (l × w × h) and ports with radii of 603 µm and 1200 µm. Polydimethylsiloxane (PDMS) (Sylgard 184, Dow Corning, Midland, MI) was prepared by mixing a 10 : 1 ratio of base/curing agent (w/w) followed by degassing for ~30 mins. The degassed mixture was cast over the mold and cured for 4 h at 85 °C. Following curing, PDMS devices were removed from molds and cleaned in EtOH using an overnight Soxhlet extraction. After cleaning, PDMS devices were placed in vacuo to remove residual ethanol from the Soxhlet extraction process.
A clean gold chip measuring 1″ × 1.5″ was placed in a 2 mM ethanolic 96% HS⋯EG3:4% HS–EG6–COOH solution overnight. The chip was then removed from the alkanethiolate solution, rinsed with EtOH, blown dry with nitrogen, and placed in a 250 mM EDC, 100 mM NHS solution to create a NHS-activated surface. After incubating for B10 min, the chip was removed from the NHS/EDC solution, rinsed with DI H2O, rinsed with EtOH, blown dry with nitrogen and then placed in a polystyrene Petri dish. Approximately 15 µL of EtOH was applied to the center of the chip, followed by application of the PDMS microfluidic device. The SAM chip and device were then placed in vacuo for ~30 min to remove the EtOH and create a tight seal between the microfluidic device and SAM surface. Localized peptide conjugation to the SAM was achieved by filling channels with a peptide solution containing 125 µMHEPpep or scrambled peptide and 125 µM GGLWLGGRGDSP in 1× PBS (pH 7.4) and incubating for 45 min. Channels were rinsed 4 times with 5 µL of DI H2O. The chip and attached microfluidic device were then immersed in a 1 mM solution of GGRGESP. The PDMS device was removed to expose the background to the GGRGESP peptide coupling solution, which blocks the background to prevent cell adhesion. After 45 min, the patterned substrate was rinsed with a 0.1 wt% SDS solution and then transferred to a 50 mL conical tube (BD Falcon, San Jose, CA) containing ~30 mL of SDS solution and sonicated for 3 min. Following sonication, the patterned substrate was finished with 5-second rinses of additional SDS solution, H2O, EtOH, and then dried with nitrogen.
hMSC osteogenic differentiation
Passage 5 hMSCs were seeded on SAM substrates at a density of 2000 cells/cm2. hMSCs were cultured on SAMs or polystyrene (PS) in medium supplemented with 10% fetal bovine serum, 50 µg/mL 2-phosphate ascorbic acid, 10 mMβ-glycerophosphate, and 100 nM dexamethasone (osteogenic induction medium (OM)) for 7 days, with medium changed once at day 3. At the end of the 7-day culture period, cells were either harvested from the substrates by trypsinization and processed for qPCR (see experimental details below) or harvested by scraping into lysis buffer for alkaline phosphatase (ALP) activity measurements and total DNA quantification. ALP activity was measured using the SensoLyte FDP fluorometric alkaline phosphatase assay kit (Anaspec, San Jose, CA) and total DNA was quantified using the CyQuant cell proliferation assay kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. For BMP receptor inhibition studies, cells were cultured in OM supplemented with 200 nM LDN-193189 (Stemgent, San Diego, CA).
Quantitative polymerase chain reaction
Total RNA was isolated using the PerfectPure RNA Cultured Cell Kit (5 Prime Gaithersburg, MD). cDNA synthesis was performed according to the instructions for the ThermoScript RT-PCR System (Invitrogen Carlsbad, CA). Briefly, 25 ng of RNA was combined with 1 µl of 50 µM Oligo(dT)20, and 2 µl of 10 mM dNTP Mix followed by 5 min incubation at 65 °C to denature the RNA and primer. Reverse transcription was carried out at 55 °C for 60 min and the reaction was terminated at 85 °C for 5 min. The CD73, CD90 and CD105 oligonucleotide primer sequences for qPCR were designed using Primer-Blast (NCBI Bethesda, MD). Primer sequence for OPN was kindly provided by Dr Wan-ju Li. GAPDH was used as an internal control. All primers were used at a final concentration of 50 nM. The amplification conditions were 95 °C for 10 min, 40 cycles of denaturing at 95 °C for 15 s, annealing and extension at 60 °C for 1min. Power SYBR Green PCR Master Mix was used to detect nucleotides (Applied Biosystems Foster City, CA). Primer efficiencies were extracted from StepOne(tm) Software v2.1 and verified with melting curves and gel electrophoresis (Applied Biosystems Foster City, CA). CD73, CD90 and CD105 presented as copies per unit RNA, OPN mRNA copies were normalized to OPN mRNA copies in untreated hMSC.
Statistical analysis
Statistical significance between samples was evaluated using Student’s t-tests with Prism software.
C Results
Serum-borne biomolecule sequestering by HEPpep SAMs is serum concentration-dependent
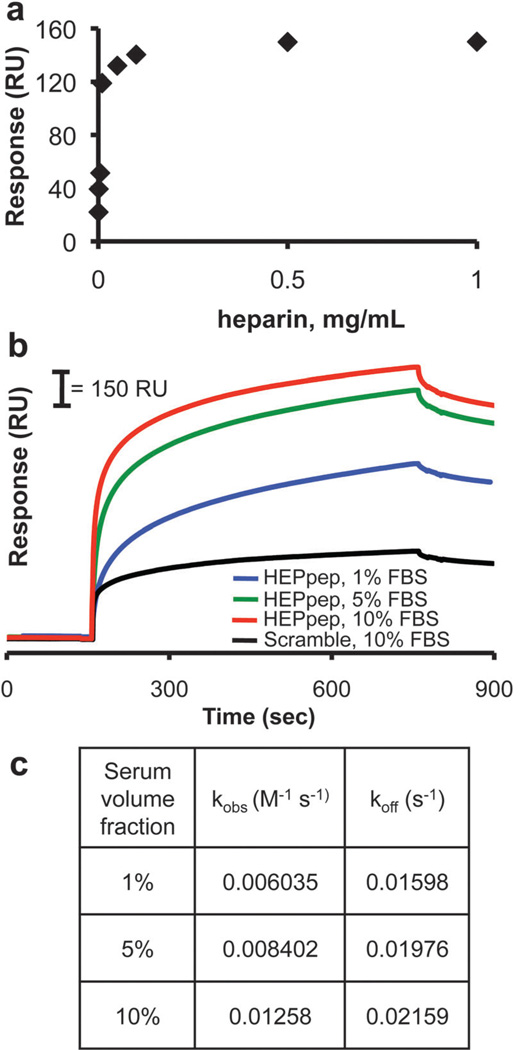
We previously demonstrated that HEPpep SAMs sequester serum-borne heparin.9 Since heparin is present as both a PG and a GAG in serum,10 and since either heparin type is likely to bind to substrates presenting a heparin-binding peptide, we will collectively refer to these serum-borne molecules as “heparin” throughout this paper. Here, we used surface plasmon resonance (SPR) to characterize the correlation between the concentration of serum-borne biomolecules in solution and the amount of biomolecule binding to HEPpep SAMs. First, we passed solutions of purified heparin glycosaminoglycan (GAG) over 1% HEPpep SAMs to characterize the correlation between heparin in solution and extent of binding to the substrate. Over the concentration range of 0.5–100 µg/mL heparin GAG, the SPR signal increased as a function of heparin GAG concentration (Fig. 2A). Above heparin GAG concentrations of 100 µg/mL, there was no further change in the SPR signal, which suggests that HEPpep ligands on a 1% SAM are saturated in the presence of heparin GAG concentrations ≥ 100 µg/mL.
Fig. 2.
(A) Characterization of SPR signal response shift as a function of heparin GAG concentration in PBS. (B) SPR sensorgrams of 2.5% HEPpep SAMs exposed to 1% (blue), 5% (green) or 10% (red) FBS or a 2.5% scramble SAM exposed to 10% FBS (black). (C) Table of observed binding rate constant (kobs) and dissociation rate constant (koff) as a function of serum volume fraction passed over 2.5% HEPpep SAMs.
Next, we characterized sequestering of serum-borne biomolecules on 1% HEPpep SAMs using SPR. Passing 1–10% fetal bovine serum (FBS) solutions over a SAM presenting 2.5% HEPpep resulted in a significant increase in the SPR signal (Fig. 2B), with the total RU shift dependent on the volume fraction of serum. Assuming that a 1000 RU shift corresponds to 1 ng/mm2 bound biomolecule,11 the density of biomolecules bound to 2.5% HEPpep SAMs, which includes heparin and heparin-binding proteins, was between 780–1210 pg/mm2 for substrates exposed to 1–10% FBS. The positive slope at the end of the serum-binding phase on 2.5% HEPpep SAMs suggests that binding has not approached saturation even when 10% FBS is passed over the chip, which is consistent with our previous results demonstrating an increase in serum-borne biomolecule binding to 1% HEPpep SAMs over the range of 1–100% FBS.9
Comparing the SPR response on HEPpep SAMs to that observed when passing a 10% FBS solution over a SAM presenting 2.5% of a control “scrambled” version of HEPpep demonstrates that serum-borne biomolecules also bind to this substrate (Fig. 2B). However, the amount of serum-borne biomolecules bound to the control SAM exposed to 10% FBS (400 pg/mm2) was significantly less than that observed on any of the HEPpep SAMs, and the relatively flat profile suggests that binding is nearly saturated under this condition. The relatively high charge density of the peptide (+ 3/peptide) suggests that the binding observed on the control SAM is due to non-specific electrostatic interactions, which is commonly observed when charged SAMs are exposed to proteins.12 However, the extent of non-specific binding is consistent with previous reports demonstrating low non-specific adsorption onto binary mixed SAMs, and is well within the range used to classify surfaces as ‘bio-inert’.13
The observed binding rate constant (kobs) and dissociation rate constant (koff) were calculated using a non-linear regression analysis of the change in RU over time for 2.5% HEPpep SAMs exposed to 1–10% FBS. kobs and koff increased as a function of serum volume fraction (Fig. 2C). This observation is consistent with our previous results demonstrating a correlation between serum volume fraction and kobs or koff for 1% HEPpep SAMs,9 which suggests that the increased surface density of HEPpep does not change the serum-borne biomolecule binding properties of the SAM.
Sequestered heparin enhances stem cell proliferation in standard growth medium
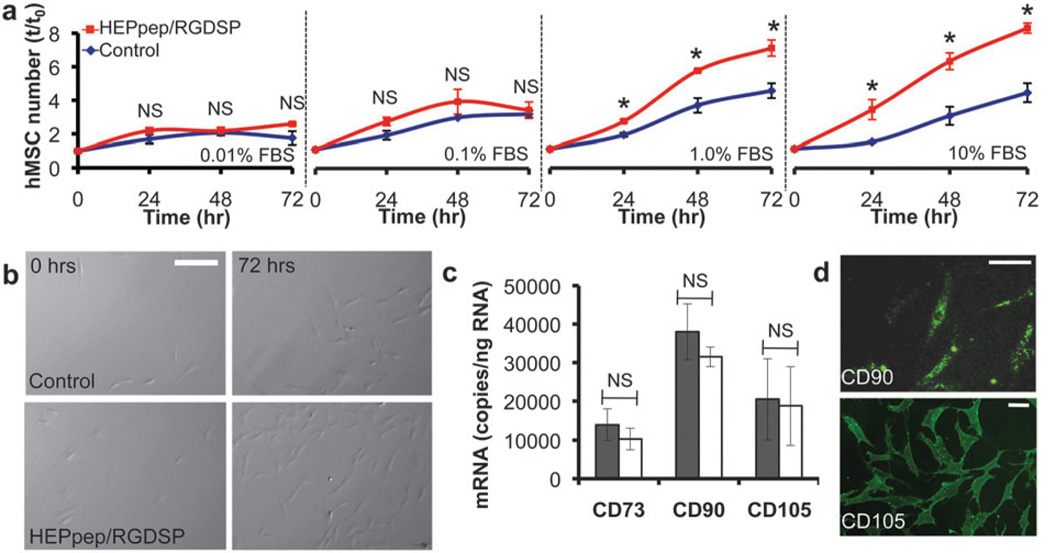
We next characterized the influence of HEPpep SAMs on human mesenchymal stem cell (hMSC) proliferation to address the hypothesis that serum-borne biomolecules localized at the cell-material interface can influence stem cell proliferation. We have recently demonstrated that SAMs patterned to include a peptide-decorated, cell-adhesive region and a surrounding bio-inert region are stable during MSC culture for up to 14 days.14 In addition, Mrksich and colleagues have recently used patterned SAMs over 1 week to characterize correlations between MSC adhesion and differentiation.15 Here, we have limited our culture studies to 7 days or less to remain within the expected window of SAM stability during MSC culture. hMSCs were cultured on SAMs presenting 2% RGDSP (a fibronectin-derived cell adhesion peptide16) and either 2% HEPpep (“HEPpep/RGDSP”) or 2% scrambled version of HEPpep (“control”) for 72 h in standard serum-containing medium. RGDSP was included because cells do not adhere to well-defined SAM surfaces presenting HEPpep or scramble alone, which is consistent with previous reports from our lab demonstrating weak adhesion of hMSCs to SAMs presenting a heparin-binding peptide,5b as well as a report from Healy and co-workers demonstrating weak adhesion of osteoprogenitor cells to materials presenting a heparin-binding peptide against an otherwise bio-inert background.17 hMSC number was significantly greater on HEPpep/RGDSP SAMs when compared to control SAMs after 24, 48, and 72 h of culture in medium supplemented with either 1% or 10% FBS (Fig. 3A and B). Interestingly, however, no increase in hMSC number was observed on HEPpep/RGDSP SAMs during culture in medium supplemented with less than 1% serum (Fig. 3A). The correlation between proliferation and the volume fraction of serum, which is known to contain heparin10 and numerous heparin-binding growth factors,18 suggests that the influence of heparin-binding SAMs on stem cell function is sensitive to the concentration of soluble biomolecules present within the culture environment. Of note, the enhanced hMSC proliferation observed on SAMs presenting a heparin-binding peptide and an RGD binding peptide is in contrast to previous work from Healy and co-workers demonstrating that osteoprogenitor cell proliferation is not enhanced when a heparin-binding peptide is co-immobilized with an RGD ligand.5a However, the work of Healy and co-workers relied on heat-inactivated serum as the culture supplement, while our work used serum that had not been heat-inactivated. Thus, the relative activity of serum-borne biomolecules is likely to be different in each study, which may explain the observed differences in the influence of a heparin-binding peptide on cell proliferation in the presence of serum.
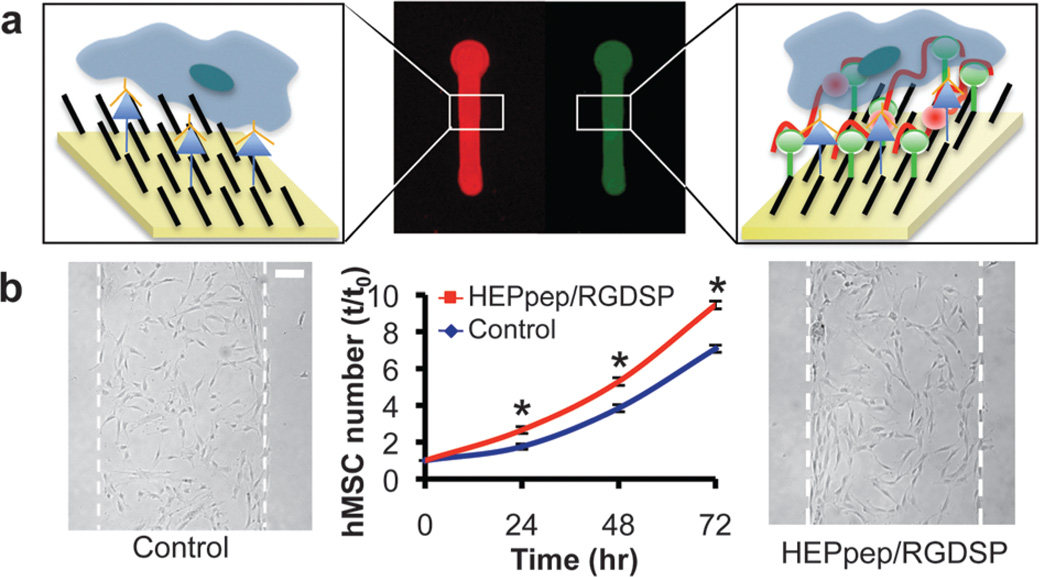
Fig. 3.
(A) hMSC number on SAMs presenting 2% RGDSP and either 2% HEPpep (red) or 2% scramble (blue) in medium supplemented with 0.01–10% FBS. (B) Bright-field photomicrographs of hMSCs on SAMs presenting 2% RGDSP and either 2% HEPpep or 2% scramble (control) at t=0 and 72 h in medium supplemented with 10% FBS (scale bar = 100 µm). (C) qPCR analysis of hMSC marker expression by cells cultured for 72 h on SAMs presenting 2% RGDSP and 2% HEPpep (white) in medium supplemented with 10% FBS or 2% RGDSP and 2% scramble (gray) in medium supplemented with 10% serum and 5 ng/mL FGF-2. (D) Fluorescent photomicrographs of cells stained with an anti-CD90 or anti-CD105 antibody after 72 h culture on HEPpep/RGDSP SAMs in 10% FBS medium (scalebar = 50 µm). * represents significant difference compared to ‘control’ and NS represents no significant difference between samples (P < 0.05).
We were also interested in the influence that endogenous heparin and growth factors localized at the cell-material interface have on hMSC phenotype over multiple population doublings. Expression of CD73, CD90, and CD105 is a common minimal requirement for classifying cells as hMSCs.19 We compared the expression of these markers after 72 h of culture on HEPpep/RGDSP SAMs in standard hMSC medium, or control SAMs in hMSC medium supplemented with 5 ng/mL recombinant FGF-2. Quantitative polymerase chain reaction (qPCR) demonstrated that CD73, CD90, and CD105 mRNA expression levels were similar (Fig. 3C). Additionally, cells cultured on HEPpep/RGDSP SAMs stained positively for CD90 and CD105 (Fig. 3D) expression after 72 h of culture. Together, these results demonstrate that culturing hMSCs on heparin-binding SAMs in medium supplemented with serum does not favor early shifts toward a specific lineage.
Enhanced hMSC proliferation on heparin-binding SAMs is dependent on FGF signaling and not direct cell-material interactions
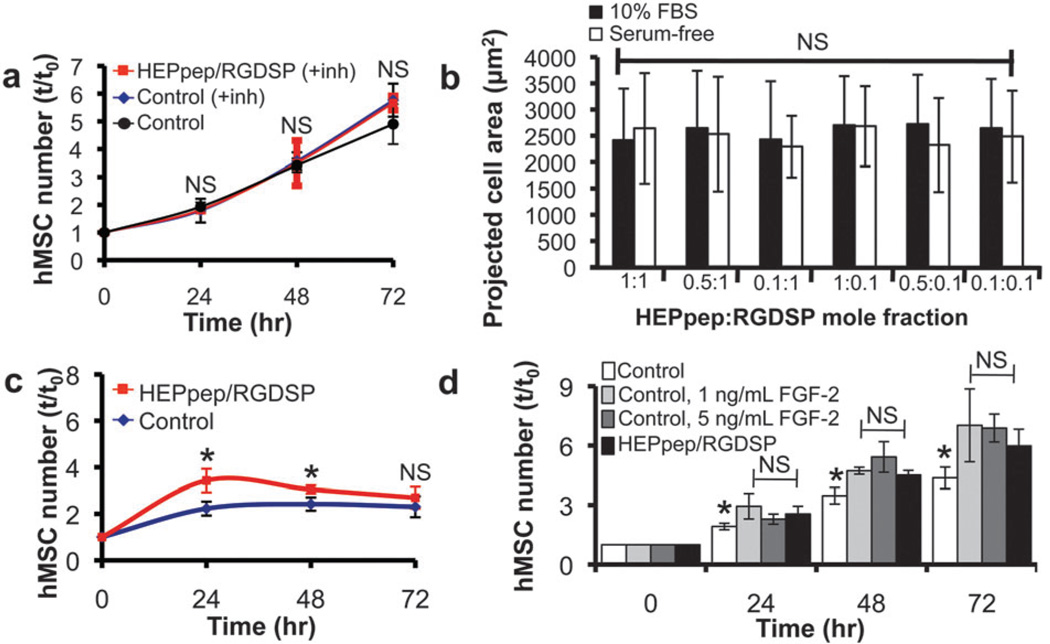
Since FGFs are heparin-binding growth factors20 and are also known to promote hMSC proliferation,1 we next used a soluble FGF receptor inhibitor to address the hypothesis that up-regulated hMSC proliferation on heparin-binding SAMs is FGF-dependent. hMSC proliferation on HEPpep/RGDSP SAMs in the presence of PD 173074, a soluble inhibitor of FGF receptor-1 and receptor-3,21 was similar to hMSC proliferation on control SAMs in the presence or absence of the FGF signaling inhibitor (Fig. 4A). These observations indicate that FGF signaling is a key mediator of the enhanced hMSC proliferation on HEPpep SAMs, and also suggest that when heparin is absent from the cell-material interface, cell-secreted or serum-borne FGFs have minimal influence on hMSC proliferation.
Fig. 4.
(A) hMSC number on SAMs presenting 2% RGDSP and either 2% HEPpep (red) or 2% scramble (blue) in medium supplemented with 10% FBS and 200 nM PD173074, an FGF receptor inhibitor, added to the medium at t = 0, or 2% RGDSP and 2% scramble in medium supplemented with 10% FBS (black). (B) Projected cell area of hMSCs on SAMs presenting RGDSP and HEPpep at co-varying surface densities in serum-free medium (white) or medium supplemented with 10% FBS (black). (C) hMSC number on SAMs presenting 2% RGDSP and either 2% HEPpep (red) or 2% scramble (blue) after overnight attachment in medium containing 10% FBS, followed by culture in serum-free medium supplemented with 10 ng/mL FGF-2 for 72 h. (D) hMSC number on SAMs presenting 2% RGDSP and 2% HEPpep in medium supplemented with 10% FBS (black) or on SAMs presenting 2% RGDSP and 2% scramble in medium supplemented with 10% FBS and 0 (white), 1 (light gray) or 5 (dark gray) ng/mL FGF-2. For (A) and (C) NS represents no significant difference compared to ‘control’ and * represents significant difference compared to ‘control’ (P < 0.05). For (D) NS represents no significant difference compared to ‘HEPpep/RGDSP’ and * represents significant difference compared to ‘HEPpep/RGDSP’ (P < 0.05).
In a previous study we observed that a heparin-binding peptide and an integrin-binding peptide can work synergistically to influence hMSC-substrate adhesion, measured as projected cell area.5b Here, we again relied on projected cell area as a means to characterize the importance of direct cell-material interactions in the enhanced hMSC proliferation observed on HEPpep/RGDSP SAMs. The projected cell area of hMSCs on HEPpep/RGDSP SAMs was similar, regardless of whether the cells were seeded in serum-free medium or medium supplemented with 10% FBS (Fig. 4B). This suggests that HEPpep does not mediate direct cell-material interactions, as the presence of soluble, serum-borne heparin does not inhibit or diminish cell adhesion onto the substrate. This observation is in contrast to previous work from our lab demonstrating that the presence of serum inhibits the synergistic influence of a different heparin-binding peptide on hMSC adhesion when co-immobilized with an RGD ligand,5b as well as studies demonstrating that soluble heparin can inhibit bone cell adhesion mediated by the binding of cell-surface heparin to adsorbed heparin-binding domain of fibronectin.22 Therefore, these observations indicate that direct cell-material interactions mediated by HEPpep binding to cell-surface heparin are unlikely, and, as such, are not a key mediator of the enhanced hMSC proliferation on HEPpep SAMs.
Together, these results indicate that FGF signaling is the prominent mechanism governing enhanced hMSC proliferation on HEPpep SAMs. However, since both FGF-2 and FGF-4 up-regulate hMSC proliferation1,23 and each signals via FGFR1 and FGFR3,24 we can infer that one or both of these growth factors are involved in up-regulating hMSC proliferation on HEPpep SAMs. In addition, although not demonstrated here, it is likely that multiple signaling mechanisms beyond those activated by FGFs are working in concert to enhance hMSC proliferation on heparin-binding SAMs. For example, early FGF signaling events may promote the expression of other heparin-binding growth factors that can enhance hMSC proliferation. Thus, when an FGFR inhibitor is added at t = 0, expression of these secondary growth factors may be attenuated, which results in a loss of amplified hMSC proliferation on heparin-binding substrates.
To further characterize the specific role of FGF-2 signaling, we seeded hMSCs overnight in medium supplemented with 10% FBS, then cultured the cells in serum-free medium supplemented with 10 ng/mL recombinant FGF-2. hMSC proliferation under these conditions was significantly greater on HEPpep/RGDSP SAMs when compared to control SAMs at 24 and 48 h (Fig. 4C). However, there was an inflection point in the growth curve between 24 and 48 h, with no further increase in cell number observed beyond this point. This observation indicates that heparin bound to the substrate during overnight incubation in serum-supplemented medium can enhance FGF-2-mediated hMSC proliferation for a short period of time, even when heparin and other serum-borne growth factors are absent from the culture medium. However, this observation suggests that the amount of heparin and/or heparin-binding growth factors, such as FGFs, secreted by hMSCs during culture are insufficient to maintain enhanced hMSC proliferation, even if a mechanism is provided to prevent diffusion of these molecules away from the cell-material interface. This observation of a relatively high required dose of soluble, serum-borne heparin and growth factors is not surprising in light of the observation that hMSC proliferation was not enhanced during culture in medium supplemented with 0.01 or 0.1% FBS (Fig. 2A). Taken together, these observations also further support the conclusion that enhanced hMSC proliferation on heparin-binding SAMs is mediated by soluble heparin sequestered at the cell-material interface, rather than direct cell-material interactions between HEPpep and cell-surface heparin, as we would not expect cell-material interactions to be disrupted in the absence of serum. In addition, these observations also suggest that the synergistic influence of FGF-2 and FGF-4 signaling, or FGF-2 signaling with other serum-borne, heparin-binding growth factors, may support long-term hMSC proliferation more effectively than FGF-2 signaling alone.
Next, we characterized the correlation between heparin at the cell-material interface and the concentration of recombinant FGF-2 required to enhance hMSC proliferation. In addition to 10% FBS, a recombinant FGF-2 supplement concentration ≥ 1 ng/mL was needed to observe a similar hMSC proliferation rate on control SAMs that do not bind heparin when compared to hMSCs on HEPpep/RGDSP SAMs in medium supplemented only with 10% serum (Fig. 4D). Importantly, this result demonstrates that enhancing hMSC proliferation when heparin is absent from the cell-material interface requires a supraphysiologic dose of FGF-2, presumably to establish a local concentration that is sufficiently high to activate cell surface FGFRs, while the high affinity of heparin-FGF binding (KD ~ 470 nM)25 can establish a local concentration of endogenous FGFs at the cell-material interface that is sufficient to activate cell surface FGFRs. Together, these data indicate that serum-borne heparin sequestered at the cell-material interface can non-covalently localize and, in turn, harness the biological activity of endogenous growth factors.
It is noteworthy that serum-borne heparin PGs and GAGs may also adsorb onto traditional poly(styrene) cell culture substrates, either through non-specific electrostatic interactions or by binding specifically to adsorbed proteins, such as fibronectin7 or laminin,8 and amplify cell responses to heparin-binding growth factors. However, these effects would be difficult to elucidate due to the complex biomolecule adsorption profile observed on traditional cell culture substrates after serum exposure.26 Therefore, chemically well-defined cell culture substrates that can sequester specific serum-borne biomolecules at the cell-material interface provide an enabling new mechanism to study the effects of serum-borne biomolecules on cell behavior by eliminating the confounding factor of non-specifically adsorbed biomolecules.
The influence of heparin sequestering can be spatially controlled
Using a SAM patterning approach recently developed by our group,14 it was also possible to spatially control the effects of heparin PG sequestering. Specifically, hMSC proliferation in medium supplemented with 10% serum was significantly greater within patterned regions containing HEPpep/RGDSP at 24, 48, and 72 h (Fig. 5), and this spatially patterned enhancement in hMSC number was similar to the enhancement observed on non-patterned SAMs (Fig. 2A). The observed spatial control over up-regulated hMSC proliferation indicates that stem cell behaviors can be differentially regulated within a single population of cells by patterning heparin-binding peptides. Importantly, this spatial control cannot be achieved by simply adding supraphysiologic concentrations of growth factor to the culture medium, and may therefore be of interest for developing therapeutic biomaterials that can direct neighboring cells toward different lineages. Patterned heparin PG sequestering may also provide high throughput tools to study fundamental questions related to extracellular matrix biology and crosstalk between neighboring stem cell populations.
Fig. 5.
(A) Schematic representation of HEPpep patterning on a single SAM substrate. (B) hMSC number over time and bright-field photomicrographs at t = 72 h of hMSCs in regions presenting 2% RGDSP and 2% scramble (control, blue), or 2% RGDSP and 2% HEPpep (red) (scale bar = 100 µm). * represents significant difference compared to ‘control’ (P < 0.05).
HEPpep SAMs enhance hMSC osteogenic differentiation
Heparin and heparin-binding growth factors have been implicated in osteogenic differentiation processes, although the role of each has been controversial to date. For example, binding of heparan sulfate GAGs to bone morphogenetic protein 2 (BMP-2) down-regulates C2C12 cell osteogenic differentiation,27 while heparan sulfate GAG chains attached to glypican-3 up-regulate BMP-mediated MC3T3-E1 cell osteogenic differentiation.28 To understand the influence of soluble, endogenous heparin and growth factors on hMSC osteogenic differentiation, we characterized alkaline phosphatase (ALP) activity and osteopontin (OPN) mRNA expression of cells cultured on different substrates in osteogenic induction medium. Alkaline phosphatase (ALP) activity was 3-fold to 5-fold higher in cells cultured on HEPpep/RGDSP SAMs when compared to cells cultured on control SAMs or polystyrene substrates in osteogenic medium (OM) (Fig. 6A). Interestingly, the enhanced ALP activity of hMSCs cultured on HEPpep/RGDSP SAMs was completely abolished in the presence of LDN-193189, a BMP receptor inhibitor.29 This observation suggests that signaling by endogenous BMPs, another family of heparin-binding growth factors,30 is involved in up-regulating ALP activity of hMSCs cultured on heparin-binding SAMs. Importantly, this observation further supports the assertion that culture substrates that sequester specific endogenous biomolecules may provide more appropriate in vitro models to understand cell differentiation processes than traditional culture substrates by eliminating the complications associated with random, non-specific biomolecule adsorption.
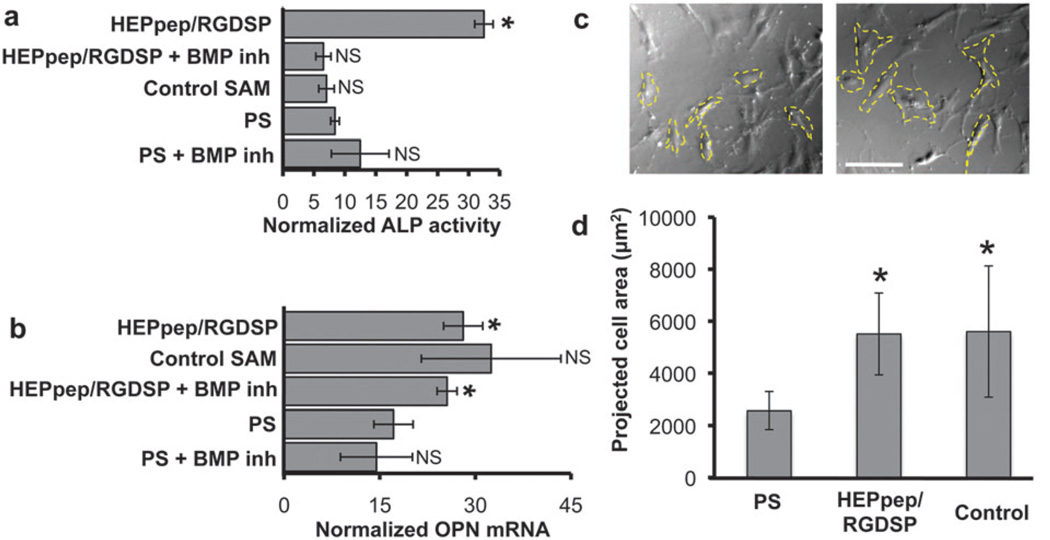
Fig. 6.
(A) ALP activity and (B) OPN mRNA expression of hMSCs cultured on different substrates in osteogenic induction medium with or without 200 nM LDN-193189, a BMP receptor inhibitor, for 7 days. (C) Representative bright-field photomicrographs (scale bar = 150 µm) and (D) quantification of hMSC projected cell area on different substrates after 5 days of culture in osteogenic induction medium. * represents significant difference compared to ‘PS’ condition, NS represents no significant difference compared to ‘PS’ condition (P < 0.05).
Cells cultured on heparin-binding SAM substrates also demonstrated higher osteopontin (OPN) mRNA expression when compared to cells cultured on PS, but this effect was not influenced by BMP receptor inhibition (Fig. 6B). This observation suggests that BMP signaling does not regulate OPN mRNA expression to the extent that it regulates ALP activity (Fig. 6A). Similarly, the OPN mRNA expression level of cells cultured on control SAMs was on average higher, although ultimately not significantly different than cells cultured on PS in OM (Fig. 6B). The observed increase in OPN mRNA expression in cells cultured on SAMs may be explained by differential hMSC spreading, as a well-defined correlation has recently been shown between hMSC spreading and fate determination15,31 and hMSC projected cell area during culture in OM was significantly increased on HEPpep/RGDSP and control SAMs when compared to cells cultured on PS (Fig. 6C and D). This result suggests that interactions between hMSCs and the RGDSP cell adhesion ligand, which was present on all SAMs explored in this study, may be involved in up-regulating OPN mRNA expression. This assertion is consistent with a recent report from Hubbell and co-workers demonstrating that fibronectin fragments—including RGDSP-with varying integrin-binding specificity differentially influence hMSC osteogenic differentiation.32
Together, these results provide early evidence that heparin and, in turn, endogenous BMPs localized at the cell-material interface enhance hMSC osteogenic differentiation. However, the discrepancy between BMP signaling involvement in ALP activity and OPN mRNA expression suggests that multiple signaling mechanisms likely influence hMSC osteogenic differentiation. Therefore, these preliminary results provide a basis for future studies using HEPpep/RGDSP SAMs to characterize the influence of BMP signaling on the expression of other osteogenesis related markers and the role of additional heparin-binding growth factors in hMSC osteogenic differentiation.
D Discussion
Cell interaction with common culture substrates, such as polystyrene or glass, is mediated by non-specifically adsorbed serum-borne biomolecules. Heparin PGs and GAGs are present in serum,10 and it is likely that heparin in either form adsorbs to common cell culture substrates and influences cell behavior to some extent. However, due to the random, non-specific nature of biomolecule adsorption onto these substrates,33 it is difficult to separate the influence of heparin from the influence of other non-specifically adsorbed biomolecules. Here, we used model substrates that sequester heparin and resist non-specific adsorption to demonstrate that serum-borne heparin sequestered at the cell-material interface is an important mediator of growth factor-dependent stem cell behavior. In particular, our results demonstrated that sequestered heparin enhances hMSC proliferation in an FGF-dependent manner. In addition, we observed preliminary evidence that sequestered heparin enhances hMSC osteogenic differentiation in a BMP-dependent manner. This approach eliminated the confounding factor of non-specifically adsorbed biomolecules to identify a correlation between serum-borne heparin at the cell-material interface and stem cell responses to serum-borne or cell-secreted growth factors. Importantly, this approach is likely to be broadly applicable to the study of other serum-borne or cell-secreted biomolecules by varying the binding specificity of the ligand immobilized onto the substrate.
The fundamental hypothesis that biomaterials decorated with heparin/heparan sulfate glycosaminoglycans can influence cell behavior by localizing growth factor activity is well-established.4 In addition, there are numerous examples that an immobilized heparin-binding peptide can influence cell behavior by promoting direct interaction between cell-surface heparin GAGs and the underlying material. For example, Healy has demonstrated that the presence of a heparin-binding peptide enhances osteoblast precursor adhesion and mineralization but does not influence proliferation of these cells.5a,17 Similarly, Kiessling demonstrated that substrates presenting a heparin-binding peptide can support the expansion of pluripotent stem cells in defined culture medium.6 In each of these examples, it is proposed that the heparin-binding peptide influences cell behavior by interacting directly with heparin glycosaminoglycans that are attached to the cell surface. Similarly, we have also previously demonstrated that an immobilized heparin-binding peptide can influence human mesenchymal stem cell adhesion under serum-free conditions,5b presumably by interacting with cell-surface proteoglycans. Here, we hypothesized that soluble heparin GAGs/PGs (i.e. heparin that is not linked to the cell surface) present in culture medium can modulate hMSC behavior by locally amplifying the activity of serum-borne or cell-secreted growth factors. To test this hypothesis, we used a SAM-based culture platform that can non-covalently sequester serum-borne heparin, which serves to mimic non-covalent heparin adsorption onto polystyrene culture substrates, while preventing non-specific adsorption of other serum-borne biomolecules. We observed enhanced hMSC proliferation on HEPpep SAMs in medium supplemented only with serum, which is known to contain soluble heparin/heparan sulfate GAGs/PGs,10 and this response was dependent on serum concentration. Taken together with the observation that HEPpep does not influence hMSC adhesion (Fig. 4B), this data suggests that sequestered, serum-borne heparin is responsible for enhancing hMSC proliferation, rather than direct interaction between the underlying material and heparin present on the cell surface.
The importance of heparin in stem cell biology has recently been described in other seminal reports. Thomson and co-workers used a conditioned medium approach to demonstrate that PGs secreted into medium by mouse embryonic feeder layers mediate human embryonic stem cell (hESC) adhesion to a substrate and enhance hESC response to FGF-2.34 In addition, Rao and co-workers demonstrated that ECMs produced by embryonic feeder layers contain proteoglycans as key components that promote hESC expansion.35 Our results, which identified serum-borne heparin as a key mediator of hMSC response to different growth factors, extend upon these previous studies and identify a broader role of heparin in stem cell biology. By providing a platform to isolate soluble heparin from other serum-borne biomolecules, the substrates developed in this study may be enabling in future studies involving both hMSCs and pluripotent stem cell types, such as hESCs or induced pluripotent stem cells (iPS).
Many previous studies have reported conflicting results on the importance of heparin and BMPs in osteogenic differentiation.27,28,30a,36 Here, our results suggest that BMP signaling is amplified by heparin sequestered at the cell-material interface, and is integral to certain aspects of hMSC osteogenic differentiation (i.e. ALP activity), but less important in others (i.e. OPN mRNA expression). These observations were only possible due to the control over the type of molecules sequestered by well-defined cell culture substrates. Future efforts can rely on the control afforded by these substrates to study the influence of BMP signaling on expression of other osteogenesis-related markers, as well as to characterize the role of other heparin-binding factors in hMSC differentiation.
Growth factor activity in natural microenvironments can be locally amplified or inhibited through specific, non-covalent interactions between growth factors and ECM proteoglycans. For example, BMP-4 spatial gradients formed via specific, non-covalent interactions with heparan sulfate PGs are key regulators of dorsoventral patterning of the vertebrate neural tube,37 and BMP-4/PG interactions also limit the range of BMP activity within the Xenopus animal cap.38 Efforts to study the local influence of proteins or spatial protein gradients on cell behavior in vitro commonly rely on proteins covalently immobilized onto a synthetic cell culture substrate. For example, Jiang and colleagues prepared counter-gradients of VEGF and fibronectin to study the influence of these two proteins on endothelial cell migration,39 while Ito and colleagues prepared an immobilized gradient of epidermal growth factor (EGF) to characterize the concentration-dependent effects of EGF on the growth of cells expressing an EGF receptor.40 Although covalently immobilized growth factors are often bioactive—that is, they can activate intracellular signaling cascades—they cannot be internalized into the cell. However, soluble growth factors bound to their cognate receptors are often co-internalized into the cell, and it has been demonstrated that such multi-molecular constructs can play a key role in mediating cell response to a growth factor.41 Here, we demonstrated that patterning a ligand that sequesters PGs allows for the bioactivity of endogenous PGs and growth factors to be harnessed within spatially defined regions. Therefore, this study mimics a mechanism of growth factor localization observed within natural microenvironments, and establishes a method to spatially regulate growth factor signaling that cannot be achieved by simply adding recombinant growth factors to the culture medium. It also directly addresses the potential issues associated with covalently immobilized growth factors, including limited internalization and potentially decreased biological activity. Although not demonstrated in this report, the substrates described herein are likely to enable studies in which growth factor signaling is amplified within specific subsets of cells of a larger population, as well as provide bio-mimetic microenvironments to characterize the influence of soluble factor gradients on cell behavior.
Understanding the factors that govern hMSC behavior is important in therapeutic applications, such as tissue engineering, that strive to replace damaged or diseased tissue with healthy, physiologically-functional tissue. In this study, the poorly defined culture additive fetal bovine serum provided a mixture of biomolecules that includes heparin PGs/GAGs and growth factors, and is intended as a model for the complex biomolecule milieu that biomaterials may be exposed to upon implantation in vivo. The in vitro model developed in this study has been designed to demonstrate the general concept that biomaterials can modulate cell behavior by sequestering specific, soluble biomolecules, while preventing the non-specific adsorption of others. Importantly, these observations may provide the basis for a new biomaterials design paradigm: materials that can non-covalently harness the bioactivity of specific endogenous biomolecules present at in vivo implant sites may eliminate the need for co-delivery of recombinant or xenogeneic biomolecules along with a stem cell-laden biomaterial to promote tissue regeneration or wound healing. In addition, substrates that can sequester specific biomolecules from complex mixtures, such as blood serum, may enable approaches wherein host-derived biomolecules can be sequestered and concentrated by the material, and subsequently re-implanted alongside a biomaterial for tissue engineering and wound healing applications, analogous to existing autologous cell transplantation therapies.
Conclusions
The substrates described in this work provide a 2-D culture platform to characterize the influence of serum-borne molecules at the cell-material interface. By eliminating the complications associated with random, non-specific biomolecule adsorption onto culture substrates, the substrates identified a correlation between serum-borne heparin and hMSC response to endogenous growth factors. Our results suggest that this strategy may be amenable to characterizing the influence of other serum-borne biomolecules on cell behavior by varying the binding specificity of the immobilized ligand. Of note, the chemistries employed in this approach are broadly applicable and have been widely used to impart specific bioactivity into a variety of otherwise bio-inert biomaterials (e.g. alginate42 and poly(ethylene glycol)43). Thus, future efforts may rely on substrates analogous to those developed here to identify ligands that can harness endogenous biomolecules to influence and study stem cell behavior, and then translate these findings to develop biomaterials for therapeutic applications.
Insight, innovation, integration.
INSIGHT: Heparin sequestered at the cell-material interface regulates mesenchymal stem cell behavior (proliferation and differentiation) by amplifying endogenous growth factor signaling. INNOVATION: This report relied on self-assembled monolayer-based cell culture substrates that sequester heparin and resist non-specific biomolecule adsorption. Eliminating the confounding factor of non-specifically adsorbed biomolecules allowed us to directly test the hypothesis that heparin sequestered at the cell-material interface enhances stem cell response to endogenous growth factors. INTEGRATION: Chemically well-defined cell culture substrates that can sequester particular subsets of serum-borne biomolecules via non-covalent interactions represent a unique platform to study the influence of endogenous biomolecules on cell behavior.
Acknowledgements
The authors would like to thank Professor Wan-Ju Li (University of Wisconsin) for providing quantitative polymerase chain reaction primer sequences and Dr Michael Schwartz (University of Wisconsin) for helpful discussions. This work was supported by the National Institutes of Health (R01HL093282 and R01EY017367) and the National Science Foundation (DMR 0906123) and the University of Wisconsin (UW) Stem Cell and Regenerative Medicine Center. SPR was performed in the UW Biophysics Instrumentation Facility.
Notes and references
- 1.Tsutsumi S, Shimazu A, Miyazaki K, Pan H, Koike C, Yoshida E, Takagishi K, Kato Y. Retention of multilineage differentiation potential of mesenchymal cells during proliferation in response to FGF. Biochem. Biophys. Res. Commun. 2001;288:413–419. doi: 10.1006/bbrc.2001.5777. [DOI] [PubMed] [Google Scholar]
- 2.Lavery K, Swain P, Falb D, Alaoui-Ismaili MH. BMP-2/4 and BMP-6/7 differentially utilize cell surface receptors to induce osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. J. Biol. Chem. 2008;283:20948–20958. doi: 10.1074/jbc.M800850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlodavsky I, Bar-Shavit R, Ishai-Michaeli R, Bashkin P, Fuks Z. Extracellular sequestration and release of fibroblast growth factor: a regulatory mechanism? Trends Biochem. Sci. 1991;16:268–271. doi: 10.1016/0968-0004(91)90102-2. [DOI] [PubMed] [Google Scholar]
- 4.Hudalla GA, Murphy WL. Biomaterials that regulate growth factor activity via bioinspired interactions. Adv. Funct. Mater. 2011 doi: 10.1002/adfm.201002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Rezania A, Healy KE. Biomimetic peptide surfaces that regulate adhesion, spreading, cytoskeletal organization, and mineralization of the matrix deposited by osteoblast-like cells. Biotechnol. Prog. 1999;15:19–32. doi: 10.1021/bp980083b. [DOI] [PubMed] [Google Scholar]; (b) Hudalla GA, Murphy WL. Immobilization of peptides with distinct biological activities onto stem cell culture substrates using orthogonal chemistries. Langmuir. 2010;26:6449–6456. doi: 10.1021/la1008208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klim JR, Li L, Wrighton PJ, Piekarczyk MS, Kiessling LL. A defined glycosaminoglycan-binding substratum for human pluripotent stem cells. Nat. Methods. 2010;7:989–994. doi: 10.1038/nmeth.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winterton LC, Andrade JD, Feijen J, Kim SW. Heparin interaction with protein-adsorbed surfaces. J. Colloid Interface Sci. 1986;111:314–342. [Google Scholar]
- 8.Charonis AS, Skubitz AP, Koliakos GG, Reger LA, Dege J, Vogel AM, Wohlhueter R, Furcht LT. A novel synthetic peptide from the B1 chain of laminin with heparin-binding and cell adhesion-promoting activities. J. Cell Biol. 1988;107:1253–1260. doi: 10.1083/jcb.107.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudalla GA, Murphy WL. Sequestering heparin from serum amplifies recombinant growth factor activity. 2011 submitted. [Google Scholar]
- 10.Lu H, McDowell LM, Studelska DR, Zhang L. Glycosaminoglycans in human and bovine serum: detection of twenty-four heparan sulfate and chondroitin sulfate motifs including a novel sialic acid-modified chondroitin sulfate linkage hexasaccharide. Glycobiol. Insights. 2010;2010:13–28. [PMC free article] [PubMed] [Google Scholar]
- 11.Fagerstam LG, Frostell-Karlsson A, Karlsson R, Persson B, Ronnberg I. Biospecific interaction analysis using surface plasmon resonance detection applied to kinetic, binding site and concentration analysis. J. Chromatogr., A. 1992;597:397–410. doi: 10.1016/0021-9673(92)80137-j. [DOI] [PubMed] [Google Scholar]
- 12.Holmlin RE, Chen X, Chapman RG, Takayama S, Whitesides GM. Zwitterionic SAMs that resist nonspecific adsorption of protein from aqueous buffer. Langmuir. 2001;17:2841–2850. doi: 10.1021/la0015258. [DOI] [PubMed] [Google Scholar]
- 13.Chapman RG, Ostuni E, S T, RE H, Yan L, Whitesides GM. Surveying for surfaces that resist the adsorption of proteins. J. Am. Chem. Soc. 2000;122:8303–8304. [Google Scholar]
- 14.Koepsel JT, Murphy WL. Patterning discrete stem cell culture environments via localized self-assembled monolayer replacement. Langmuir. 2009;25:12825–12834. doi: 10.1021/la901938e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kilian KA, Bugarija B, Lahn BT, Mrksich M. Geometric cues for directing the differentiation of mesenchymal stem cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:4872–4877. doi: 10.1073/pnas.0903269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 17.Harbers GM, Healy KE. The effect of ligand type and density on osteoblast adhesion, proliferation, and matrix mineralization. J. Biomed. Mater. Res., Part A. 2005;75:855–869. doi: 10.1002/jbm.a.30482. [DOI] [PubMed] [Google Scholar]
- 18.Maurer H. Towards chemically-defined, serum-free media for mammalian cell culture. Oxford: IRL Press; 1986. [Google Scholar]
- 19.Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 20.Ornitz DM, Yayon A, Flanagan JG, Svahn CM, Levi E, Leder P. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol. Cell Biol. 1992;12:240–247. doi: 10.1128/mcb.12.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohammadi M, Froum S, Hamby JM, Schroeder MC, Panek RL, Lu GH, Eliseenkova AV, Green D, Schlessinger J, Hubbard SR. Crystal structure of an angiogenesis inhibitor bound to the FGF receptor tyrosine kinase domain. EMBO J. 1998;17:5896–5904. doi: 10.1093/emboj/17.20.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalton BA, McFarland CD, Underwood PA, Steele JG. Role of the heparin binding domain of fibronectin in attachment and spreading of human bone-derived cells. J. Cell Sci. 1995;108(Pt 5):2083–2092. doi: 10.1242/jcs.108.5.2083. [DOI] [PubMed] [Google Scholar]
- 23.Farre J, Roura S, Prat-Vidal C, Soler-Botija C, Llach A, Molina CE, Hove-Madsen L, Cairo JJ, Godia F, Bragos R, Cinca J, Bayes-Genis A. FGF-4 increases in vitro expansion rate of human adult bone marrow-derived mesenchymal stem cells. Growth Factors. 2007;25:71–76. doi: 10.1080/08977190701345200. [DOI] [PubMed] [Google Scholar]
- 24.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 25.Thompson LD, Pantoliano MW, Springer BA. Energetic characterization of the basic fibroblast growth factor-heparin interaction: identification of the heparin binding domain. Biochemistry. 1994;33:3831–3840. doi: 10.1021/bi00179a006. [DOI] [PubMed] [Google Scholar]
- 26.(a) Bale MD, Wohlfahrt LA, Mosher DF, Tomasini B, Sutton RC. Identification of vitronectin as a major plasma protein adsorbed on polymer surfaces of different copolymer composition. Blood. 1989;74:2698–2706. [PubMed] [Google Scholar]; (b) Horbett TA. The role of adsorbed proteins in animal cell adhesion. Colloids Surf., B. 1994;2:225–240. [Google Scholar]
- 27.Jiao X, Billings PC, O’Connell MP, Kaplan FS, Shore EM, Glaser DL. Heparan sulfate proteoglycans (HSPGs) modulate BMP2 osteogenic bioactivity in C2C12 cells. J. Biol. Chem. 2007;282:1080–1086. doi: 10.1074/jbc.M513414200. [DOI] [PubMed] [Google Scholar]
- 28.Haupt LM, Murali S, Mun FK, Teplyuk N, Mei LF, Stein GS, van Wijnen AJ, Nurcombe V, Cool SM. The heparan sulfate proteoglycan (HSPG) glypican-3 mediates commitment of MC3T3-E1 cells toward osteogenesis. J. Cell Physiol. 2009;220:780–791. doi: 10.1002/jcp.21825. [DOI] [PubMed] [Google Scholar]
- 29.Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT. Structure–activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg. Med. Chem. Lett. 2008;18:4388–4392. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.(a) Ruppert R, Hoffmann E, Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur. J. Biochem. 1996;237:295–302. doi: 10.1111/j.1432-1033.1996.0295n.x. [DOI] [PubMed] [Google Scholar]; (b) Choi YJ, Lee JY, Park JH, Park JB, Suh JS, Choi YS, Lee SJ, Chung CP, Park YJ. The identification of a heparin binding domain peptide from bone morphogenetic protein-4 and its role on osteogenesis. Biomaterials. 2010;31:7226–7238. doi: 10.1016/j.biomaterials.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 31.Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, Chen CS. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat. Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martino MM, Mochizuki M, Rothenfluh DA, Rempel SA, Hubbell JA, Barker TH. Controlling integrin specificity and stem cell differentiation in 2D and 3D environments through regulation of fibronectin domain stability. Biomaterials. 2009;30:1089–1097. doi: 10.1016/j.biomaterials.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.(a) Horbett TA. Mass action effects on competitive adsorption of fibrinogen from hemoglobin solutions and from plasma. Thromb. Haemost. 1984;51:174–181. [PubMed] [Google Scholar]; (b) Brash JL, ten Hove P. Effect of plasma dilution on adsorption of fibrinogen to solid surfaces. Thromb. Haemost. 1984;51:326–330. [PubMed] [Google Scholar]
- 34.Levenstein ME, Berggren WT, Lee JE, Conard KR, Llanas RA, Wagner RJ, Smith LM, Thomson JA. Secreted proteoglycans directly mediate human embryonic stem cell-basic fibroblast growth factor 2 interactions critical for proliferation. Stem Cells. 2008;26:3099–3107. doi: 10.1634/stemcells.2007-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abraham S, Riggs MJ, Nelson K, Lee V, Rao RR. Characterization of human fibroblast-derived extracellular matrix components for human pluripotent stem cell propagation. Acta Biomater. 2010;6:4622–4633. doi: 10.1016/j.actbio.2010.07.029. [DOI] [PubMed] [Google Scholar]
- 36.Zhao B, Katagiri T, Toyoda H, Takada T, Yanai T, Fukuda T, Chung UI, Koike T, Takaoka K, Kamijo R. Heparin potentiates the in vivo ectopic bone formation induced by bone morphogenetic protein-2. J. Biol. Chem. 2006;281:23246–23253. doi: 10.1074/jbc.M511039200. [DOI] [PubMed] [Google Scholar]
- 37.Hu Q, Ueno N, Behringer RR. Restriction of BMP4 activity domains in the developing neural tube of the mouse embryo. EMBO Rep. 2004;5:734–739. doi: 10.1038/sj.embor.7400184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohkawara B, Iemura S, ten Dijke P, Ueno N. Action range of BMP is defined by its N-terminal basic amino acid core. Curr. Biol. 2002;12:205–209. doi: 10.1016/s0960-9822(01)00684-4. [DOI] [PubMed] [Google Scholar]
- 39.Liu L, Ratner BD, Sage EH, Jiang S. Endothelial cell migration on surface-density gradients of fibronectin, VEGF, or both proteins. Langmuir. 2007;23:11168–11173. doi: 10.1021/la701435x. [DOI] [PubMed] [Google Scholar]
- 40.Chen G, Ito Y. Gradient micropattern immobilization of EGF to investigate the effect of artificial juxtacrine stimulation. Biomaterials. 2001;22:2453–2457. doi: 10.1016/s0142-9612(00)00432-4. [DOI] [PubMed] [Google Scholar]
- 41.(a) Hawker JR, Jr, Granger HJ. Internalized basic fibroblast growth factor translocates to nuclei of venular endothelial cells. Am. J. Physiol. 1992;262:H1525–H1537. doi: 10.1152/ajpheart.1992.262.5.H1525. [DOI] [PubMed] [Google Scholar]; (b) Sperinde GV, Nugent MA. Heparan sulfate proteoglycans control intracellular processing of bFGF in vascular smooth muscle cells. Biochemistry. 1998;37:13153–13164. doi: 10.1021/bi980600z. [DOI] [PubMed] [Google Scholar]
- 42.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 43.Liu SQ, Ee PL, Ke CY, Hedrick JL, Yang YY. Biodegradable poly(ethylene glycol)-peptide hydrogels with well-defined structure and properties for cell delivery. Biomaterials. 2009;30:1453–1461. doi: 10.1016/j.biomaterials.2008.11.023. [DOI] [PubMed] [Google Scholar]