Abstract
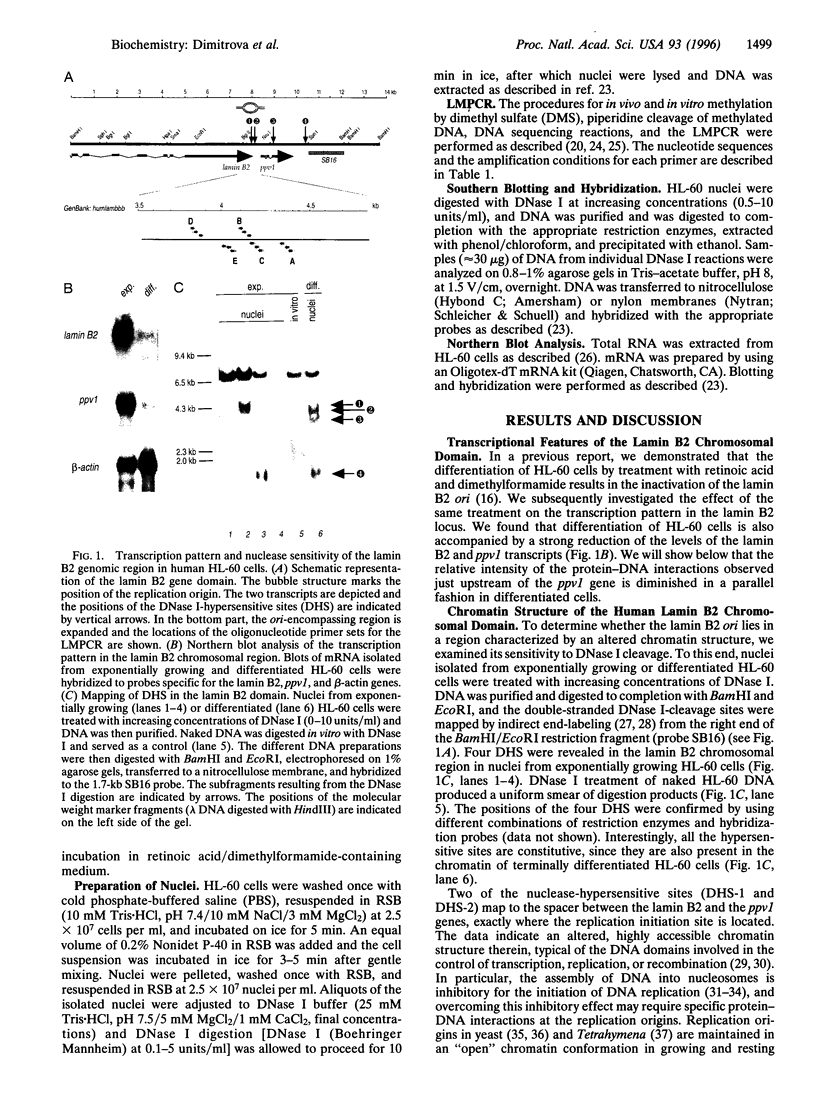
Protein-DNA interactions were studied in vivo at the region containing a human DNA replication origin, located at the 3' end of the lamin B2 gene and partially overlapping the promoter of another gene, located downstream. DNase I treatment of nuclei isolated from both exponentially growing and nonproliferating HL-60 cells showed that this region has an altered, highly accessible, chromatin structure. High-resolution analysis of protein-DNA interactions in a 600-bp area encompassing the origin was carried out by the in vivo footprinting technique based on the ligation-mediated polymerase chain reaction. In growing HL-60 cells, footprints at sequences homologous to binding sites for known transcription factors (members of the basic-helix-loop-helix family, nuclear respiratory factor 1, transcription factor Sp1, and upstream binding factor) were detected in the region corresponding to the promoter of the downstream gene. Upon conversion of cells to a nonproliferative state, a reduction in the intensity of these footprints was observed that paralleled the diminished transcriptional activity of the genomic area. In addition to these protections, in close correspondence to the replication initiation site, a prominent footprint was detected that extended over 70 nucleotides on one strand only. This footprint was absent from nonproliferating HL-60 cells, indicating that this specific protein-DNA interaction might be involved in the process of origin activation.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bell S. P., Kobayashi R., Stillman B. Yeast origin recognition complex functions in transcription silencing and DNA replication. Science. 1993 Dec 17;262(5141):1844–1849. doi: 10.1126/science.8266072. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Learned R. M., Jantzen H. M., Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988 Sep 2;241(4870):1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Marahrens Y., Rao H., Stillman B. The replicon model and eukaryotic chromosomes. Cold Spring Harb Symp Quant Biol. 1993;58:435–442. doi: 10.1101/sqb.1993.058.01.050. [DOI] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992 May 14;357(6374):128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Biamonti G., Giacca M., Perini G., Contreas G., Zentilin L., Weighardt F., Guerra M., Della Valle G., Saccone S., Riva S. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol Cell Biol. 1992 Aug;12(8):3499–3506. doi: 10.1128/mcb.12.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biamonti G., Perini G., Weighardt F., Riva S., Giacca M., Norio P., Zentilin L., Diviacco S., Dimitrova D., Falaschi A. A human DNA replication origin: localization and transcriptional characterization. Chromosoma. 1992;102(1 Suppl):S24–S31. doi: 10.1007/BF02451782. [DOI] [PubMed] [Google Scholar]
- Bozzoni I., Baldari C. T., Amaldi F., Buongiorno-Nardelli M. Replication of ribosomal DNA in Xenopus laevis. Eur J Biochem. 1981 Sep 1;118(3):585–590. doi: 10.1111/j.1432-1033.1981.tb05559.x. [DOI] [PubMed] [Google Scholar]
- Briggs M. R., Kadonaga J. T., Bell S. P., Tjian R. Purification and biochemical characterization of the promoter-specific transcription factor, Sp1. Science. 1986 Oct 3;234(4772):47–52. doi: 10.1126/science.3529394. [DOI] [PubMed] [Google Scholar]
- Brown J. A., Holmes S. G., Smith M. M. The chromatin structure of Saccharomyces cerevisiae autonomously replicating sequences changes during the cell division cycle. Mol Cell Biol. 1991 Oct;11(10):5301–5311. doi: 10.1128/mcb.11.10.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Kelly T. J. Transcriptional activator nuclear factor I stimulates the replication of SV40 minichromosomes in vivo and in vitro. Cell. 1989 Nov 3;59(3):541–551. doi: 10.1016/0092-8674(89)90037-8. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- DePamphilis M. L. Eukaryotic DNA replication: anatomy of an origin. Annu Rev Biochem. 1993;62:29–63. doi: 10.1146/annurev.bi.62.070193.000333. [DOI] [PubMed] [Google Scholar]
- Demarchi F., D'Agaro P., Falaschi A., Giacca M. In vivo footprinting analysis of constitutive and inducible protein-DNA interactions at the long terminal repeat of human immunodeficiency virus type 1. J Virol. 1993 Dec;67(12):7450–7460. doi: 10.1128/jvi.67.12.7450-7460.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H., Dowell S. J., Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994 Jul 29;78(2):303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H. Protein-DNA interactions at a yeast replication origin. Nature. 1992 May 14;357(6374):169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Diller J. D., Raghuraman M. K. Eukaryotic replication origins: control in space and time. Trends Biochem Sci. 1994 Aug;19(8):320–325. doi: 10.1016/0968-0004(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Dutta A. Trans-plication factors? Curr Biol. 1993 Oct 1;3(10):709–712. doi: 10.1016/0960-9822(93)90076-z. [DOI] [PubMed] [Google Scholar]
- Elgin S. C. The formation and function of DNase I hypersensitive sites in the process of gene activation. J Biol Chem. 1988 Dec 25;263(36):19259–19262. [PubMed] [Google Scholar]
- Falaschi A., Biamonti G., Cobianchi F., Csordas-Toth E., Faulkner G., Giacca M., Pedacchia D., Perini G., Riva S., Tribioli C. Presence of transcription signals in two putative DNA replication origins of human cells. Biochim Biophys Acta. 1988 Dec 20;951(2-3):430–442. doi: 10.1016/0167-4781(88)90117-0. [DOI] [PubMed] [Google Scholar]
- Falaschi A., Giacca M. The quest for a human ori. Genetica. 1994;94(2-3):255–266. doi: 10.1007/BF01443439. [DOI] [PubMed] [Google Scholar]
- Falaschi A., Giacca M., Zentilin L., Norio P., Diviacco S., Dimitrova D., Kumar S., Tuteja R., Biamonti G., Perini G. Searching for replication origins in mammalian DNA. Gene. 1993 Dec 15;135(1-2):125–135. doi: 10.1016/0378-1119(93)90057-a. [DOI] [PubMed] [Google Scholar]
- Giacca M., Zentilin L., Norio P., Diviacco S., Dimitrova D., Contreas G., Biamonti G., Perini G., Weighardt F., Riva S. Fine mapping of a replication origin of human DNA. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):7119–7123. doi: 10.1073/pnas.91.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D. S., Garrard W. T. Nuclease hypersensitive sites in chromatin. Annu Rev Biochem. 1988;57:159–197. doi: 10.1146/annurev.bi.57.070188.001111. [DOI] [PubMed] [Google Scholar]
- Huberman J. A., Riggs A. D. On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol. 1968 Mar 14;32(2):327–341. doi: 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- Ishimi Y. Preincubation of T antigen with DNA overcomes repression of SV40 DNA replication by nucleosome assembly. J Biol Chem. 1992 May 25;267(15):10910–10913. [PubMed] [Google Scholar]
- Li J. J., Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993 Dec 17;262(5141):1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Li R., Yang L., Fouts E., Botchan M. R. Site-specific DNA-binding proteins important for replication and transcription have multiple activities. Cold Spring Harb Symp Quant Biol. 1993;58:403–413. doi: 10.1101/sqb.1993.058.01.047. [DOI] [PubMed] [Google Scholar]
- Lohr D., Torchia T. Structure of the chromosomal copy of yeast ARS1. Biochemistry. 1988 May 31;27(11):3961–3965. doi: 10.1021/bi00411a011. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Bustin M., Miller O. L., Jr Electron microscopic analysis of chromosome metabolism in the Drosophila melanogaster embryo. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):741–754. doi: 10.1101/sqb.1978.042.01.075. [DOI] [PubMed] [Google Scholar]
- Meier R. W., Chen T., Mathews S., Niklaus G., Tobler A. The differentiation pathway of HL60 cells is a model system for studying the specific regulation of some myeloid genes. Cell Growth Differ. 1992 Oct;3(10):663–669. [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Palen T. E., Cech T. R. Chromatin structure at the replication origins and transcription-initiation regions of the ribosomal RNA genes of Tetrahymena. Cell. 1984 Apr;36(4):933–942. doi: 10.1016/0092-8674(84)90043-6. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Rowley A., Cocker J. H., Harwood J., Diffley J. F. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995 Jun 1;14(11):2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. T. Nucleosome positioning can affect the function of a cis-acting DNA element in vivo. Nature. 1990 Jan 25;343(6256):387–389. doi: 10.1038/343387a0. [DOI] [PubMed] [Google Scholar]
- Tribioli C., Biamonti G., Giacca M., Colonna M., Riva S., Falaschi A. Characterization of human DNA sequences synthesized at the onset of S-phase. Nucleic Acids Res. 1987 Dec 23;15(24):10211–10232. doi: 10.1093/nar/15.24.10211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth E. C., Marusic L., Ochem A., Patthy A., Pongor S., Giacca M., Falaschi A. Interactions of USF and Ku antigen with a human DNA region containing a replication origin. Nucleic Acids Res. 1993 Jul 11;21(14):3257–3263. doi: 10.1093/nar/21.14.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virbasius C. A., Virbasius J. V., Scarpulla R. C. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993 Dec;7(12A):2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]







