Abstract
MicroRNA-210 (miR-210) is a signature microRNA of hypoxia. We found robust increase (>100-fold) of miR-210 abundance in activated T cells, especially in the TH17 lineage. Hypoxia synergized with T cell receptor (TCR)–CD28 stimulation to accelerate and increase the magnitude of Mir210 expression. Mir210 was directly regulated by HIF-1α, a key regulator of TH17 polarization. Surprisingly, Hif1a was identified as a miR-210-target, suggesting negative-feedback by miR-210 to inhibit HIF-1α protein expression. Deletion of Mir210 promoted TH17 differentiation under conditions with limited oxygen. In experimental colitis, miR-210 reduced Hif1a transcript abundance, reduced the proportion of cells producing inflammatory cytokines and controlled disease severity. Our study identifies miR-210 as an important regulator of T cell differentiation in hypoxia, which can limit immunopathology.
Introduction
We usually study in vitro T cell responses at the atmospheric O2 concentration of 21%. However, oxygen tensions in lymphoid tissues are markedly lower (<5%) than those found in peripheral arterial blood (13%)1, 2. Immune cells are exposed to varying oxygen tensions in lymphoid organs2. T cells are activated during antigen recognition at inflammatory or tumor sites, both of which are hypoxic2. Moreover, hypoxic extracellular environments exist in healthy and diseased non-lymphoid tissues, including adipose tissue3, skin4 and the gastrointestinal tract5. Therefore, activated T cells are dependent on an intracellular machinery that enables them to adapt to changes in oxygen tension and execute their functions in situ. The transcription factor hypoxia-inducible factor 1 (HIF-1), mediates at least some of the required metabolic switches from oxidative phosphorylation to aerobic glycolysis in response to hypoxia6.
The HIF-1 complex consists of two subunits, HIF-1α and HIF-1β7. Under normoxic conditions, HIF-1α is proline hydroxylated by prolyl hydroxylases (PHDs) that promotes its binding to the von Hippel Lindau protein (VHL) E3 ligase complex, followed by HIF-1α ubiquitination and proteasomal degradation8, 9. Hypoxic conditions inhibit HIF-1α degradation and lead to its stabilization. Recently, a further regulatory layer in non-lymphocytes was described, revealing that in intestinal epithelial cells Hif1a is negatively regulated by microRNA-155 (miR-155) during prolonged hypoxia10.
In addition to the cellular response to hypoxia, HIF-1 plays an important role in regulating TH17 differentiation. TH17 cells mount responses against extracellular bacterial and fungal infections in the intestine and the airways11. Despite the benefit of such immune responses, TH17 cells can also play immunopathologic roles in experimental as well as naturally occurring autoimmune settings, including collagen-induced arthritis, experimental autoimmune encephalomyelitis (EAE) or inflammatory bowel diseases (IBD)12-14. HIF-1 promotes TH17 differentiation by directly inducing Rorc transcription and subsequently collaborates with RORγt to regulate downstream TH17 genes and inhibiting regulatory T cell (Treg) differentiation through an active process that targets Foxp3 protein for degradation15. In addition, deficiency of Hif1a in T cells diminishes the expression of glycolytic molecules and alters the dichotomy between these two T cell subsets, demonstrating that HIF-1α induces metabolic reprogramming and orchestrates lineage differentiation of T cells16.
miRNAs are noncoding single-stranded RNAs of about 22 nucleotides that mediate sequence-dependent posttranscriptional negative regulation of gene expression17. Various stresses, including hypoxia, regulate miRNA expression and function18. For example, a subset of miRNAs induced by hypoxia, referred to as “hypoxamiRs”, contribute to the regulation of the broad spectrum of genes regulated by hypoxia. Among these miRNAs, miR-210 is the master hypoxamiR and regulates a variety of cellular events in non-lymphoid tissues19-22.
Recent work has identified miRNAs as pivotal regulators of helper T cell differentiation and function23, 24. Two genome-wide miRNA-profiling studies in activated T cells revealed that miR-210 is highly expressed after T cell stimulation25, 26. However, Mir210-regulation and its role in T cell activation and differentiation have not been studied.
Results
T cell activation robustly induces Mir210
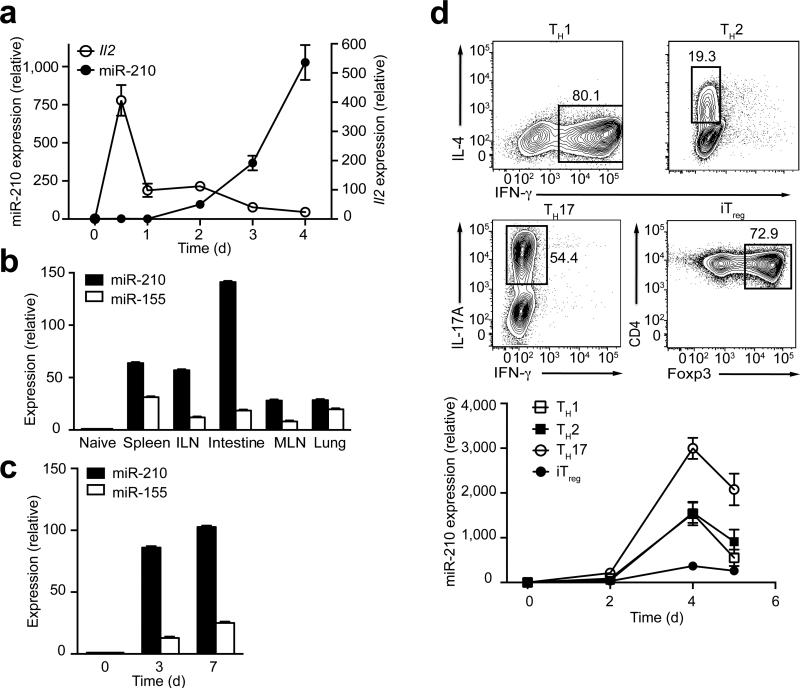
Two independent studies of global miRNA expression profiling in lymphocytes reported that miR-210 is highly expressed in activated mouse T cells (Supplementary Fig. 1)25, 26. To address the miR-210 expression pattern in more detail, we assessed the expression of miR-210 during T cell activation by real-time PCR (RT-PCR) following the stimulation of naive mouse CD4+ T cells with anti-CD3 and anti-CD28. TCR-CD28 stimulation led to an over 500-fold increase in miR-210 abundance in CD4+ T cells that were activated for 4 d compared to unstimulated cells (Fig. 1a). In contrast to interleukin 2 (Il2), which was rapidly upregulated by 12 h post stimulation, miR-210 abundance increased more slowly and reached highest amounts at 4 d after T cell activation, suggesting a possible regulatory role of miR-210 in later stages of T cell activation and function (Fig. 1a,d). We tested whether induction of Mir210 also occurs in vivo in activated CD4+ T cells using two different approaches. First, we examined miR-210 expression in homeostatically expanded CD4+ T cells isolated from various lymphoid tissues. To this end, naive CD4+ T cells were sorted and adoptively transferred into congenic Rag2−/− mice. After 3 weeks, we harvested cells from the spleen, inguinal lymph nodes (ILNs), mesenteric lymph nodes (MLNs), lung and small intestine. After their homeostatic expansion, the transferred CD4+ T cells isolated from the different tissues displayed a uniform, memory-like (CD44hi CD62L–) phenotype and all of the samples moderately upregulated miR-155, an antigen stimulation-induced microRNA27 (Fig. 1b and Supplementary Fig. 2a). miR-210 was also upregulated in these T cells, but unlike miR-155, miR-210 showed a distinct expression pattern, with much higher expression in CD4+ T cells isolated from hypoxic tissues like the small intestine, compared to CD4+ T cells isolated from relatively normoxic tissues like the lung. This suggests that miR-210 might play a more critical role in T cells residing within hypoxic sites. We also tested the in vivo induction of Mir210 following ovalbumin (OVA) immunization. We transferred naive CD4+ T cells isolated from OT-II TCR-transgenic mice (specific for the OVA-peptide 323–339) into congenic recipient mice, followed by OVA challenge. Compared to naive T cells, miR-210 was markedly upregulated in these T cells (Fig. 1c). Similarly, Mir210 was robustly induced during CD8+ T cell activation both by in vitro stimulation and by using a mouse model of lymphocytic choriomeningitis virus (LCMV) infection (Supplementary Fig. 2b,c). Collectively, these data suggest that T cell activation leads to markedly increased miR-210 expression both in vitro and in vivo.
Figure 1.
Mir210 is induced after T cell activation and regulated during T cell differentiation. (a) The expression of miR-210 or Il2 in activated T cells was assessed by RT-PCR. The data were normalized by miR-210 expression in naive T cells (n=3 independent biological replicates per data point). (b) Homeostatically expanded CD4+ T cells were sorted from various tissues 3 weeks after adoptive transfer. The expression of miR-210 and miR-155 were assessed by RT-PCR. (c) miR-210 and miR-155 expression within in vivo activated OTII CD4+ T cells were determined by RT-PCR. (d) After 4 d polarizing naive CD4+ T cells towards the TH1, TH2, TH17 or iTreg lineage, cells with selective expression of IFN-γ, IL-4, IL-17A or Foxp3 were assessed by flow cytometry with the percentages of gated cells depicted (top). The time dependency of miR-210 expression in polarized T cells was measured by RT-PCR (bottom). Relative expression is normalized to sno202. Data are from one experiment representative of two (b,c) or three (d) independent experiments (mean and s.d. in a–d).
To study how Mir210 is regulated during T cell differentiation, naive CD4+ T cells were in vitro polarized into TH1, TH2, TH17 and Treg cells and the time-dependent appearance of miR-210 was measured during the process of polarization. The highest increase in miR-210 was observed in TH17 cells, suggesting a preferential role of miR-210 in TH17 polarization or function (Fig. 1d).
CD28– but not IL-2–signaling controls Mir210 expression
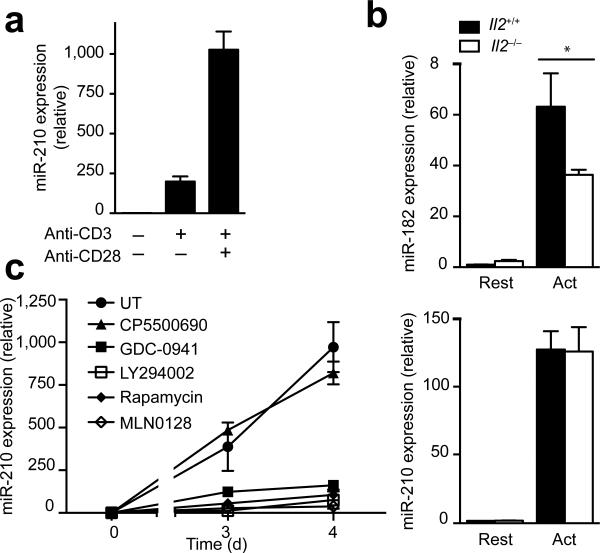
To examine whether CD28-mediated costimulation is involved in the upregulation of Mir210, we stimulated naive CD4+ T cells using anti-CD3 in the absence or presence of anti-CD28. miR-210 abundance in samples costimulated with anti-CD28 was 6-fold higher than in samples that were stimulated with anti-CD3 alone (Fig. 2a). Since CD28 costimulation is known to promote IL-2 production28 and the induction of Il2 transcripts preceded the induction of Mir210 (Fig. 1a), we investigated whether CD28-mediated costimulation might indirectly induce Mir210 by assessing miR-210 abundance in IL-2–deficient, CD4+ T cells (Fig. 2b). Whereas the expression of the IL-2-regulated miR-182 in IL-2–deficient CD4+ T cells was markedly reduced following TCR stimulation (Fig. 2b top)29, IL-2–deficiency had no effect on miR-210 expression, suggesting that IL-2 signaling is not required for the induction of Mir210 (Fig. 2b bottom).
Figure 2.
CD28-costimulation controls Mir210 expression via the PI(3)K-mTOR pathway. (a) Naive CD4+ T cells were activated by anti-CD3 in the presence or absence of anti-CD28 for 4 d and miR-210 expression was analyzed by RT-PCR. (b) RT-PCR analysis of miR-182 and miR-210 expression in activated T cells from wild-type or Il2−/− DO11.10 Rag2−/− mice. (c) CD4+ naive T cells were stimulated in the presence of the kinase inhibitors CP550 690 (100 nM), GDC-0941 (1 μM), LY294002 (5 μM), Rapamycin (20 nM) or MLN0128 (200 nM). miR-210 expression was measured by RTPCR. Relative expression is normalized to sno202 (n=3 independent biological replicates per data point). Data are from one experiment representative of three (a) independent experiments (mean and s.d. in a–c, * P<0.05, unpaired t-test in b, abbreviations: Rest, resting; Act, activated; UT, untreated).
Since phosphatidylinositol-3-OH kinase (PI(3)K) signaling is a major CD28 downstream signaling pathway30, we addressed the requirement of PI(3)K for the induction of Mir210 with small molecule inhibitors of enzymes involved in the PI(3)K signaling pathways. Whereas CP550690, an inhibitor of Jak1 and Jak3, showed no inhibitory effect on the expression of miR-210 in TCR–CD28-stimulated T cells, both PI(3)K (LY294002 and GDC-0941) and mTOR (rapamycin and MLN0128) inhibitors potently suppressed miR-210 expression (Fig. 2c). Thus, the induction of Mir210 in activated T cells is dependent upon the PI(3)K–mTOR pathway.
HIF-1α is required for Mir210 induction
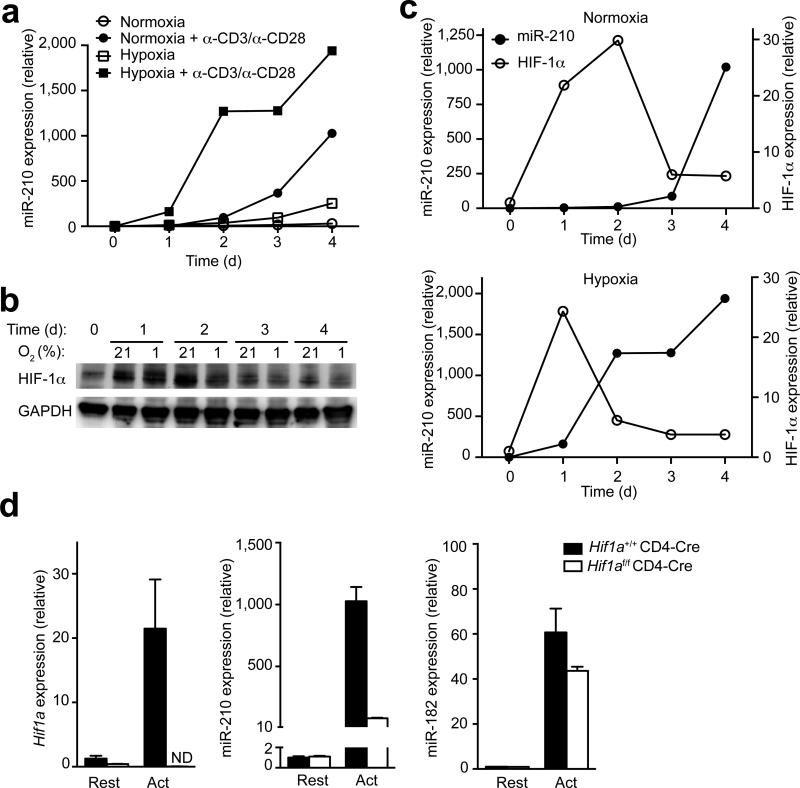
To study the influence of hypoxia on the regulation of Mir210 in T lymphocytes, we compared the induction of Mir210 in resting versus anti-CD3 and anti-CD28 stimulated primary CD4+ T cells under normoxic (21% O2) or hypoxic (1% O2) conditions. TCR stimulation under normoxia resulted in a robust but delayed upregulation of miR-210 (Fig. 3a). Unstimulated, T cells increased miR-210 abundance only modestly under hypoxic conditions, suggesting that in T cells, unlike other cell types, hypoxia alone is insufficient to induce robust miR-210 expression. However, stimulated CD4+ and CD8+ T cells exhibited rapid and markedly increased expression of miR-210 in hypoxia, suggesting a synergistic effect of TCR-stimulation and hypoxia (Fig. 3a and Supplementary Fig. 2b). Notably, this observation was specific for miR-210 since the abundance of miR-155 was not affected by oxygen tension during T cell activation (Supplementary Fig. 2d).
Figure 3. HIF-1α is required for Mir210 induction.
(a) Quantitative RT-PCR analysis of miR-210 abundance in unstimulated or anti-CD3– anti-CD28 stimulated CD4+ T cells. Cells were induced for a period of 4 d under normoxic (21% O2) or hypoxic (1% O2) conditions and RNA was harvested at the indicated time points. Relative miR-210 expression is normalized to its expression in naive T cells. (b) Immunoblot analysis with a HIF-1α-specific or GAPDH-specific antibody in total protein lysates of unstimulated or anti-CD3–anti-CD28 stimulated CD4+ T cells. Cells were induced for a period of 4 d under normoxic (21% O2) or hypoxic (1% O2) conditions and protein was harvested at the indicated time points. (c) Densitometric analysis of HIF-1α protein levels shown in (b) in comparison to miR-210 expression levels presented in (a). (d) Quantitative RT-PCR analysis of Hif1a, miR-210, and miR-182 transcripts in resting or stimulated wild-type (Hif1a+/+CD4-cre) or HIF-1α–deficient (Hif1af/fCD4-cre) CD4+ T cells. Relative expression is normalized to β2 (Hif1a) or sno202 (miR-210 and miR-182) and n=3 independent biological replicates per data point. Data are from one experiment representative of three (a–c) independent experiments. (mean and s.d. in a,c,d; abbreviations: Rest, resting; Act, activated; ND not detectable).
Since HIF-1α is the master regulator of hypoxic gene expression, we reasoned that HIF-1α might regulate Mir210 in T lymphocytes, as shown for other cell types31. Therefore, we compared HIF-1α protein abundance in resting versus TCR-stimulated CD4+ T cells under normoxic or hypoxic conditions. TCR-stimulation under both settings resulted in the accumulation of HIF-1α protein by 24 h (Fig. 3b,c). Consistent with miR-210 expression, hypoxia also led to more HIF-1α protein than in normoxia on day 1. Similar results were also found after CD8+ T cell activation (Supplementary Fig. 3a). The observations that both miR-210 and HIF-1α expression were regulated by oxygen tension and that the expression of HIF-1α preceded the induction of Mir210 support the notion that HIF-1 acts upstream of miR-210. Furthermore, we found that CD28– deficiency resulted in substantial reductions of HIF-1α protein in activated T cells (Supplementary Fig. 3b), indicating that TCR-CD28 signaling may induce Mir210 via HIF-1α.
To test this hypothesis, we studied TCR signaling-induced Mir210 expression under normoxic conditions in the presence or absence of HIF-1α. Whereas in HIF-1α–sufficient T cells anti-CD3–anti-CD28 stimulation resulted in robust upregulation of both Hif1a transcripts and miR-210, the induction of Mir210 was markedly attenuated in a HIF-1α–deficient background (Fig. 3d left and middle), indicating that HIF-1α is required for Mir210 induction in activated T cells. The deficiency of HIF-1α did not affect global activation of CD4+ T cells since miR-182 expression was largely unaffected by deletion of Hif1a (Fig. 3d right). Consequently, we reanalyzed a ChIP-seq dataset, performed in a recent study32 with TH17 polarized cells, which revealed that after T cell activation HIF-1α directly bound the promoter region of Mir210, with a binding peak at about 400 bp upstream of the Mir210 sequence (Supplementary Fig. 3c), thereby demonstrating that Mir210 is a direct downstream target of HIF-1α in activated TH17 cells.
Whereas HIF-1α protein abundance was sustained until 2 d post stimulation in normoxia, HIF-1α protein amounts markedly decreased between d 1 and d 2 under hypoxic conditions (Fig. 3b,c). Under normoxic conditions, the decrease in HIF-1α protein was detected later, between d 2 and d 3 after TCR stimulation. In both cases, the decrease in HIF-1α protein correlated with increases in miR-210 abundance, raising the possibility that HIF-1α could be both an upstream Mir210 regulator and a miR-210 target (Fig. 3b,c). Similar results were observed in CD8+ T cells activated under normoxic or hypoxic conditions (Supplementary Fig. 2b, 3a).
T cell-specific deletion of Mir210
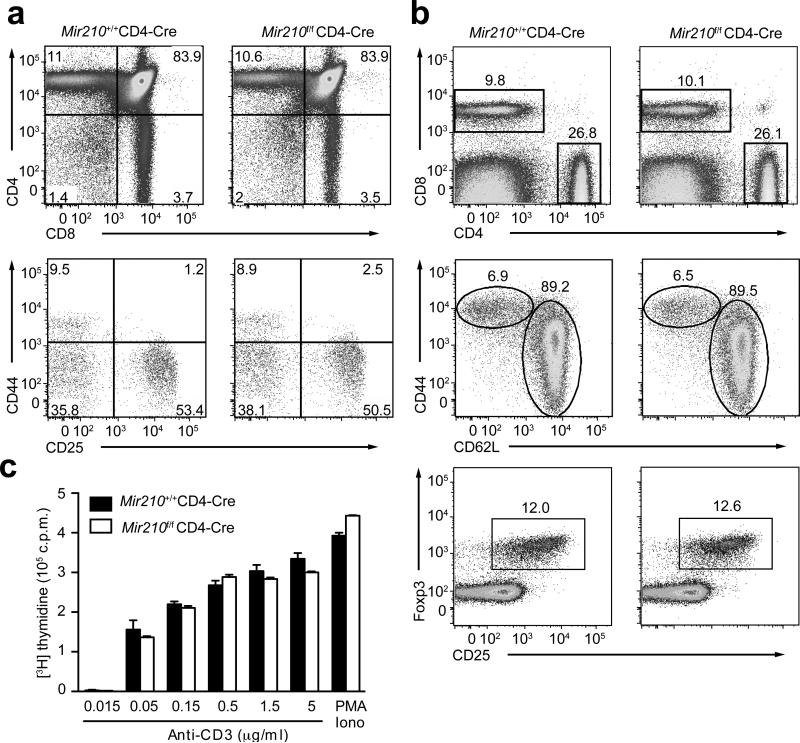
To investigate the role of miR-210 in T cells, mice with loxP-flanked Mir210 alleles (Mir210f/f)33 were crossed with CD4-Cre transgenic mice to delete the floxed Mir210 alleles in T cells (hereafter referred to as Mir210–/– mice). RT-PCR analysis indicated efficient deletion (>98%) of Mir210 in activated T cells (Supplementary Fig. 4a). Consistent with low miR-210 abundance in immature thymocytes (Supplementary Fig. 4a), miR-210–deficiency had little effect on T cell development (Fig. 4a). Moreover, wild-type and Mir210–/– mice had comparable numbers and distributions of CD4+ and CD8+ T cells in the periphery and the expression of CD62L and CD44 on peripheral CD4+ T cells was unaffected by deletion of Mir210 in 6-8 week old mice (Fig. 4b and data not shown). The development of natural regulatory T cells in the periphery appeared to be unaltered (Fig. 4b). T cell proliferation in response to TCR–CD28 or PMA–ionomycin stimulation was also comparable in wild-type or Mir210–/– T cells (Fig. 4c). Therefore, miR-210 is not required for T cell development, homeostasis and proliferation.
Figure 4. miR-210–deficiency has no apparent effect on T cell development and proliferation.
(a) Flow cytometry analysis of thymocytes (top panels) or gated double negative thymocytes (bottom panels) from 7 weeks old wild-type or Mir210–/– mice, stained with anti-CD4 and anti-CD8 or anti-CD44 and anti-CD25. Numbers adjacent to (or in) outlined areas indicate percent cells in each throughout. (b) The surface expression of CD4, CD8 (top), CD44, CD62L (middle) or CD25 and the intracellular expression of Foxp3 (bottom) in splenocytes derived from 7 weeks old wild-type or Mir210–/– mice were assessed by flow cytometry. (c) T cell proliferation was analyzed in triplicate samples by 3H-thymidine uptake 72 h after stimulation with either anti-CD3 or PMA plus ionomycin (mean and s.d.). Data are representative of three (a–c) independent experiments.
Hif1a is a miR-210 target gene
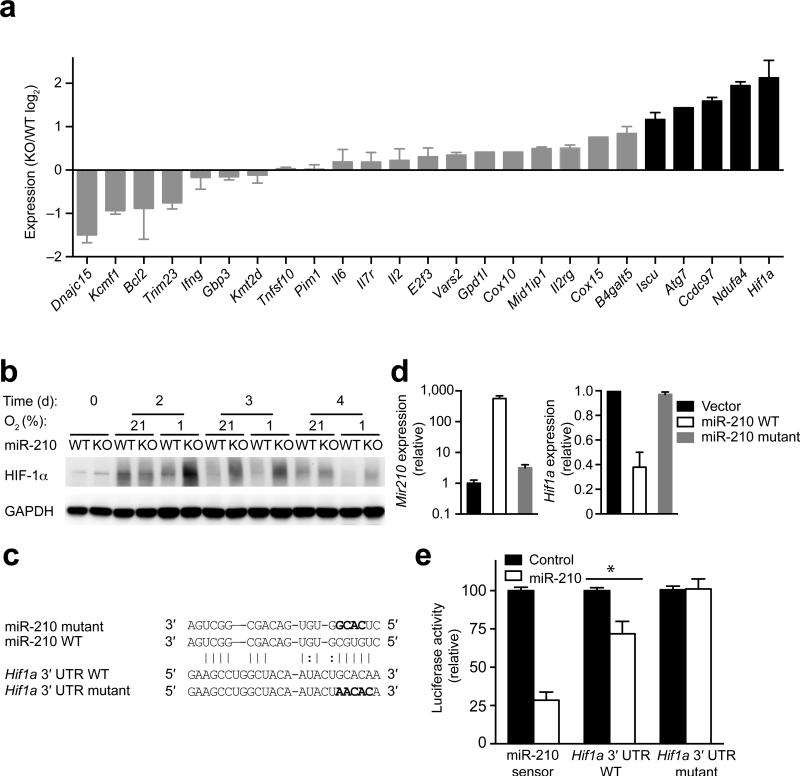
To explore the role of miR-210 in T cell differentiation, we used an in silico analysis to identify potential target genes of miR-210. We identified miR-210 potential target genes in mouse T cells by performing the following two-step selection process. First, we used four algorithms, Target scan, PicTar, miRDB and miRanda34-37, to predict miR-210 target genes. All targets predicted by miRanda were scored for an empirical probability of target inhibition using mirSVR scores38and a stringent mirSVR score cutoff of -1.1. We combined this list with previously reported miR-210 targets, resulting in 69 potential miR-210 target genes (Supplementary Fig. 4b and Supplementary Table 1). Next, we selected for T cell-expressed target genes according to the Immgen data-base (www.immgen.org), resulting in 21 genes (Supplementary Fig. 4b and Supplementary Table 1). We compared their expression by RT-PCR in wild-type or Mir210–/– CD4+ T cells, which were activated for 4 d. Assuming that miR-210–deficiency resulted in a higher expression of direct miR-210 targets, we identified five candidate genes that exhibited more than two-fold increased expression in Mir210–/– CD4+ T cells (Fig. 5a and Supplementary Fig. 4b). Two of these genes, Iscu and Ndufa4, were previously reported as miR-210 targets31, 39, 40. Surprisingly, Hif1a, was the highest upregulated (>4 fold) miR-210 target gene (Fig. 5a). To validate this observation, we compared HIF-1α protein in resting and TCR-stimulated wild-type or Mir210–/– CD4+ T cells under normoxic or hypoxic conditions. Under hypoxia, we detected a marked and sustained increase in HIF-1α protein abundance in activated Mir210–/– T cells (Fig. 5b). Moreover, a moderate increase in HIF-1α protein was even detected in Mir210–/– T cells activated for 3 or 4 d in normoxia (Fig. 5b). These results support the notion that Hif1a is a miR-210 target gene, and that miR-210 adds an additional layer of regulation by curbing HIF-1α activity under hypoxia. The different kinetics of HIF-1α protein accumulation and degradation in stimulated wild-type CD4+ T cells under normoxia or hypoxia (Fig. 3b, 3c), likely accounts for the fact that wild-type cells activated for 2 d under hypoxia had slightly lower HIF-1α protein abundance than cells activated under normoxia (Fig. 5b). We determined whether ectopic expression of miR-210 decreased Hif1a transcript abundance. We reconstituted miR-210 expression in TCR-CD28–activated Mir210–/– CD4+ T cells by transduction with bicistronic retroviruses expressing GFP and wild-type pri-miR-210 or GFP and a pri-miR-210 variant whose seed region was replaced by a scrambled control sequence (Fig. 5c). We sorted GFP+ CD4+ T cells and found that Hif1a expression was markedly decreased by wild-type but not mutant miR-210 (Fig. 5d).
Figure 5. miR-210 directly targets Hif1a.
(a) RT-PCR analysis of the expression of predicted miR-210 target and control gene transcripts in anti-CD3–anti-CD28–stimulated wild-type or Mir210–/– CD4+ T cells. The genes with more than 2-fold upregulation were highlighted. (b) Naive CD4+ T cells from wild-type (WT) or Mir210–/– (KO) mice were activated by anti-CD3–anti-CD28 under normoxic (21% O2) or hypoxic (1% O2) conditions for 4 d and HIF-1α as well as GAPDH protein abundance was detected by immunoblot analysis. (c) Sequence alignment between miR-210 and the 3′ UTR of Hif1a. The mutated sequences in seed regions of miR-210 and Hif1a are highlighted in bold figures. (d) Mir210–/– CD4+ T cells were anti-CD3 plus anti-CD28 stimulated for 36 h, transduced with bicistronic retroviruses expressing GFP (Vector), GFP and wild-type pri-miR-210 (miR-210 WT) or GFP and mutated pri-miR-210 (miR-210 mutant) followed by RT-PCR analysis for Hif1a and miR-210 expression in GFP+, CD4+ T cells at 84 h. (e) Mir210–/– CD4+ T cells were cotransfected with luciferase reporters and miR-210 or control miRNA mimics. Luciferase activity was measured 24 h after transfection and was normalized to control transfected cells. (* P<0.05, n=4 independent biological replicates per data point t-test). Data are from one experiment representative of two (a,b) or three (d) independent experiments. (mean and s.d. in a,d,e).
Hif1a contains a non-canonical miR-210 targeting site in its 3′ UTR, containing one G:U wobble at position 7 of the 6-mer seed region. Recent HITS-CLIP studies revealed that a sizeable fraction of miRNA binding sites do not obey the seed pairing rule suggesting that non-canonical sites are widespread41, 42. One of these studies also provides a T cell-specific genome-wide argonaute (AGO) binding data set (CLIP Base), which enables analysis of target interactions with all miRNAs expressed in activated T cells42. CLIP Base shows a large peak of AGO binding to the Hif1a 3′ UTR at around 150 bp from the beginning of the 3′ UTR where the predicted miR-210 target site is located (Supplementary Fig. 5a). This potential miR-210 target site is highly conserved among multiple species, stressing its evolutionary importance (Supplementary Fig. 5b). To determine whether miR-210 binds this site directly, we cloned the full-length Hif1a 3′ UTR (>1800 nucleotides) into a luciferase reporter construct, and cotransfected this reporter with miR-210 mimics into Mir210–/– CD4+ T cells. A control luciferase reporter with a tandem stretch of four perfect miR-210 binding sites (miR-210 sensor) exhibited markedly reduced luciferase activity of the miR-210 sensor, indicating the miR-210 mimics do function in vivo. Interestingly, miR-210 expression also suppressed by about 30% the cotransfected Hif1a 3′ UTR reporter that contains only a single copy of the putative miR-210 binding (Fig. 5e). Importantly, this suppression of Hif1a 3′ UTR by miR-210 was abolished by mutation of the miR-210 binding site. Thus, Hif1a is a bona fide target gene of miR-210.
miR-210 regulates TH17 polarization under reoxygenation
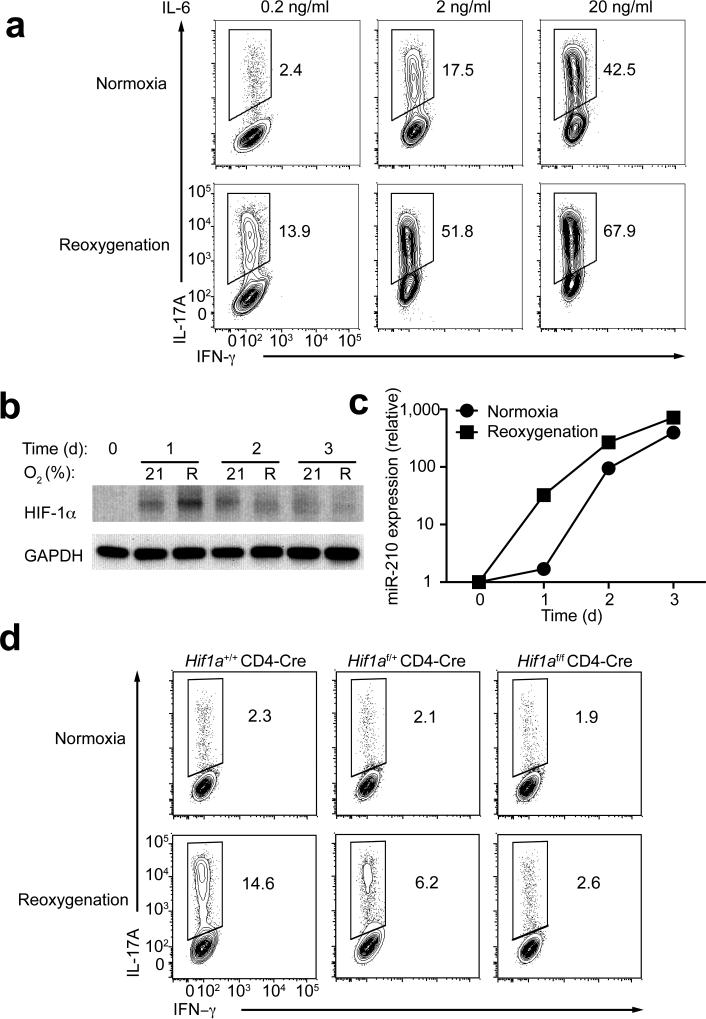
Since HIF-1 promotes TH17 differentiation15, 16, we investigated the potential role of miR-210 in TH17 cell differentiation in vitro. We compared the efficiency of wild-type or Mir210–/– naive T cells to differentiate into TH17 cells in normoxia or an oxygen-limited environment. Under hypoxic conditions (1% O2), the expansion of activated T cells was severely impaired (Supplementary Fig. 5c). Therefore, reoxygenation conditions have been used to study TH17 cell differentiation43. Reoxygenation has been used to reflect the changes of O2 concentration experienced by migrating T cells in vivo and involves the differentiation of TH17 cells by a priming step under a low O2 concentration (5% O2) followed by transfer to normoxic conditions43 (Supplementary Fig. 6a). TH17 differentiation was promoted under reoxygenation versus normoxia under suboptimal (0.2 ng/ml IL-6), intermediate (2.0 ng/ml IL-6) or maximal (20.0 ng/ml IL-6) TH17 skewing conditions (Fig. 6a). We used suboptimal IL-6 concentration (0.2 ng/ml) in subsequent analyses, since it showed the largest net increase in TH17 differentiation. Monitoring HIF-1α protein in TH17 cells polarized under normoxia or reoxygenation, we detected more HIF-1α protein in cells that were activated for 1 d under reoxygenation conditions, and a moderate increase in HIF-1α protein in cells polarized for 1, 2 or 3 d in normoxia (Fig. 6b), suggesting a critical role for HIF-1α in TH17 differentiation in normoxia and reoxygenation conditions15, 16, 43. Consequently, CD4+ T cells polarized under reoxygenation conditions exhibited a more rapid and increased expression of miR-210 (Fig. 6c), which was associated with a rapid decrease in HIF-1α protein between d 1 and d 2 (Fig. 6b, 6c). Similar results were observed in activated T cells in nonpolarizing conditions (Supplementary Figure 6b,c). Notably, the increase of TH17 differentiation under reoxygenation, compared to normoxia, was reduced by about 50% in Hif1af/+ CD4-Cre and was abolished in Hif1af/f CD4-Cre T cells, indicating that HIF-1α protein is limiting for the differentiation of TH17 cells under reoxygenation conditions (Fig. 6d).
Figure 6. Priming of naive T cells under reoxygenation increases miR-210 abundance.
(a) Naive CD4+ T cells were polarized towards TH17 cells with titrated doses of IL-6 under normoxic or reoxygenation conditions (Supplementary Fig. 6a), followed by intracellular staining of IL-17A and IFN-γ. (b–c) Immunoblot analysis of HIF-1α and GAPDH and RT-PCR analysis of miR-210 in TH17 cells polarized under normoxic or reoxygenation conditions (0.2 ng/ml of IL-6). Relative miR-210 abundance was normalized to its expression in naive T cells. (d) Naive CD4+ T cells from Hif1a+/+CD4-Cre, Hif1a f/+CD4-Cre or Hif1af/fCD4-Cre mice were differentiated under TH17 skewing conditions with 0.2 ng/ml of IL-6 under normoxic or reoxygenation conditions, followed by intracellular staining of IL-17A and IFN-γ. Data are representative of two (b,c,d) or three (a) independent experiments. (mean and s.d. in c).
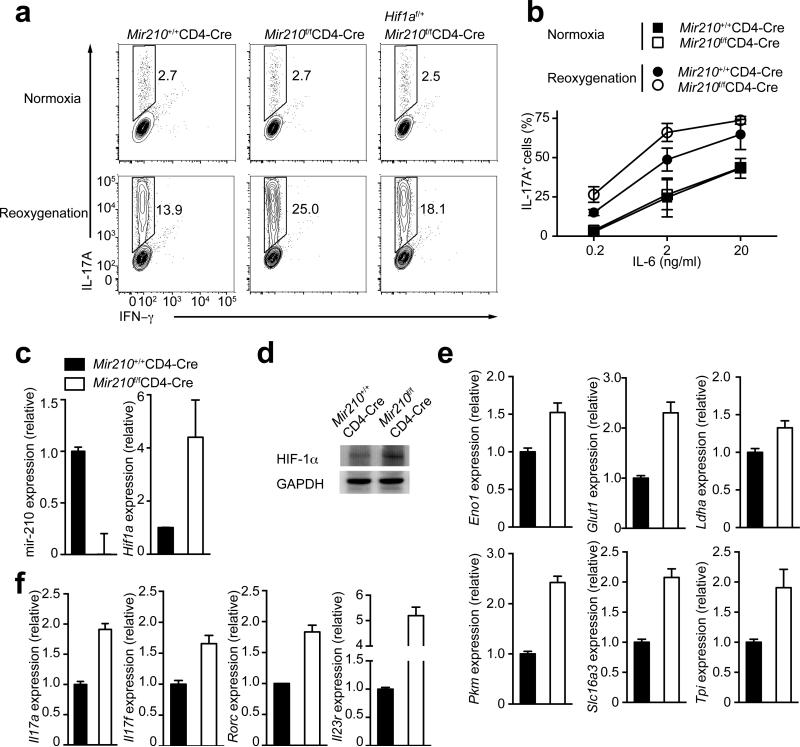
Whereas deficiency in miR-210 had subtle effects on TH17 differentiation under normoxic conditions, Mir210 deletion markedly increased the proportion of TH17 cells under reoxygenation, especially if low or intermediate IL-6 concentrations were used (Fig. 7a,b). By comparing wild-type or Mir210–/– CD4+ T cells which were TH17 polarized under reoxygenation conditions, we observed that Hif1a transcript and HIF-1α protein amounts were increased in Mir210–/– cells (Fig. 7c,d). The increased HIF-1α protein was associated with marked up-regulation of genes encoding key regulators of the glycolytic pathway (Fig. 7e), promoting TH17 differentiation16. This correlated well with increased expression of TH17 signature genes, including Il17a, Il17f, Rorc and Il23r (Fig. 7f). Intriguingly, the observed increase in lineage differentiation under reoxygenation conditions, and the further polarization by deletion of Mir210 exclusively applied for the TH17 subset (Supplementary Fig. 6d,e). Next, we investigated intracellular Foxp3 abundance in TH17-polarized wild-type or Mir210–/– CD4+ T cells, skewed under suboptimal (0.2 ng/ml IL-6) TH17 conditions in normoxia or reoxygenation conditions. Normoxia-cultured cells exhibited a high proportion of Foxp3+ cells, irrespective whether they were miR-210–sufficient or –deficient (Supplementary Fig. 6f), consistent with reduced TH17 differentiation under those conditions (Fig. 6a). In contrast, wild-type CD4+ T cells, skewed under reoxygenation conditions exhibited a decreased proportion of Foxp3+ cells (Supplementary Figure 6f), correlating well with increased TH17 differentiation potential (Fig. 6a) and higher HIF-1α protein amounts (Fig. 6b). Mir210 deletion, which resulted in further increase of HIF-1α protein expression under reoxygenation (Fig. 7d), caused a further drop of Foxp3+ cells (Supplementary Fig. 6f).
Figure 7. miR-210–deficiency along with reoxygenation conditions markedly increases TH17 differentiation.
(a) Naive CD4+ T cells from wild-type (Mir210+/+CD4-Cre), Mir210–/– (Mir210f/fCD4-Cre) or Hif1af/+Mir210f/fCD4-Cre mice were differentiated under TH17 skewing conditions (0.2 ng/ml of IL-6), followed by intracellular staining of IL-17A and IFN-γ. (b) Naive CD4+ T cells from wild-type (Mir210+/+CD4-Cre) or Mir210–/– (Mir210f/fCD4-Cre) mice were differentiated under TH17 skewing conditions with varying doses of IL-6 under normoxic or reoxygenetion conditions, followed by intracellular staining of IL-17A and IFN-γ. (c–d) Wild-type or Mir210–/– T cells were TH17 differentiated under reoxygenetion conditions as described in (a) for 3 d. Transcripts of miR-210 and Hif1a were detected by RT-PCR analysis; HIF-1α and GAPDH protein levels were detected by immunoblot analysis. (e–f) Quantitative RT-PCR analysis of HIF-1α target genes of the glycolytic pathway (Eno1, Glut1, Ldha, Pkm, Slc16a3 and Tpi) and TH17 signature genes (Il17a, Il17f, Rorc and Il23r) in wild-type or Mir210–/– CD4+ TH17 cells that were differentiated under reoxygenetion conditions. Relative expression is normalized to sno202 (miR-210) or β-2m (the rest of genes). Data are from one experiment representative of two (a right, d,e), three (c,f) or four (a left and middle, b) independent experiments. (mean and s.d. in b,c,e,f).
To substantiate the model that miR-210 attenuates TH17 differentiation by targeting Hif1a, we generated Mir210–/–Hif1af/+mice, and compared the TH17 polarization potential of their naive T cells in normoxia or under reoxygenation to naive T cells of wild-type or Mir210–/– mice (Fig. 7a). Corroborating our previous results, the enhanced TH17 cell differentiation under reoxygenation caused by miR-210–deficiency was considerably suppressed in Mir210–/– Hif1af/+ T cells, indicating that miR-210 regulates TH17 polarization mainly by targeting Hif1a.
miR-210 controls IBD severity
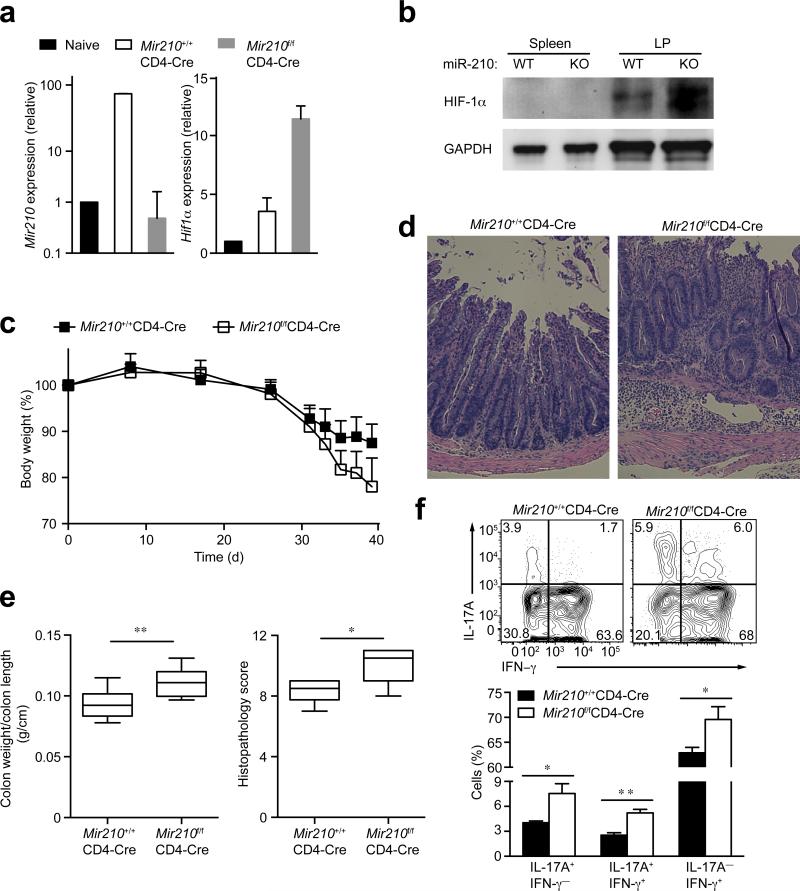
The gut is a hypoxic environment5 and contains a high proportion of IL-17A-producing cells15. To test whether miR-210 influences the differentiation of TH17 cells during an inflammatory bowel disease (IBD) model of colitis, we transferred naive, Treg-depleted (CD45.2+) T cells from wild-type or Mir210–/– mice into CD45.1+ Rag2−/− mice. Five weeks after T cell transfer, wild-type T cells from the lamina propria contained a marked increase in miR-210 abundance compared to naive T cells (Fig. 8a). Consistent with the reoxygenation in vitro model, miR-210–deficiency in vivo resulted in a markedly increased transcript and protein expression of HIF-1α in Mir210–/– intestinal T cells (Fig. 8a,b). In contrast, splenic T cells isolated from the same animals did not upregulate HIF-1α protein (Fig. 8b). Both groups, the CD45.2+ wild-type T cell-transferred control and the CD45.2+ Mir210–/– T cell-transferred group, started to lose body weight about 3-4 weeks after T cell transfer. However, mice reconstituted with Mir210–/– T cells exhibited substantially more weight loss, associated with increased mucosal leukocyte infiltration (Fig. 8c,d). Moreover, the Mir210–/– T cell-transferred group had higher weight/length ratios of the colon, which correlated with worse histopathological scores (Fig. 8e), evidence of more severe disease in the absence of miR-210. We measured the percentages of IL-17A-producing cells among CD4+ T cells, isolated from the lamina propria of the sick animals and observed a marked increase in the proportion of IL-17A+ T cells in Mir210–/– T cell-transferred mice, indicating that miR-210 controls TH17 differentiation in vivo (Fig. 8f). Furthermore, the proportion of IL-17A+ interferon-γ (IFN-γ+) double-positive cells and IFN-γ+ single-positive cells were increased, indicating that miR-210–deficiency may also facilitate the conversion of TH1-like cells from TH17 cells (Fig. 8f). Collectively, these results support an important role for miR-210 in controlling TH17 T cell differentiation and the development of immunopathology in vivo.
Figure 8. miR-210 in T cells ameliorates IBD disease in a CD4+ T cell transfer model of colitis.
(a) Wild-type (Mir210+/+CD4-Cre) or Mir210–/– (Mir210f/fCD4-Cre) naive T cells were transferred into Rag2−/− recipient mice. Five weeks later, the colons from recipient mice were excised and lamina propria CD4+ T cells were isolated from the colon. Naive CD4+ T cells were included as a control. miR-210 abundance and Hif1a transcripts were analyzed by RT-PCR analysis. (b) HIF-1α as well as GAPDH protein abundance in CD4+ T cells isolated from spleen or lamina propria of Rag2−/− recipient mice described in (a) was detected by immunoblot analysis. (c) Change in body weight of Rag2−/− recipient mice given wild-type or Mir210–/– naive T cells (n=9 per group). (d) Histopathology of distal colon at 5.5 weeks after transfer. Original magnification, X100. (e) Ratio of colon weight vs colon length (n=8–9 per group) and histopathological score (n=6–8 per group) of each group are shown. (f) Expression of IL-17A and IFN-γ in CD4+ T cells isolated from lamina propria (n=3–4 mice per group). * P<0.05, ** P<0.001, unpaired t-test. Data are from one experiment representative of two (a–f) independent experiments.
Discussion
In two independent global microRNA expression profiling studies in lymphocytes, miR-210 was shown to be induced in effector T cells25, 26. Our studies support and extend these findings by suggesting miR-210 plays a particularly important role in regulating the TH17 T cell lineage. We found that TCR stimulation led to a robust induction of Mir210 in T cells under normoxic conditions. Moreover, hypoxia-challenged and antigen-stimulated T cells exhibited a more rapid and increased induction of Mir210, suggesting that multiple signals are required for optimal Mir210 induction. Signals provided by antigen and costimulatory stimulation synergize with signals provided by the hypoxic environment. Immune cells encounter rapid changes in oxygen tension as they develop and migrate into different compartments of the body44. Therefore, a hypoxia-mediated signal to properly induce Mir210 is an intriguing and additional layer of regulation for microRNAs critical in the immune system. Mir210 is only fully induced when T cells are activated at inflammatory sites or in cancerous tissues, both of which are considered hypoxic45. What might be a possible biological function of this HIF-1α-induced microRNA under hypoxic conditions? miR-210 is not only upregulated by HIF-1α, but also negatively regulates Hif1a transcript and HIF-1α protein abundance in peripheral T cells, providing the first example of desensitizing HIF-1 activity by a microRNA in lymphocytes under hypoxic conditions.
In contrast to normoxia, hypoxic conditions inhibit HIF-1α degradation and lead to its stabilization8, 9. However, the mechanism by which HIF-1α activity is turned off to prevent its excessive activation in hypoxia was obscure. Two principal mechanisms have been proposed. First, although the hydroxylation activity of PHD is limited in hypoxia, the HIF-1α-driven increased expression of PHD2 and PHD3 under hypoxic conditions may compensate for the loss of their activities and consequently represents a negative-feedback loop targeting HIF-1α expression at the protein level46-48. Second, hypoxia-induced miR-155 directly targets Hif1a in intestinal epithelial cells, desensitizing HIF-1 activity at the transcript level10. However, we found that miR-155 is not a hypoxia-regulated microRNA in human or mouse T cells and suggest that the “hypoxamiR” miR-210 acts to specifically negatively regulate HIF-1α activity in T cells at later stages of activation.
Priming of naive T cells under O2-limiting (5%) conditions, significantly increases HIF-1α protein and thereby accelerates TH17 differentiation43. T cell-specific deletion of VHL, results in more HIF-1α and further increases TH17 differentiation under reoxygenation conditions43. We showed that HIF-1α-induced Mir210 is highly expressed in TH17 cells and that miR-210 directly suppresses Hif1a transcript abundance. Therefore, miR-210 appears to play a role in feedback inhibition to curb HIF-1 activity in T cells under hypoxia and consequently restraining TH17 cell differentiation and associated immunopathology. This is consistent with less Hif1a in response to ectopic expression of miR-210 and a marked upregulation of Hif1a mRNA and protein in activated, Mir210–/– T cells. This effect was most evident under reoxygenation conditions in vitro and in hypoxic environments in vivo suggesting that miR-210 plays its most important tuning role in low-oxygen conditions.
Consistent with many miRNA knockouts exhibiting phenotypes only under stress conditions49, Mir210–/– T cells revealed enhanced TH17 polarization under reoxygenation, but not normoxia. Low oxygen tension increases the described HIF-1α–miR-210 regulatory feedback loop in two ways. Firstly, hypoxia synergizes with TCR-CD28 stimulation to prompt more rapid and robust induction of Mir210, which secondly leads to a more profound negative regulation of Hif1a mRNA, resulting in curbed HIF-1α activity in hypoxia. Therefore, miR-210 is a crucial regulator of HIF-1 activity and TH17 differentiation in low-oxygen conditions, which is further corroborated by our IBD studies.
HIF-1α has been suggested to be a potential therapeutic target for the treatment of a variety of TH17 cell-mediated autoimmune diseases15. Our in vitro and in vivo studies demonstrate that miR-210 is a potent regulator of Hif1a mRNA and protein in T cells under hypoxic conditions. Therefore, targeting Hif1a by administrating miR-210 mimics in vivo might be an effective therapeutic strategy for TH17-dependent autoimmune diseases.
Online Methods
Reagents
Rabbit polyclonal antibody against HIF-1α and mouse monoclonal antibody against GAPDH (Abcam, clone 6C5) were purchased from Cayman Chemical and Abcam, respectively. Anti–IL-4 (clone 11B11), anti–IFN-γ (clone XMG1.2), anti-CD3ε (clone 2C11) and anti-CD28 (clone 37.51) were purchased from UCSF Cell Culture Facility. CP550 690 was a gift from J.J. O'Shea (NIH). GDC-0941 and MLN0128 were gifts from K. Shokat (UCSF). LY294002 and Rapamycin were purchased from Millipore. PMA and ionomycin were purchased from Sigma-Aldrich.
Mice
The Mir210 conditional knockout mice have been generated as described previously33. Briefly, Mir210f/f mice were generated by crossing miR-210 (lacZ-neor-flox) mice33 with the actin-Flp deleter mouse strain (Jax) to delete the lacZ–neomycin-resistance cassette, followed by backcrossing to the C57BL/6 (Jax) background for more than 9 generations. The C57BL/6 (Jax), Cd28−/− (Jax), BoyJ (Taconic), CD45.1+CD45.2–Rag2−/− (Taconic), CD4-Cre (Taconic), OTII (Taconic) and P14 TCR transgenic (Jax) mouse strains were purchased from the indicated vendors. Il2–/– D011.10 mice and Hif1af/f mice have been described previously50, 51. All mice were housed and bred in specific-pathogen-free conditions in the Animal Barrier Facility at the University of California, San Francisco. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of California, San Francisco.
RNA Analysis
For microRNA assays, total RNA was extracted with a mirVana miRNA Isolation Kit (Applied Biosystems) and then reverse transcribed with TaqMan Reverse Transcription reagents (Applied Biosystems). TaqMan miRNA assays were performed in an ABI Quantstudio machine (Applied Biosystems). miRNA assay results were normalized to the abundance of snoRNA202. The expression of each miRNA is presented as the fold change relative to its expression in wild-type naive T cells. For RT-PCR of non-miRNA genes, cDNA was synthesized with Superscript III/II reverse transcription (Invitrogen). The information of all primers is listed in Supplementary Table 2.
OVA Immunization and LCMV Infection
For the activation of CD4+ T cells, splenic and lymph node-derived naive T cells (CD4+CD62LhiCD44loCD25–) were sorted by flow cytometry from OT-II TCR-transgenic mice. After CFSE labeling, the sorted cells were transferred into congenic recipient mice. 24 h later, the recipients were immunized s.c. with the OVA323-339 peptide (100 μg/mouse Genscript) in the presence of Complete Freund's Adjuvant (CFA; DIFCO). Mice were sacrificed 7 d after immunization, and activated CD4+CFSElo T cells were isolated from the draining LN, followed by RNA isolation for miRNA expression analysis. For the in vivo activation of CD8+ T cells, P14 transgenic T cells were transferred into congenic recipient mice. The recipients were infected by lymphocytic choriomeningitis virus (LCMV). Naive (d 0), effector (d 8 post infection) and memory (d 202 post infection) LCMV-specific CD8+ T cells were sorted by flow cytometry, followed by RNA isolation for miRNA expression analysis.
In vitro T Cell Activation and Polarization Assays
Sorted naive T cells (CD4+CD62LhiCD44loCD25–) were used for in vitro culture in RPMI medium (plus β-mercaptoethanol) supplemented with 10% (vol/vol) FBS, 100 μM HEPES pH 7.5 and antibiotics. T cells were activated with plate-bound 2 μg/ml anti-CD3 (clone 2C11; UCSF Cell Culture Facility) and 2 μg/ml anti-CD28 (clone 37.51 UCSF Cell Culture Facility) for 4 d under conditions with various oxygen concentrations. For TH17 conditions, 10 μg/ml anti–IL-4, 10 μg/ml anti–IFN-γ, 2 ng/ml TGF-β, and 20, 2 or 0.2 ng/ml IL-6 were added to the cultures, and cells were stimulated for 3 d–5 d. Cells were then harvested and stimulated for a period of 5 h with PMA (50 ng/ml) and ionomycin (1 nM) in the presence of GolgiStop (BD-Biosciences), and the differentiated cells were analyzed by flow cytometry for cytokine production by intracellular staining using a LSRFortessa (BD-Biosciences). For the reoxygenation assay, T cells were firstly primed under a low O2 concentration (5% O2) for 36 h, and then the cells were transferred into a normoxic incubator for the rest of the culture.
Primary T cell Transfection, Luciferase Reporter Assays and Retroviral Transduction
The lucifierase reporter assays and T cell transfection of miRNA mimics have been described previously52. Briefly, the full-length 3′ UTR of Hif1a was cloned into the pMIR-REPORT™ luciferase reporter construct (Invitrogen). Isolated naive CD4+ T cells were stimulated with anti-CD3 and anti-CD28 for 4 d. The activated T cells were cotransfected with reporter constructs and miRNA mimics (Dharmacon) by the Neon electroporation transfection system (Invitrogen) with an optimized version of the manufactures’ recommended protocol. Luciferase activity was measured 24 h after transfection with the pMIR-REPORT™ miRNA Expression Reporter Vector System (Invitrogen) and a Mithras LB 940 (Berthold Technologies) platereader. Retroviral transduction was performed as described previously53.
Adoptive Transfer and Chronic Colitis model
The inflammatory bowel disease model has been described previously54. Briefly, 1 × 106 flow-sorted naive T cells (CD4+CD25–CD44–CD62L+) were adoptively transferred i.p. into 8–12 week-old CD45.1+CD45.2–Rag2−/− recipient mice (no specific randomization protocol was used). The body weight of each mouse was monitored after adoptive transfer. After mice were sacrificed, the colonic specimens taken from proximal and middle colons were subjected to histopathological assessment. Tissue samples were fixed in 10% neutral buffered formalin (pH7.0). Paraffin-embedded sections were stained with hematoxylin and eosin. These sections were analyzed and the severity of the inflammation was scored in a blinded manner. The degree of inflammation in the colon was graded according to a modification of the previously described scoring system54. Briefly, for mucosal damage, 0 points were assigned to normal appearance, 1 point to discrete lymphoepithelial lesions, 2 points to diffuse crypt elongation or goblet depletion, or 3 points to extensive crypt elongation, mucosal erosion/ulceration. For cell infiltration, 0 points were assigned for normal or for the presence of occasional leukocytes, 1 point for widely scattered leukocytes or focal aggregates of leukocytes, 2 points for the confluence of leukocytes extending into the submucosa with focal effacement of the muscularis, or 3 points for transmural extension of leukocyte infiltration. For crypt abscess, the assigned points were: 0 for no crypt abscess; 1 point for the presence of rare crypt abscess; or, 2 points for multiple crypt abscesses.
Statistical Analysis
Prism software was used for statistical analysis. Differences between groups were compared by unpaired two-tailed t-test. A P value of less than 0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank A. Roque for animal husbandry. We thank R. Locksley and Z. Wang for access and assistance with cell sorting; R. Wang (St Jude Children's Research Hospital) for providing organs from Hif1af/f Cd4-Cre+ mice; M. Matloubian for providing RNA from LCMV infected mice; A. Abbas for providing Il2–/– DO11.10 mice; K. Shokat (UCSF) and J. J. O'shea (US National Institutes of Health) for providing kinase inhibitors; K. Mark Ansel and L. Jeker for critical reading of the manuscript. H.W. is a recipient of an Arthritis Foundation Postdoctoral Fellowship and H.F. is a recipient of a DFG Fellowship. L.W. was supported by the National Basic Research Program of China (2013CB967002). This work was also supported in part by a grant from the Keck Foundation.
Footnotes
AUTHOR CONTRIBUTIONS
H.W. and H.F. planned and conducted experiments, analyzed and interpreted data and wrote the manuscript. M.O. performed histology analysis. L.W. contributed critical pre-publication data. M.T.M. provided guidance and direction and the Mir210 targeted mice. A.W. supervised the work, helped conceive the experiments, and edited the manuscript.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- 1.Caldwell CC, et al. Differential effects of physiologically relevant hypoxic conditions on T lymphocyte development and effector functions. J. Immunol. 2001;167:6140–6149. doi: 10.4049/jimmunol.167.11.6140. [DOI] [PubMed] [Google Scholar]
- 2.McNamee EN, Korns Johnson D, Homann D, Clambey ET. Hypoxia and hypoxia-inducible factors as regulators of T cell development, differentiation, and function. Immunol. Res. 2013;55:58–70. doi: 10.1007/s12026-012-8349-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye J. Emerging role of adipose tissue hypoxia in obesity and insulin resistance. Int. J. Obes. 2009;33:54–66. doi: 10.1038/ijo.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedogni B, et al. The hypoxic microenvironment of the skin contributes to Akt-mediated melanocyte transformation. Cancer Cell. 2005;8:443–454. doi: 10.1016/j.ccr.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Taylor CT, Colgan SP. Hypoxia and gastrointestinal disease. J. Mol. Med. 2007;85:1295–1300. doi: 10.1007/s00109-007-0277-z. [DOI] [PubMed] [Google Scholar]
- 6.Semenza GL. Hypoxia-inducible factor 1 (HIF-1) pathway. Sci. STKE. 20072007:cm8. doi: 10.1126/stke.4072007cm8. [DOI] [PubMed] [Google Scholar]
- 7.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 9.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 10.Bruning U, et al. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol. Cell. Biol. 2011;31:4087–4096. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu. Rev. Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 12.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu. Rev. Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 13.Fife BT, et al. Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nat. Immunol. 2009;10:1185–1192. doi: 10.1038/ni.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu S, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leung AK, Sharp PA. MicroRNA functions in stress responses. Mol. Cell. 2010;40:205–215. doi: 10.1016/j.molcel.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan YC, Banerjee J, Choi SY, Sen CK. miR-210: the master hypoxamir. Microcirculation. 2012;19:215–223. doi: 10.1111/j.1549-8719.2011.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Camps C, et al. hsa-miR-210 Is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin. Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 21.Giannakakis A, et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol. Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulshreshtha R, et al. A microRNA signature of hypoxia. Mol. Cell. Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010;10:111–122. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 24.Baumjohann D, Ansel KM. MicroRNA-mediated regulation of T helper cell differentiation and plasticity. Nat. Rev. Immunol. 2013;13:666–678. doi: 10.1038/nri3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuchen S, et al. Regulation of microRNA expression and abundance during lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chong MM, et al. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 28.Fraser JD, Irving BA, Crabtree GR, Weiss A. Regulation of interleukin-2 gene enhancer activity by the T cell accessory molecule CD28. Science. 1991;251:313–316. doi: 10.1126/science.1846244. [DOI] [PubMed] [Google Scholar]
- 29.Stittrich AB, et al. The microRNA miR-182 is induced by IL-2 and promotes clonal expansion of activated helper T lymphocytes. Nat. Immunol. 2010;11:1057–1062. doi: 10.1038/ni.1945. [DOI] [PubMed] [Google Scholar]
- 30.Kane LP, Andres PG, Howland KC, Abbas AK, Weiss A. Akt provides the CD28 costimulatory signal for up-regulation of IL-2 and IFN-gamma but not TH2 cytokines. Nat. Immunol. 2001;2:37–44. doi: 10.1038/83144. [DOI] [PubMed] [Google Scholar]
- 31.Huang X, et al. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park CY, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang X, El Naqa IM. Prediction of both conserved and nonconserved microRNA targets in animals. Bioinformatics. 2008;24:325–332. doi: 10.1093/bioinformatics/btm595. [DOI] [PubMed] [Google Scholar]
- 35.Krek A, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 36.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 38.Betel D, Koppal A, Agius P, Sander C, Leslie C. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 2010;11:R90. doi: 10.1186/gb-2010-11-8-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuchiya S, et al. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1). J. Biol. Chem. 2011;286:420–428. doi: 10.1074/jbc.M110.170852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Favaro E, et al. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loeb GB, et al. Transcriptome-wide miR-155 binding map reveals widespread noncanonical microRNA targeting. Mol. Cell. 2012;48:760–770. doi: 10.1016/j.molcel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ikejiri A, et al. Dynamic regulation of Th17 differentiation by oxygen concentrations. Int. Immunol. 2012;24:137–146. doi: 10.1093/intimm/dxr111. [DOI] [PubMed] [Google Scholar]
- 44.Westermann J, Bode U. Distribution of activated T cells migrating through the body: a matter of life and death. Immunol. Today. 1999;20:302–306. doi: 10.1016/s0167-5699(99)01474-7. [DOI] [PubMed] [Google Scholar]
- 45.Vaupel P, Thews O, Kelleher DK, Hoeckel M. Current status of knowledge and critical issues in tumor oxygenation. Results from 25 years research in tumor pathophysiology. Adv. Exp. Med. Biol. 1998;454:591–602. doi: 10.1007/978-1-4615-4863-8_70. [DOI] [PubMed] [Google Scholar]
- 46.Henze AT, et al. Prolyl hydroxylases 2 and 3 act in gliomas as protective negative feedback regulators of hypoxia-inducible factors. Cancer Res. 2010;70:357–366. doi: 10.1158/0008-5472.CAN-09-1876. [DOI] [PubMed] [Google Scholar]
- 47.Minamishima YA, et al. A feedback loop involving the Phd3 prolyl hydroxylase tunes the mammalian hypoxic response in vivo. Mol. Cell. Biol. 2009;29:5729–5741. doi: 10.1128/MCB.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stiehl DP, et al. Increased prolyl 4-hydroxylase domain proteins compensate for decreased oxygen levels. Evidence for an autoregulatory oxygen-sensing system. J. Biol. Chem. 2006;281:23482–23491. doi: 10.1074/jbc.M601719200. [DOI] [PubMed] [Google Scholar]
- 49.Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum. Mol. Genet. 2010;19:R169–175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoyer KK, Kuswanto WF, Gallo E, Abbas AK. Distinct roles of helper T-cell subsets in a systemic autoimmune disease. Blood. 2009;113:389–395. doi: 10.1182/blood-2008-04-153346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Steiner DF, et al. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H, et al. Tonic ubiquitylation controls T-cell receptor:CD3 complex expression during T-cell development. EMBO J. 2010;29:1285–1298. doi: 10.1038/emboj.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamaji O, et al. The development of colitogenic CD4(+) T cells is regulated by IL-7 in collaboration with NK cell function in a murine model of colitis. J. Immunol. 2012;188:2524–2536. doi: 10.4049/jimmunol.1100371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.