Abstract
The Kes1 OSBP (oxysterol-binding protein) is a key regulator of membrane trafficking through the TGN (trans-Golgi network) and endosomal membranes. We demonstrated recently that Kes1 acts as a sterol-regulated rheostat for TGN/endosomal phosphatidylinositol 4-phosphate signalling. Kes1 utilizes its dual lipid-binding activities to integrate endosomal lipid metabolism with TORC1 (target of rapamycin complex 1)-dependent proliferative pathways and transcriptional control of nutrient signalling.
Keywords: Golgi, Kes1, lipid-transfer protein, nuclear signalling, oxysterol-binding protein (OSBP), Sec14, target of rapamycin complex 1 (TORC1)
Introduction
Golgi and endosomal systems are highly dynamic organelles that undergo organized renewal and maturation [1]. These organelles are central sites of intracellular signalling and membrane sorting. The co-ordination of signalling and trafficking functions of the Golgi/endosomes is not understood, but could potentially act through a lipid signalling interface. Lipid-transfer proteins help to co-ordinate lipid metabolism with core protein components of the membrane trafficking machinery through the TGN (trans-Golgi network) and endosomal membranes [2–5]. Specifically, the yeast PtdIns/PtdCho (phosphatidylcholine)-transfer protein Sec14 and the OSBP (oxysterol-binding protein) family member Kes1/Osh4 regulate trafficking through the yeast TGN/endosomal system [2,3,6]. Sec14 acts as a coincidence sensor that couples PtdCho metabolism with production of PtdIns4P [7,8]. Kes1 dampens Sec14-regulated PtdIns4P signalling in the TGN/endosomes [9,10]. It remains unclear why cells utilize an opposing pair of lipid transfer proteins in the TGN/endosomes. In our studies, we have demonstrated that Kes1 uses its dual lipid-binding activities to integrate multiple aspects of lipid metabolism in late stages of the secretory pathway with TORC1 (target of rapamycin complex 1) and nutrient signalling in yeast [11]. In the present paper, we review the primary findings of that work.
Effects of sterol and lipid binding on Kes1 function
Kes1 is a PtdIns4P- and sterol-binding protein [9,12], and Kes1 localization to the TGN/endosomal membranes requires a pool of PtdIns4P derived from the phosphoinositide 4-kinase Pik1 [9,10]. However, how Kes1 co-ordinates its dual lipid-binding activities was previously unknown. Several studies suggest that Kes1 acts as a diffusible carrier for sterols at sites of membrane apposition where the diffusion space between sterol `donor' and `acceptor' membranes is limited [12–15]. To better study the effects of PtdIns4P and sterol binding on Kes1 function, alleles were generated to specifically inhibit binding of either lipid or to ablate binding to both. The kes1R236E,K242E,K243E triple mutant (kes13E) is defective for PtdIns4P binding, but not sterol binding [11]. This mutant is biologically inactive, in part because it cannot associate with the TGN/endosomal membranes [9]. Sterol-binding mutants of Kes1 were also generated [11,12,16–18]. The kes1Y97F and kes1T185V mutants prevent sterol binding by disrupting a critical network of ordered water molecules deep within the sterol-binding pocket. Kes1 is non-essential in vegetative cells; however, it inhibits cell growth when overexpressed in yeast cells [3,11,16]. Our studies show that expression of Kes1 and the sterol-binding mutants kes1Y97F and kes1T185V (kes1Y97F is reported to be non-functional [12]) inhibit cell growth, whereas the PtdIns4P-binding mutant kes13E has no effect on cell growth [11]. In fact, kes1Y97F and kes1T185V are even more toxic to yeast than is Kes1 itself. These findings contradict reports that ablation of sterol-binding activity results in simple inactivation of Kes1 [12,13].
That Kes1 sterol-binding mutants represent hyperactive forms of Kes1 is of interest in that kes1Y97F shows enhanced association with the TGN/endosomal membranes relative to Kes1 itself [11]. As with Kes1, the kes13E PtdIns4P-binding and TGN/endosome-targeting defects are sufficient to disrupt kes1Y97F super-recruitment to endosomal membranes and neutralize kes1Y97F-mediated growth inhibition. Together, these findings are inconsistent with models describing a physiologically significant role for Kes1 in non-vesicular sterol transfer. Rather, it is more likely that Kes1 functions as a sterol sensor on the TGN/endosomal membranes (see below).
Kes1 super-recruitment to the TGN/endosomal membrane impairs vesicular trafficking
Kes1 regulates Golgi-derived vesicular transport by clamping Pik1-generated Golgi PtdIns4P [9–11]. We and others have shown that increased levels of Kes1 impair trafficking through the TGN/endosomal system. Specifically, trafficking of CPY (carboxypeptidase Y), the v-SNARE (vesicular soluble N-ethylmaleimide-sensitive fusion protein-attachment protein receptor) Snc1, and the bulk endocytic tracer FM4-64 is perturbed by ectopic expression of Kes1 and the sterol-binding mutants kes1Y97F and kes1T185V [3,11,16]. We found that Kes1 sterol-binding mutants had greater inhibitory effects on CPY trafficking than did Kes1 itself [11].
These data suggest that Kes1 acts as a rheostat for TGN/endosomal PtdIns4P signalling whose activity depends on PtdIns4P binding to Kes1. Sterol binding to Kes1 promotes Kes1 release from PtdIns4P in the TGN/endosomal membrane (Figure 1). The dual-binding activity of Kes1 allows it to act as a sterol-regulated `brake' on PtdIns4P-dependent trafficking through the endosome. This is demonstrated by increased association of the sterol-binding mutant kes1Y97F to the TGN/endosomal membrane, ultimately resulting in inhibition of cell growth and diminished trafficking through TGN/endosomes. These data suggest that Kes1 is a sterol sensor in the TGN/endosomal system, rather than a sterol-transfer protein.
Figure 1. A PtdIns4P rheostat regulates Kes1 membrane association.
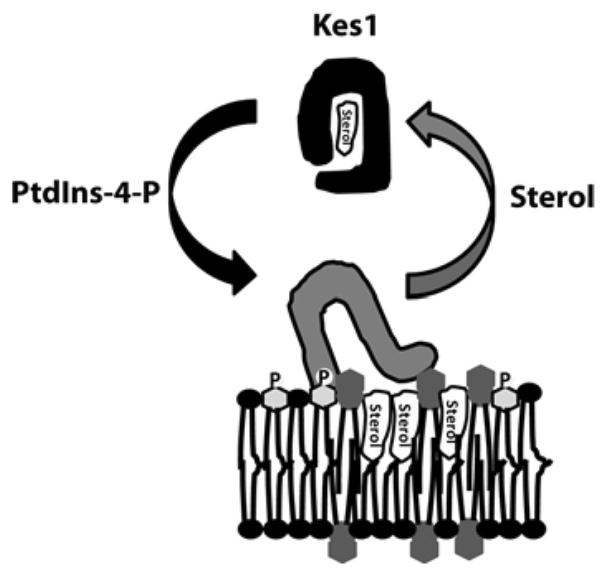
Kes1 is recruited to membranes by its ability to bind to PtdIns4P. Sterol binding at the TGN/endosome releases Kes1 from membranes.
Kes1 and nutrient signalling
Kes1 intoxication causes growth inhibition and defects in trafficking without significant loss of viability. In the following sections, we summarize the effects of Kes1/kes1Y97F overexpression on nutrient signalling.
Effects of Kes1 overexpression on autophagy
Although yeast expressing Kes1/kes1Y97F exhibit membrane trafficking defects through the TGN/endosomal system, thin-section electron microscopy analyses did not find the expected accumulation of aberrant toroid-shaped membrane systems, called Berkeley bodies, that are indicative of cargo-engorged secretory compartments [19,20]. Instead, we observed intravacuolar vesicles in Kes1- and kes1Y97F-intoxicated cells, suggesting that autophagic processes are induced in these cells despite their growth in nutrient-sufficient medium [11]. This was further confirmed using both an in vivo reporter assay for monitoring the autophagic-dependent processing of the otherwise cytosolic Pho8Δ60 alkaline phosphatase, as well as visualizing the mobilization of Atg18 from the cytosol to pre-autophagosomal compartments in Kes1- and kes1Y97F-overbearing cells [11]. Interestingly, an earlier preliminary study claimed that Kes1 inhibits autophagy [16], which we find is clearly not the case.
Kes1 and GAAC (general amino acid control) pathway
The onset of precocious autophagic responses diagnoses nutrient-sensing defects in cells with elevated Kes1/kes1Y97F. Metabolomic profiling found a decrease in intracellular amino acid pools in Kes1/kes1Y97F yeast [11]. The most prominent decreases were of arginine, asparagine, aspartate, glutamate, glutamine, threonine and tryptophan. We found that spiking the medium with a concentrated NEQR (asparagine/glutamate/glutamine/arginine) cocktail rescued growth of Kes1/kes1Y97F-arrested cells. However, the NEQR cocktail did not rescue defects in TGN/endosomal trafficking, suggesting that Kes1 intoxication primarily result from nutrient-sensing defects rather than the membrane trafficking defects alone.
Supplementation with asparagine was found to be necessary, although not sufficient, to alleviate Kes1-dependent growth defects. We infer from this asparagine requirement that ammonium (NH4+) starvation is the core nutritional insult to Kes1-intoxicated cells. This is supported by the observations that (i) the expression of genes encoding the ammonium permeases MEP1 and MEP2 is greatly diminished in Kes1/kes1Y97F-arrested cells, and (ii) ASP1 deletion prevents the NEQR cocktail from rescuing growth inhibition of Kes1-intoxicated cells. ASP1 encodes an asparaginase which produces glutamine by transferring an amino group from asparagine to glutamate. Glutamine acts as a biosynthetic NH4+ pool and has other roles in amino acid uptake, TOR (target of rapamycin) activation and maintenance of mitochondrial integrity [21]. Interestingly, supplementation of glutamine alone cannot rescue the Kes1/kes1Y97F growth defect, suggesting that asparagine may have another role. It was shown previously that NH4+ starvation blocks translation of GCN4 mRNA in an eIF2α (eukaryotic initiation factor 2α)-independent manner, attenuating the GAAC pathway [22]. Kes1 intoxication recapitulates this effect. The mammalian GCN (general control non-derepressible) 2/ATF4 (activating transcription factor 4) (homologous with yeast Gcn2/Gcn4) pathway specifically requires asparagine for tumour cell survival [23].
We also found defects in TORC1 signalling in Kes1/kes1Y97F cells. Amino acids promote TORC1 signalling by modulating activity of the Rag GTPases Gtr1 and Gtr2 [24–27]. Neither casamino acids nor the NEQR cocktail can rescue growth of Kes1-intoxicated gtr1Δ or gtr2Δ cells [11]. This suggests that the NEQR cocktail rescues growth of Kes1-intoxicated cells by restoring Gtr-mediated activation of TORC1.
The GAAC pathway is activated by Gcn2-mediated phosphorylation of eIF2α which decreases eIF2α activity and specifically promotes translation of the Gcn4 transcription factor open reading frame [28,29]. Kes1/kes1Y97F cells exhibit enhanced levels of phospho-eIF2α, but do not induce transcription of the Gcn4-dependent target genes HIS4 and ARG1 [11,30]. We found that induction of the GAAC fails at two distinct points upon Kes1 intoxication: (i) the amount of Gcn4 translated is reduced, resulting in less Gcn4 protein, and (ii) even when Gcn4 is constitutively expressed, Gcn4 remains non-functional. Overall, we found that defects in Gcn4 transcriptional activation cause GAAC failure in Kes1/kes1Y97F-intoxicated cells.
Effects of sphingolipid homoeostasis and large SRB (suppressor of RNA polymerase B) Mediator on the GAAC
Lipidomic analyses of Kes1/kes1Y97F-intoxicated cells demonstrated bulk ceramide, sphingoid base and sphingoid base phosphate levels to be elevated. These data suggest that Kes1 activity directly impinges on sphingolipid homoeostasis. Might defects in sphingolipid homoeostasis contribute to defects in Gcn4-dependent GAAC activation in Kes1/kes1Y97F cells? If so, sphingolipid derangements evoked by independent means should recapitulate Kes1/kes1Y97F intoxication. The growth of wild-type yeast challenged with sub-growth-inhibitory levels of PHS (phytosphingosine) or with 3-AT (3-aminotriazole) treatment (histidine starvation) to induce the GAAC pathway was unaffected. However, if cells were dually challenged, cell growth was inhibited and cells lost Gcn4-dependent induction of target genes, recapitulating the results observed in Kes1/kes1Y97F cells. These data indicate that activity of the GAAC pathway is modulated by sphingolipid metabolism; the disruption of normal sphingolipid metabolism (presumably in the TGN/endosomal membranes) initiates signalling events that are interpreted in the nucleus and antagonize Gcn4 activity.
We have identified that both Kes1/kes1Y97F-dependent and PHS-dependent attenuation of Gcn4 transcriptional activation of GAAC target genes requires the CDK8 (cyclin-dependent kinase 8) module of the Mediator complex. Previous studies show that the CDK8 module of Mediator inhibits RNA polymerase II-dependent transcription initiation and elongation [31]. Since PHS-treated cells fail to induce Gcn4-dependent target genes, we asked whether Gcn4 can enter the nucleus and bind to enhancers in PHS-treated cells. ChIP (chromatin immunoprecipitation) showed enhanced binding of Gcn4 to the ARG1 UAS (upstream activating sequence) in cells treated with 3-AT and only a slight reduction (~20%) in the amount of Gcn4 bound to ARG1 UAS in PHS-treated cells. The ChIP data suggest that yet another level of control must exist downstream of pre-initiation complex assembly to account for inhibition of ARG1 transcription in response to dual 3-AT and PHS treatment of cells.
The identities of the nuclear effectors remain unknown. The catalytic subunit of the CDK8 module (Srb10) may be activated by sphingolipids at specific (Gcn4-dependent?) enhancers. Alternatively, epigenetic effects may also be involved. Recently, mammalian HDACs (histone deacetylases) were shown to be inhibited by sphingosine 1-phosphate [32]. Perhaps yeast HDACs or histone acetyltransferases are direct targets of sphingolipid-mediated regulation in this Kes1-dependent signalling pathway.
Future perspectives
We have identified a novel signal transduction pathway that is relayed from distal compartments of the secretory pathway to the nucleus where the OSBP homologue Kes1 inhibits Gcn4 activity by coupling lipid metabolism with nutrient signalling in yeast (Figure 2). Collectively, these events down-regulate TOR signalling and subsequently halt cell-cycle progression. The executioners of this pathway are highly conserved. In mammals, Kes1-like ORPs (OSBP-related proteins) are most highly expressed in terminally differentiated non-mitotic tissues, such as skeletal muscle, heart and brain [33]. We speculate that this modality helps to guide cell entry into post-mitotic states or maintains post-mitotic cell physiology. A model where Kes1 co-ordinates Golgi/endosome function with proliferative signalling and cell-cycle control raises interesting possibilities for these proteins in differentiation and organogenesis.
Figure 2. Integration of TGN/endosomal trafficking disruption to transcriptional competence of the GAAC pathway.
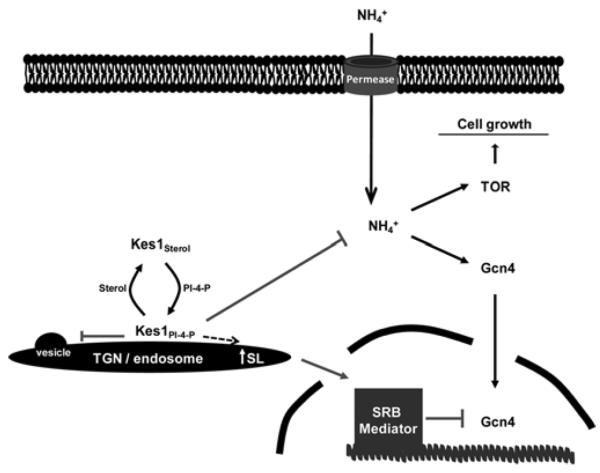
Kes1-mediated trafficking arrest leads to manipulation of TGN/endosomal sphingolipid homoeostasis which in turn controls cell proliferation, TOR signalling and NH4+ availability. In addition, the sphingolipid signal inhibits transcription initiation/elongation of genes loaded with Gcn4 and RNA polymerase II via action of the CDK8 module of the large SRB Mediator complex. PI-4-P, PtdIns4P.
That Kes1 antagonizes ATF4-dependent transcriptional programmes may have major implications in cancer biology. The mammalian GCN2/ATF4 (homologous with yeast Gcn2/Gcn4) signalling pathway is a highly conserved nutrient, oxidative and hypoxia stress pathway important for tumour survival. Recently, it was demonstrated that activation of the GCN2/ATF4 pathway promotes tumour cell survival and proliferation under amino acid starvation conditions [23]. ATF4 is required to maintain amino acid metabolism in tumour cells [23], and ORPs are implicated in tumour metastasis and cell-cycle progression [34,35]. An association between increased ORP4L (ORP4 long) expression and metastatic cancers is shown. Increased ORP4L mRNA is observed in several cancer cell lines, including cervical-cancer-derived cell lines (HeLa, C33A and SiHa) and a prostate cancer cell line (LNCaP), as well as in breast cancer tissues and lung cancer blood samples [35]. There are 19 human ORPs (encoded by 12 ORP genes) compared with seven ORPs in yeast, all of which bind sterols [36,37]. We proffer what we believe to be an attractive hypothesis that Kes1-like ORPs antagonize ATF4 activity in mammals, and that enhancing ORP activity in tumours will inhibit their vigour.
Acknowledgments
Funding The primary work summarized in the present paper was supported by the National Institutes of Health [grant number GM44530 (to V.A.B.)]. M.L.V. is supported by the National Institutes of Health [training grant number T32 CA009156-37].
Abbreviations used
- 3-AT
3-aminotriazole
- ATF4
activating transcription factor 4
- CDK8
cyclin-dependent kinase 8
- ChIP
chromatin immunoprecipitation
- CPY
carboxypeptidase Y
- eIF2α
eukaryotic initiation factor 2α
- GAAC
general amino acid control
- GCN
general control non-derepressible
- HDAC
histone deacetylase
- OSBP
oxysterol-binding protein
- ORP
OSBP-related protein
- PHS
phytosphingosine
- PtdCho
phosphatidylcholine
- SRB
suppressor of RNA polymerase B
- TGN
trans-Golgi network
- TOR
target of rapamycin
- TORC1
target of rapamycin complex 1
- UAS
upstream activating sequence
References
- 1.Glick BS, Nakano A. Membrane traffic within the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 2009;25:113–132. doi: 10.1146/annurev.cellbio.24.110707.175421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bankaitis VA, Aitken JR, Cleves AE, Dowhan W. An essential role for a phospholipid transfer protein in yeast Golgi function. Nature. 1990;347:561–562. doi: 10.1038/347561a0. [DOI] [PubMed] [Google Scholar]
- 3.Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MK, Bankaitis VA. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 4.Bard F, Malhotra V. The formation of TGN-to-plasma membrane transport carriers. Annu. Rev. Cell Dev. Biol. 2006;22:439–455. doi: 10.1146/annurev.cellbio.21.012704.133126. [DOI] [PubMed] [Google Scholar]
- 5.Graham TR, Burd CG. Coordination of Golgi functions by phosphatidylinositol 4-kinases. Trends Cell Biol. 2011;21:113–121. doi: 10.1016/j.tcb.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cleves AE, McGee TP, Whitters EA, Champion KM, Aitken JR, Dowhan W, Goebl M, Bankaitis VA. Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell. 1991;64:789–800. doi: 10.1016/0092-8674(91)90508-v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaaf G, Ortlund EA, Tyeryar KR, Mousley CJ, Ile KE, Garrett TA, Ren J, Woolls MJ, Raetz CR, Redinbo MR, Bankaitis VA. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 2008;29:191–206. doi: 10.1016/j.molcel.2007.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bankaitis VA, Mousley CJ, Schaaf G. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem. Sci. 2010;35:150–160. doi: 10.1016/j.tibs.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Rivas MP, Fang M, Marchena J, Mehrotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fairn GD, Curwin AJ, Stefan CJ, McMaster CR. The oxysterol binding protein Kes1p regulates Golgi apparatus phosphatidylinositol 4-phosphate function. Proc. Natl. Acad. Sci. U.S.A. 2007;104:15352–15357. doi: 10.1073/pnas.0705571104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mousley CJ, Yuan P, Gaur NA, Trettin KD, Nile AH, Deminoff SJ, Dewar BJ, Wolpert M, Macdonald JM, Herman PK, et al. A sterol binding protein integrates endosomal lipid metabolism with TOR signaling and nitrogen sensing. Cell. 2011 doi: 10.1016/j.cell.2011.12.026. doi:10.1016/j.cell.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raychaudhuri S, Im YJ, Hurley JH, Prinz WA. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J. Cell Biol. 2006;173:107–119. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J. Cell Biol. 2009;187:889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz TA, Prinz WA. Sterol transport in yeast and the oxysterol binding protein homologue (OSH) family. Biochim. Biophys. Acta. 2009;1771:769–780. doi: 10.1016/j.bbalip.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeBlanc MA, McMaster CR. Lipid binding requirements for oxysterol-binding protein Kes1 inhibition of autophagy and endosome-trans-Golgi trafficking pathways. J. Biol. Chem. 2010;285:33875–33884. doi: 10.1074/jbc.M110.147264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Canagarajah BJ, Hummer G, Prinz WA, Hurley JH. Dynamics of cholesterol exchange in the oxysterol binding protein family. J. Mol. Biol. 2008;378:737–748. doi: 10.1016/j.jmb.2008.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh RP, Brooks BR, Klauda JB. Binding and release of cholesterol in the Osh4 protein of yeast. Proteins. 2009;75:468–477. doi: 10.1002/prot.22263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novick P, Fields C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- 20.Esmon B, Novick P, Schekman R. Compartmentalized assembly of oligosaccharides on exported glycoproteins in yeast. Cell. 1981;25:451–460. doi: 10.1016/0092-8674(81)90063-5. [DOI] [PubMed] [Google Scholar]
- 21.Wise DR, Thompson CB. Glutamine addiction: a new therapeutic target in cancer. Trends Biochem. Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grundmann O, Mösch HU, Braus GH. Repression of GCN4 mRNA translation by nitrogen starvation in Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:25661–25671. doi: 10.1074/jbc.M101068200. [DOI] [PubMed] [Google Scholar]
- 23.Ye J, Kumanova M, Hart LS, Sloane K, Zhang H, De Panis DN, Bobrovnikova-Marjon E, Diehl JA, Ron D, Koumenis C. The GCN2-ATF4 pathway is critical for tumour cell survival and proliferation in response to nutrient deprivation. EMBO J. 2010;29:2082–2096. doi: 10.1038/emboj.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J. Cell Sci. 1998;111:11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- 25.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 26.Nakashima N, Noguchi E, Nishimoto T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics. 1999;152:853–867. doi: 10.1093/genetics/152.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binda M, Peli-Gulli MP, Bonfils G, Panchaud N, Urban J, Sturgill TW, Loewith R, De Virgilio C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell. 2009;35:563–573. doi: 10.1016/j.molcel.2009.06.033. [DOI] [PubMed] [Google Scholar]
- 28.Hinnebusch J, Tilly K. Linear plasmids and chromosomes in bacteria. Mol. Microbiol. 1993;10:917–922. doi: 10.1111/j.1365-2958.1993.tb00963.x. [DOI] [PubMed] [Google Scholar]
- 29.Hinnebusch AG. Translational regulation of yeast Gcn4: a window on factors that control initiator t-RNA binding to the ribosome. J. Biol. Chem. 1997;272:21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 30.Natarajan K, Meyer MR, Jackson BM, Slade D, Roberts C, Hinnebusch AG, Marton MJ. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol. Cell. Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem. Sci. 2010;35:315–322. doi: 10.1016/j.tibs.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hait NC, Allegood J, Maceyka M, Strub GM, Harikumar KB, Singh SK, Luo C, Marmorstein R, Kordula T, Milstien S, Spiegel S. Regulation of histone acetylation in the nucleus by sphingosine 1-phosphate. Science. 2009;325:1254–1257. doi: 10.1126/science.1176709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson M, Bocher V, Lehto M, Chinetti G, Kuismanen E, Ehnholm C, Staels B, Olkkonen VM. The two variants of oxysterol binding protein-related protein-1 display different tissue expression patterns, have different intracellular localization, and are functionally distinct. Mol. Biol. Cell. 2003;14:903–915. doi: 10.1091/mbc.E02-08-0459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alphey L, Jimenez J, Glover D. A Drosophila homologue of oxysterol binding protein (OSBP): implications for the role of OSBP. Biochim. Biophys. Acta. 1998;1395:159–164. doi: 10.1016/s0167-4781(97)00159-0. [DOI] [PubMed] [Google Scholar]
- 35.Fournier MV, Guimarães da Costa F, Paschoal MEM, Ronco LV, Carvalho MG, Pardee AB. Identification of a gene encoding a human oxysterol-binding protein-homologue: a potential general molecular marker for blood dissemination of solid tumors. Cancer Res. 1999;59:3748–3753. [PubMed] [Google Scholar]
- 36.Fairn GD, McMaster CR. Emerging roles of the oxysterol-binding protein family in metabolism, transport, and signaling. Cell. Mol. Life Sci. 2008;65:228–236. doi: 10.1007/s00018-007-7325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olkkonen VM, Levine TP. Oxysterol binding proteins: in more than one place at one time? Biochem. Cell Biol. 2004;82:87–98. doi: 10.1139/o03-088. [DOI] [PubMed] [Google Scholar]


