SUMMARY
The vast majority of currently licensed human vaccines work on the basis of long-term protective antibody responses. It is now conceivable that an antibody-dependent HIV vaccine might be possible, given the discovery of HIV broadly neutralizing antibodies (bnAbs) in some HIV-infected individuals. However, these antibodies are difficult to develop and have characteristics indicative of a high degree of affinity maturation in germinal centers (GCs). CD4+ T follicular helper (Tfh) cells are specialized for B cell help and necessary for GCs. Therefore, the development of HIV bnAbs might depend on Tfh cells. Here, we identified in normal individuals a subpopulation of circulating memory PD-1+CXCR5+ CD4+ T cells that are resting memory cells most related to bona fide GC Tfh cells by gene expression profile, cytokine profile, and functional properties. Importantly, the frequency of these cells correlated with the development of bnAbs against HIV in a large cohort of HIV+ individuals.
INTRODUCTION
Most human vaccines currently in use rely on the generation of protective, long-lasting antibody responses (Plotkin, 2010). T follicular helper (Tfh) cells are CD4+ T cells specialized in providing help to B cells, particularly within germinal centers (GCs), which are distinct structures in secondary lymphoid organs. Tfh cells support B cell differentiation into affinity-matured long-lived plasma cells and memory B cells by colocalizing with B cells and delivering signals via costimulatory molecules and lymphokines (CD40L, interleukin-21 [IL-21], IL-4, and CXCL13) that constitute the functional signature of this specific CD4+ T cell subset (Crotty, 2011). Furthermore, Tfh cells are needed for the crucial affinity-maturation process of B cells in GCs, whereby antigen-specific B cells undergo repeated rounds of somatic hypermutation and positive selection by Tfh cells to rapidly evolve high-affinity somatically mutated B cell receptors (BCRs) (Crotty, 2011; Victora and Nussenzweig, 2012); this results in the development of memory B cells and plasma cells with greater protective efficacy. In addition to being necessary for GCs, Tfh cells are also frequently limiting for the magnitude of GCs and antibody responses (Johnston et al., 2009; Rolf et al., 2010; Victora et al., 2010). Therefore, there is widespread interest in manipulating Tfh cells for vaccine enhancement.
Because of their necessary role in the generation of protective T-cell-dependent antibody responses, there is substantial potential for an understanding of Tfh cells to facilitate better long-term antibody responses for vaccines. One case of great importance is the generation of HIV broadly neutralizing antibodies (bnAbs) in humans. Seminal studies in the past few years have shown that 5% or more of HIV+ individuals are able to develop highly potent bnAbs (Kwong and Mascola, 2012). HIV bnAbs that can neutralize 70% or more of globally circulating HIV strains (Huang et al., 2012; Scheid et al., 2011; Walker et al., 2011; 2009) and can prevent infection in passive-transfer experiments using non-human primates (Moldt et al., 2012) have been characterized. Therefore, a vaccine eliciting such antibodies might have the ability to protect immunized individuals from HIV infection (Burton et al., 2012; McMichael and Haynes, 2012). Although this is an extremely important and interesting potential HIV vaccine strategy, little is known about the cellular mechanisms involved in generating HIV bnAbs. One hypothesis is that Tfh cells are important for the development of HIV bnAbs because of the extensive somatic hypermutation observed in the vast majority of HIV bnAbs (Streeck et al., 2013).
Here, we describe a subset of blood-circulating memory CXCR5+CD4+ T cells that are characterized by stable and moderate expression of the Tfh cell marker PD-1 (PD-1+CXCR5+ cells) and that most resemble GC Tfh cells among resting memory CD4+ T cells in terms of B cell help functionality and transcriptional signature. Strikingly, a highly functional PD-1+ CXCR3−CXCR5+CD4+ T cell population is overrepresented in rare individuals who generate bnAbs against HIV.
RESULTS
Total CXCR5+CD4+ T Cells in Blood Fail to Correlate with bnAb Production in HIV+ Donors
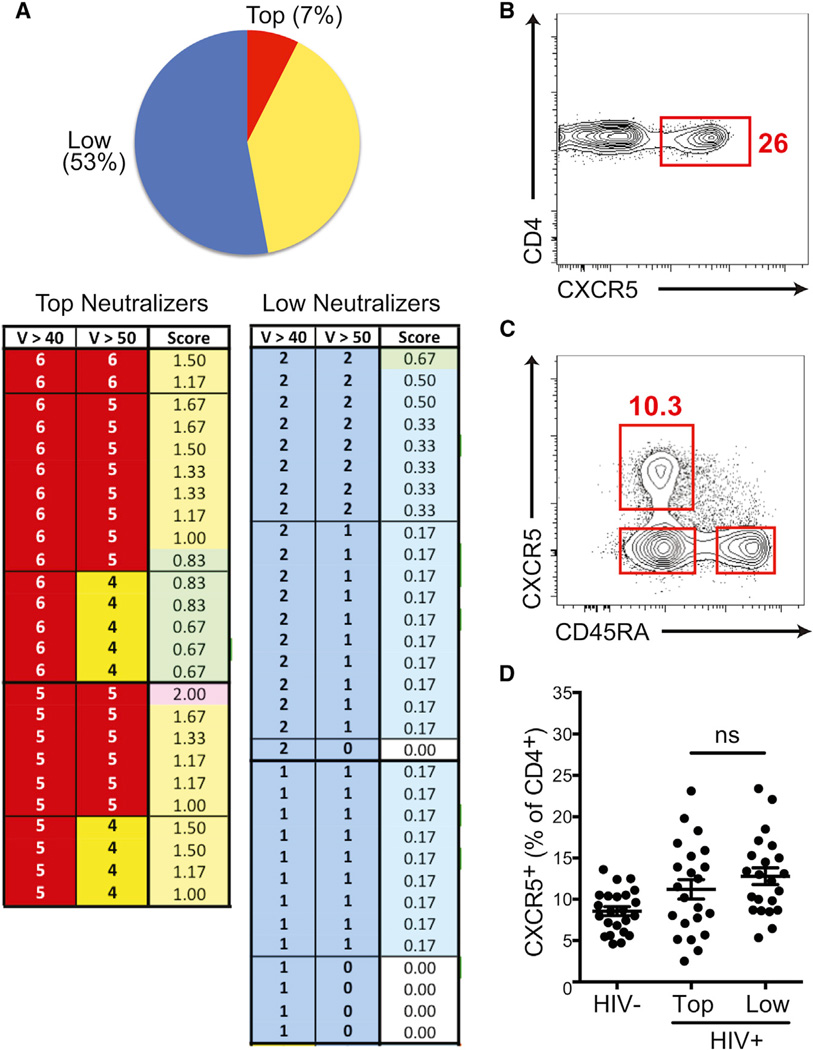
Among HIV+ individuals, only a minority is able to develop highly potent bnAbs against HIV. Importantly, those bnAbs take multiple years to develop, and virtually all have evidence of extensive affinity maturation in GCs, as indicated by the very high levels of somatic hypermutation in the immunoglobulin genes encoding those BCRs (Klein et al., 2013; Liao et al., 2013; Streeck et al., 2013). Given that Tfh cells can be a limiting factor for B cell responses (Johnston et al., 2009; Victora et al., 2010) and that Tfh cells are associated with better antibody responses to simian immunodeficiency virus (Petrovas et al., 2012) and chronic viral infections in mice (Boettler et al., 2012; Fahey et al., 2011; Harker et al., 2011), the HIV+ individuals who make bnAbs might have better Tfh cell responses. Testing this hypothesis required screening a large cohort of HIV+ individuals. The International AIDS Vaccine Initiative (IAVI) Protocol C program has continuously screened a large number of HIV− individuals across nine sites in Africa over a period of more than 7 years in order to enroll individuals into Protocol C shortly after they become HIV+ (blood draws are initially scheduled every 3 months), thus allowing for longitudinal study of the immune responses to HIV starting from early time points after infection. A total of 615 individuals were enrolled at the time of the start of this study for the examination of mechanisms of bnAb development. A total of 1,757 serum samples from 328 HIV+ individuals, who had each been tracked for more than 4 years, were tested for HIV neutralizing antibodies in over 42,000 individual HIV neutralization assays with the use of a high-throughput neutralization assay against a panel of six cross-clade viruses specifically selected to represent a larger neutralization panel (Simek et al., 2009). The majority of HIV+ individuals had antibody responses of low potency and breadth by the neutralization criteria used (“low”), as expected (Figure 1A). The top 25 neutralizing-antibody-scoring individuals (7%) were classified as the top neutralizers (“top”) (Figure 1A). Top neutralizers identified from the 6-virus cross-clade panel were subsequently confirmed with a 36-virus cross-clade panel and showed a good correlation (E.L. and P.P., unpublished data).
Figure 1. Blood Total CXCR5+CD4+ T Cells Fail to Correlate with HIV bnAb Development.
(A) Categorization of 328 HIV+ individuals enrolled in the IAVI Protocol C cohort by HIV-neutralizing ability of serum (upper panel: top neutralizers, red; intermediate neutralizers, yellow; and low neutralizers, blue). The number of HIV viruses neutralized, of a cross-clade panel (of six total viruses), at either >40% or >50% at a 1/100 serum dilution for top and low neutralizing groups is shown for each individual donor. “Score” indicates the overall bnAb score (lower panel). “V” stands for virus neutralization.
(B) Representative frequency of blood CD4+CXCR5+ cells (gated on CD45RO+CD4+ T cells).
(C) A representative flow cytometry plot of an HIV+ donor shows the gate for total CXCR5+CD4+ T cells (gated on CD3+CD4+ live cells).
(D) The percentage of CXCR5+CD4+ T cells in HIV− and HIV+ groups. Blood samples were from the earliest available time point (~4 months) after HIV infection, years before the development of bnAbs. “Top” indicates the top neutralizers, and “low” indicates the low neutralizers. Each point represents an individual donor. Mean and SEM are shown for each group. “ns” stands for not significant.
Also see Figure S1.
Fifteen to twenty-five percent of circulating antigen-experienced (CD45RO+) CD4+ T cells express CXCR5 (Figure 1B)(Breitfeld et al., 2000), the primary chemokine receptor expressed by Tfh cells. CXCR5+ cells can migrate to B cell follicles because of the CXCL13 expression within B cell follicles (Vinuesa and Cyster, 2011). We hypothesized that the frequency of circulating CXCR5+CD4+ T cells might predict subsequent HIV bnAb development in HIV+ individuals. Therefore, upon identifying the top neutralizers at ≥3.5 years postinfection, we examined the earliest available blood sample from each of the top neutralizers (4 months postinfection on average) and asked whether the frequency of total CXCR5+CD4+ T cells at this time point was predictive of the ability of some individuals to later develop bnAbs against HIV. There was no correlation between the capacity to develop bnAbs against HIV and total CXCR5+ CD4+ T cell percentage (top versus low, p = 0.29 [Figures 1C and 1D]). Similarly, no correlation was found at the time of bnAb generation, ~40 months postinfection (Figure S1A, available online). However, it is unclear what the biology of total CXCR5+CD4+ T cells in blood represents, given that these cells have limited similarity to Tfh cells (Streeck et al., 2013). This issue is explored in greater detail later in this study.
The vast majority of CXCR5+CD4+ T cells in blood are resting cells. However, a very small population of activated cells (median 0.33% of CXCR5+CD4+ T cells in healthy donors) that express ICOS and very high levels of PD-1 (PD-1+++) (Simpson et al., 2010; Figure S1B) are also present. This phenotype is similar to that of GC Tfh cells, which are ICOS+PD-1+++ (Crotty, 2011; Ma et al., 2009; Rasheed et al., 2006). The majority of the blood ICOS+PD-1+++CXCR5+CD4+ T cells were Ki67+ (Figure S1E). Although it has been suggested that these cells are circulating activated Tfh cells, they do not express elevated amounts of Bcl6 or Maf (Simpson et al., 2010; data not shown), which are central Tfh cell transcription factors required for Tfh cell differentiation and function (Crotty, 2011). As an additional confounding factor, it is reasonable to expect that some fraction of the ICOS+ PD-1+++CXCR5+ cells are not related to Tfh cells given that human CD4+ T cells readily express moderate levels of many chemokine receptors, including CXCR5, as activation markers for several days after T cell receptor (TCR) stimulation (Langenkamp et al., 2003). Currently there is not a definitive phenotypic marker that resolves this ambiguity. Nevertheless, we examined whether these activated ICOS+PD-1+++ cells were associated with HIV bnAb development. ICOS+PD-1+++CXCR5+CD4+ T cell frequencies in the top and low neutralizer groups were not statistically significant at the ~4-month time point (Figure S1C) or at the time of bnAb generation (Figure S1D). HIV-specific CD4+ T cells were quantitated by intracellular cytokine staining after short peptide stimulation. Whereas interferon-g [IFN-γ]+CXCR5− HIV-specific CD4+ T cells were detectable in many of the HIV+ individuals (25/60), no HIV-specific CXCR5+ CD4+ T cells were detected (data not shown). This might be due to assay limitations, given that even in mice, it is difficult to induce antigen-specific Tfh cells to secrete cytokines in conventional short-term peptide-stimulation assays. Alternatively, it is plausible that HIV-specific Tfh cells are not present in blood but remain sequestered in lymphoid tissue B cell follicles during the ongoing viral infection while Th1 cells recirculate and patrol infected tissues.
The failure of total CXCR5+CD4+ T cells and ICOS+PD-1+++ CXCR5+CD4+ T cells to correlate with the ability to generate HIV bnAbs might primarily speak to our lack of understanding of the relationship between blood CXCR5+CD4+ T cells and lymphoid Tfh cells. These shortcomings in our understanding impelled us to examine human blood CXCR5+CD4+ T cells in greater depth to determine whether a proportion of quiescent CXCR5+CD4+ T cells in blood has a clearer relationship to Tfh cells. Despite some functional similarities between circulating CXCR5+CD45RO+CD4+ T cells and Tfh cells (Chevalier et al., 2011; Morita et al., 2011), the transcriptional profile of the bulk CXCR5+CD4+ T cell population is substantially divergent from that of active GC Tfh cells (Chevalier et al., 2011; Chtanova et al., 2004; Rasheed et al., 2006). In addition, the functional capacity of total blood CXCR5+CD4+ T cells to help B cells in vitro is generally not much different from that of CXCR5− blood cells (Chevalier et al., 2011; Pallikkuth et al., 2012; Streeck et al., 2013). Finally, it is not clear whether any circulating CXCR5+ cells are memory Tfh cells, given that there are limited data demonstrating resting antigen-specific CXCR5+CD4+ T cells in humans. In sum, the overall biology of circulating CXCR5+CD4+ T cells is still enigmatic, and many observers have concluded that human Tfh memory cells most likely do not exist (Craft, 2012; Ma et al., 2012). We endeavored to clarify these issues.
Human Blood PD-1+CXCR5+ Quiescent CD4+ T Cells
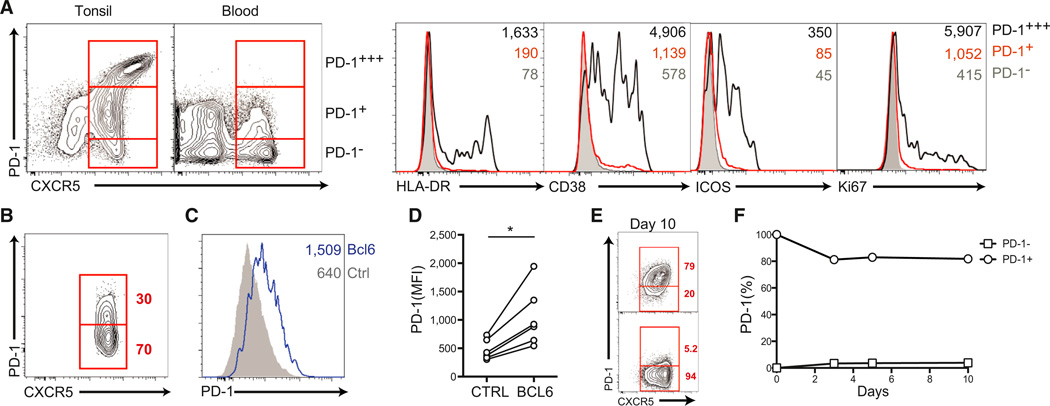
To develop a deeper understanding of human blood CXCR5+ cells and their relationship to memory, we undertook a detailed phenotypic characterization of these cells. In comparison to GC Tfh cells, human blood CXCR5+CD4+ T cells expressed intermediate levels of CXCR5 (Figure 2A). The majority of blood CXCR5+CD4+ T cells were PD-1− (Figure 2A). As noted above, a very small population of CXCR5+ cells (~0.3%) were ICOS+ and highly expressed PD-1 (Figure S1B), at levels similar to those of GC Tfh cells (PD-1+++). Unexpectedly, we found that a substantial population (~30%) of CXCR5+ cells expressed PD-1 at low to moderate levels (PD-1+ [Figures 2A and 2B]). Unlike the very small population of ICOS+PD-1+++ activated cells, these cells were Ki67− and ICOS− (Figure 2A). Herein, we refer to the population of CXCR5+ cells expressing a modest amount of PD-1 as PD-1+CXCR5+ cells to distinguish them from the far less abundant ICOS+ PD-1+++ activated CXCR5+ cells and to distinguish the low PD-1 expression level from the high PD-1 level expressed by ICOS+ PD-1+++ activated CXCR5+ cells in blood and GC Tfh cells in lymphoid tissue (Figure 2A).
Figure 2. Human Blood PD-1+CXCR5+CD4+ T Cells.
(A) Comparison of PD-1 and CXCR5 expression between tonsil and blood CD45RO+CD4+ T cells. Expression of the activation markers HLA-DR, CD38, ICOS, and Ki67 on PD1− (filled gray), PD1+ (red line), and PD1+++ (black line) populations of CXCR5+CD4+ T cells is shown. Mean fluorescence intensity (MFI) is shown in the upper left corner of each plot.
(B) Representative frequency of PD-1+ cells in blood CXCR5+CD45RO+CD4+ T cells.
(C and D) Bcl6 control of PD-1 expression. (C) Flow cytometric analysis of PD-1 expression on blood naive CD4+ T cells transduced with either a Bcl6 lentiviral vector (Bcl6-LV) or a control vector (CTRL-LV). (D) MFI of PD-1 expression from cells in (E). *p = 0.01. Data from three independent experiments are shown.
(E and F) Maintenance of PD-1 expression on sorted blood PD-1+ and PD-1−CXCR5+CD45RO+CD4+ T cells after 10 days of in vitro culture. (E) Flow cytometric analysis of PD-1. (F) Quantitation of PD-1 maintenance. Data represent six or more donors per time point.
Also see Figure S2.
PD-1 is transiently induced by TCR triggering in T cells (Keir et al., 2008). However, PD-1 is also a component of the Tfh cell gene expression program, regulated by Bcl6 (Kroenke et al., 2012). The introduction of Bcl6 expression in naive blood CD4+ T cells resulted in specific upregulation of PD-1 (p = 0.01 [Figures 2C and 2D]). The observation of PD-1 expression on a subset of circulating CXCR5+CD4+ T cells raised the question of whether the PD-1+ cells were quiescent memory cells that stably express PD-1 as part of a Tfh cell differentiation program. PD-1+CXCR5+CD4+ T cells had no evidence of recent activation when examined directly ex vivo; compared to resting memory PD12212;CXCR5+CD4+ T cells, they showed unchanged levels of activation or proliferation markers (Ki67, HLA-DR, CD38, and ICOS [Figure 2A]). Furthermore, we determined that activated human CD4+ T cells expressed PD-1 for only a few days after removal of TCR stimulation (Figure S2A). To determine whether the resting PD-1+CXCR5+CD4+ T cells stably expressed PD-1, we incubated sorted PD-1+ and PD-1−CXCR5+CD4+ T cells for up to 20 days ex vivo in the absence of stimulation. Strikingly, PD-1 expression, as well as CXCR5 expression, was stable on the surface of the cells (Figures 2E and 2F, Figure S2B, and data not shown) (Figure S2C). Overall, these data indicate that PD-1+CXCR5+CD4+ T cells in human blood are quiescent cells with a stable phenotype, consistent with their potentially being memory cells.
Blood PD-1+CXCR32212;CXCR5+CD4+ T Cells and GC Tfh Cells Share a Transcriptional Profile Signature
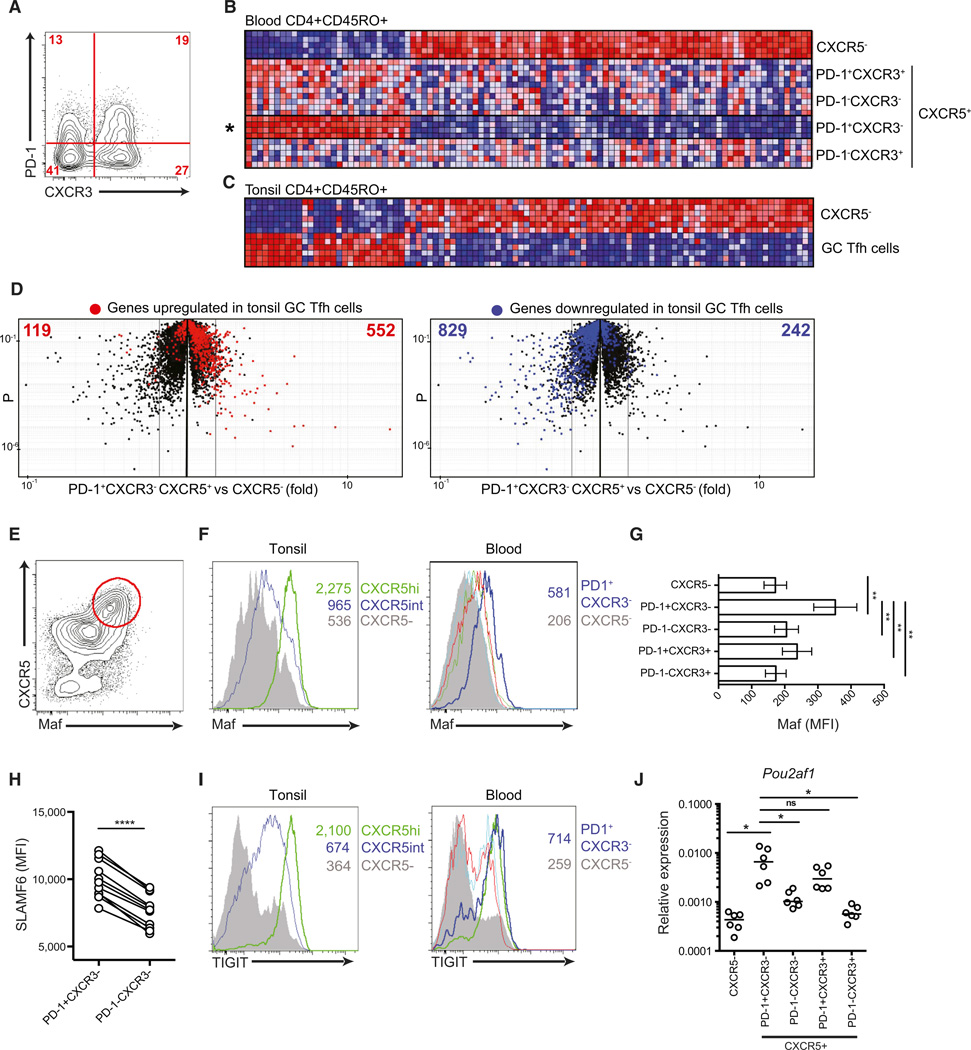
We hypothesized that resting blood PD-1+CXCR5+CD4+ T cells might have more similarities than blood PD-1−CXCR5+CD4+ T cells to GC Tfh cells. A first test of this hypothesis was to perform transcriptional profiling comparing blood PD-1+CXCR5+ cells and GC Tfh cells from tonsils, a secondary lymphoid tissue. Previous studies showed that blood CXCR3+CXCR5+CD4+ T cells are poor helpers of B cells (Morita et al., 2011), which we confirmed (not shown), and we therefore separated CXCR3− and CXCR3+ cells prior to gene expression profiling (Figure 3A). Compared to other CXCR5+CD4+ T cell subsets and CXCR5−CD4+ T cells, blood PD-1+CXCR3−CXCR5+ cells had a distinct gene expression profile (Figure 3B). We then compared the blood CXCR5+CD4+ T cells to CD4+ T cells in tonsils as a site of active GCs. Importantly, the gene signature of resting blood PD-1+CXCR3−CXCR5+ cells was clearly similar to that of GC Tfh cells; 94 of the top 100 genes were concordantly regulated (Figure 3C). Furthermore, unbiased analysis of all gene expression differences between blood PD-1+CXCR3−CXCR5+CD4+ T cells and CXCR5−CD4+ T cells revealed that most PD-1+CXCR3−CXCR5+CD4+ T cell gene expression changes matched the changes in GC Tfh cells (Figure 3D).
Figure 3. Blood PD-1+CXCR3−CXCR5+CD4+ T Cells Exhibit a Tfh-Cell-like Gene Expression Profile.
(A–D) Four populations of human blood CXCR5+CD45RO+CD4+ T cells, defined by PD-1 and CXCR3 expression, shown in the representative plot in (A) were cell sorted, and RNA was isolated for gene expression analysis. (B) Differentially expressed genes were hierarchically clustered, and the top 100 genes differentially expressed by blood PD-1+CXCR3−CXCR5+CD45RO+CD4+ T cells (PD-1+CXCR3−) versus CXCR5−CD45RO+CD4+ T cells were plotted as heatmaps for all subsets. (C) The 100-gene signature described in (B) was then examined for expression in lymphoid tissue (tonsil) GC Tfh cells (CXCR5hiPD-1+++CD45RO+CD4+ T cells) and non-Tfh cells (CXCR5−CD45RO+CD4+ T cells [CXCR5−]). In (B) and (C), blue indicates low relative expression and red indicates high relative expression. Rows represent independent subjects. (D) “Volcano” plots of the total gene expression data set from blood PD-1+CXCR3−CXCR5+CD45RO+CD4+ T cells versus CXCR5−CD45RO+CD4+ T cells obtained in (A) and (B). Each dot represents the mean for all subjects of an individual gene probe. Colors indicate genes upregulated (red) or downregulated (blue) more that 1.5-fold in lymphoid tissue GC Tfh cells (CXCR5hiPD-1+++CD45RO+CD4+ T cells) compared to CXCR5intCD45RO+CD4+ T cells. Numbers are the upregulated (right) or downregulated (left) GC Tfh cell genes in each half of the plot.
(E) Maf expression in tonsil CD45RO+CD4+ T cells.
(F) Flow cytometric analysis of Maf expression in tonsil and blood CD45RO+CD4+ T cell populations. In blood CXCR5+ populations, colors are as follows: blue, PD-1+CXCR3− cells; green, PD-1+CXCR3+ cells; red, PD1−CXCR3− cells; cyan, PD1−CXCR3+ cells; and gray, CXCR5− cells.
(G) Maf MFI quantitation of data in (F). Eight subjects are shown with mean and SEM. **p < 0.001.
(H) The graph displays individual SLAMF6 MFIs from 11 subjects. ****p < 0.0001.
(I) TIGIT expression by flow cytometry of tonsil and blood CD45RO+CD4+ T cell populations. Blood CXCR5+ populations are as in (F).
(J) Expression of POU2AF1 normalized to ACTB mRNA from six donors. Individual values and medians are depicted. *p < 0.01. “ns” stands for not significant.
Also see Figure S3.
The top-100-gene signature of blood PD-1+CXCR3−CXCR5+ CD4+ T cells included a number of Tfh cell gene products, including MAF. In both mice and humans, Maf is a critical regulator of Tfh cell differentiation and function (Bauquet et al., 2009; Ise et al., 2011; Kroenke et al., 2012). Maf was highly and selectively expressed by Tfh and GC Tfh cells in tonsils (Figure 3E). We then examined resting blood PD-1+CXCR3−CXCR5+CD4+ T cells and found that they expressed the highest Maf levels of any blood CD4+ T cell population tested (p < 0.01 [Figures 3F and 3G]). Although Bcl6 is required for Tfh cell differentiation, higher Bcl6 expression in PD-1+CXCR3−CXCR5+CD4+ T cells compared to CXCR5− memory CD4+ T cells was not detected (Figure S3). This is consistent with mouse memory Tfh cells (Choi et al., 2013; Hale et al., 2013; Liu et al., 2012). The receptor SLAMF6 (NTB-A/Ly108) regulates T cell and B cell (T:B) interactions and is expressed highly on GC Tfh cells (Kageyama et al., 2012).We observed that PD-1+CXCR3−CXCR5+CD4+ T cells expressed higher levels of SLAMF6 (p < 0.001 [Figure 3H]). TIGIT was also highly expressed on both tonsillar Tfh cells and blood PD-1+CXCR3−CXCR5+CD4+ T cells (Figure 3I). Furthermore, the transcription-factor-encoding gene POU2AF1, known to be a regulator of CXCR5 expression in B cells, was more highly expressed in PD-1+CXCR3−CXCR5+CD4+ T cells than in PD-1−CXCR5+CD4+ T cells (Figure 3J). Thus, quiescent blood PD-1+CXCR3−CXCR5+CD4+ T cells express Maf and other signature components of the Tfh cell program.
Resting PD-1+CXCR3−CXCR5+CD4+ T Cells Are a Population of Circulating Memory Tfh Cells
A fundamental question regarding CXCR5+CD4+ T cells in the blood is the matter of Tfh cell memory. Although there has been some evidence of human Tfh cell memory (Morita et al., 2011), it is frequently argued that memory Tfh cells might not exist (Breitfeld et al., 2000; Craft, 2012; Ma et al., 2012). All human blood CD45RO+CD4+ T cells expressing no activation markers are conventionally inferred to be memory cells. On the basis of this definition, PD-1+CXCR3−CXCR5+CD4+ T cells are memory cells (Figure 2A and Figure S4A). Resting CD45RO+CD4+ T cells are usually classified as central-memory T (TCM) or effector-memory T (TEM) cells (Sallusto et al., 1999), although a more sophisticated subsetting is more accurate. Whereas TEM cells lack CCR7 and migrate to inflamed tissues, TCM cells express CCR7 and circulate through secondary lymphoid organs. Most blood CXCR5+CD4+ T cells were previously identified as TCM cells by virtue of their positivity for CCR7 (Breitfeld et al., 2000; Chevalier et al., 2011). We asked whether the same is true for PD-1+CXCR3−CXCR5+CD4+ T cells. We found that this subset expressed CCR7 but at lower levels than those of CXCR5− and PD-1−CXCR5+CD4+ TCM cells (Figures 4A and 4B). To formally address whether PD-1+CXCR3−CXCR5+CD4+ T cells represent memory cells, we then tested for the presence of antigen-specific memory cells by using tetanus-specific major histocompatibility complex (MHC) class II tetramer staining (Figure 4C and Figure S4). A substantial fraction of tetanus-specific memory CD4+ T cells were CXCR5+ (Figure 4D). Furthermore, tetanus-specific PD-1+CXCR3−CXCR5+CD4+ T cells were present in most donors (Figures 4E and 4F and Figure S4). Interestingly, tetanus-specific memory CD4+ T cells exhibited heterogeneous phenotypes, consistent with the well-recognized heterogeneity of CD4+ T cells (Streeck et al., 2013) and recently recognized heterogeneity of memory CD8+ T cells (Newell et al., 2012). Together, the phenotypic and MHC class II tetramer data demonstrate that PD-1+CXCR3−CXCR5+CD4+ T cells are a population of circulating memory Tfh cells.
Figure 4. Existence of Tetanus-Specific Memory CD4+ T Cells with the PD-1+CXCR3−CXCR5+ Phenotype.
(A) Flow cytometric analysis of CCR7 expression on PD-1+CXCR3− (black) and PD-1−CXCR3− (red) CXCR5+CD45RO+CD4+ T cells in comparison to CXCR5− cells (gray).
(B) CCR7 MFIs of PD-1+CXCR3−, PD-1−CXCR3−, CCR7+CD45RA−CXCR5− (TCM),CCR7−CD45RA−CXCR5− (TEM), and CD45RA+ (naive) CD4+ T cells. Individual values and medians are depicted. **p < 0.001.
(C) Tetanus-specific CD4+ T cells were identified by HLA-DRB1*0401 tetramers loaded with tetanus peptide and enriched with magnetic beads. The FACS plot shows a representative tetanus-specific tetramer staining postenrichment.
(D) CXCR5 expression by tetanus-specific CD4+ T cells. Data are from seven independent HLA-DRB1*0401+ subjects. Individual values and means are shown.
(E) A representative flow cytometry plot of PD-1 and CXCR3 expression by tetanus-specific CXCR5+ cells.
(F) Quantitation of PD-1+CXCR3− cells within the CXCR5+ tetanus-specific CD4+ T cell population. Individual values and medians are plotted.
Also see Figure S4.
Memory PD-1+CXCR3−CXCR5+CD4+ T Cells Are Highly Functional for B Cell Help
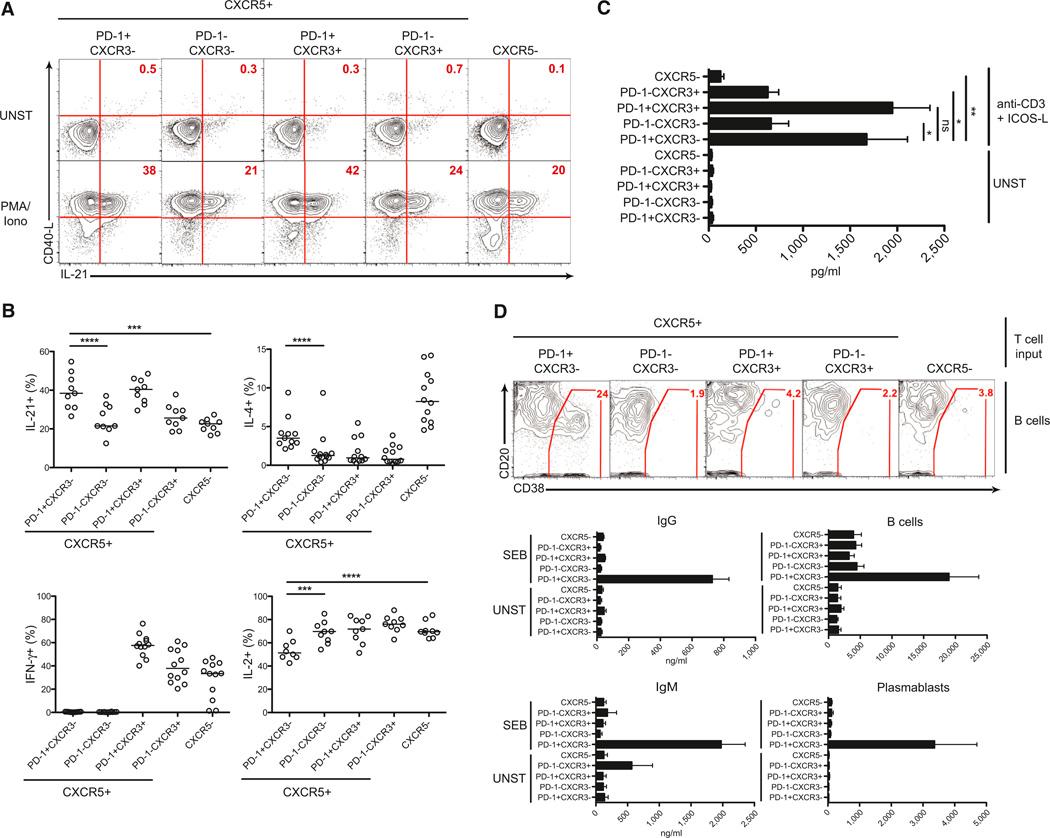
Tfh cells are specialized providers of help to B cells. Coexpression of IL-21, IL-4, and CXCL13 is the canonical functional signature of Tfh cells (Crotty, 2011). We compared IL-21 and IL-4 expression of blood PD-1+CXCR5+CD4+ T cells to that of other CXCR5+CD4+ T cell subsets and non-Tfh cells. Upon stimulation with PMA plus ionomycin, PD-1+CXCR3−CXCR5+CD4+ and PD-1+CXCR3+CXCR5+CD4+ T cells produced the highest amounts of IL-21 (Figures 5A and 5B). PD-1+CXCR3−CXCR5+CD4+ T cells expressed the most IL-4 among the CXCR5+ cells, consistent with their higher Maf expression (Figure 5B and Figure S5A). CXCL13 expression is highly restricted. Tfh cells are capable of producing large quantities of CXCL13 (Kim et al., 2004; Kroenke et al., 2012; Yu et al., 2009), and blood CXCR5+CD4+ T cells can produce CXCL13 (Chevalier et al., 2011). By contrast, blood CXCR5−CD4+ T cells did not express CXCL13 (Figure 5C). Compared to PD-1−CXCR5+CD4+ T cells, PD-1+CXCR3−CXCR5+CD4+ and PD-1+CXCR3+CXCR5+CD4+ T cells produced high levels of CXCL13 upon stimulation (Figure 5C). GC Tfh cells in lymphoid organs have low or absent expression of canonical cytokines of other effector CD4+ T cell types (IFN-γ, IL-17, and IL-13) (Kroenke et al., 2012; Ma et al., 2009; Yu et al., 2009). Blood PD-1+CXCR3−CXCR5+CD4+ T cells had no IFN-γ expression, whereas PD-1−CXCR3+CXCR5+CD4+ and PD-1−+CXCR3+CXCR5+CD4+ T cells expressed high levels of IFN-γ upon stimulation (Figure 5B). Interestingly, IL-2 is an inhibitor of Tfh cell differentiation (Johnston et al., 2012) and PD-1+CXCR3−CXCR5+CD4+ T cells had the lowest percentage of IL-2 expressors among all CD45RO+CD4+ T cells tested (Figure 5B). Thus, PD-1+CXCR3−CXCR5+CD4+ T cells have a cytokine profile comparable to that of active GC Tfh cells upon restimulation.
Figure 5. Blood PD-1+CXCR3−CXCR5+CD4+ T Cells Display the Strongest Tfh Cell Functionality.
(A and B) Blood CD4+ T cell populations (CD45RO+) were isolated by cell sorting and examined for intracellular expression of IL-21, CD40L, IL-4, IFN-γ, and IL-2 by flow cytometry with or without (UNST) short in vitro stimulation (PMA and ionomycin). (A) IL-21 and CD40L expression are shown. (B) Quantitation of IL-21, IL-4, IFN-γ, and IL-2. Each point represents cells from one subject; a composite of four independent experiments is shown. Bars indicate the median.
(C) Isolated blood CD4+ T cell populations were stimulated with anti-CD3 and ICOS-L or left unstimulated (UNST) for 5 days. CXCL13 released in the supernatant was measured by ELISA. Data are from nine donors among four independent experiments. Mean and SEM shown. “ns” stands for not significant.
(D) Purified blood CD4+ T cell populations were cultured with memory B cells either in the presence of the superantigen SEB or not (UNST) for 7 days. Plasmablast (CD20loCD38+) differentiation was analyzed by flow cytometry. Contour plots show a representative donor. The bar graphs on the right quantify B cells (CD3− cells) and plasmablasts at the end in the coculture. The bar graphs on the left show IgG and IgM production (measured by ELISA) in the T:B cocultures. Data represent the mean and SEM from four independent experiments. *p < 0.01, **p < 0.001, ***p < 0.0005, ****p < 0.0001.
Also see Figure S5.
One reason for skepticism about blood CXCR5+CD4+ T cells’ being related to Tfh cells has been that most in vitro assays have shown that compared to blood CXCR5−CD4+ T cells, bulk CXCR5+CD4+ T cells provide only marginally better B cell help (Chevalier et al., 2011; Pallikkuth et al., 2012; Streeck et al., 2013). The identification of a subset of CXCR5+CD4+ T cells that express IL-21, IL-4, and CXCL13 similarly to Tfh cells led us to examine whether blood PD-1+CXCR3−CXCR5+CD4+ T cells were functionally superior at providing help to B cells. Importantly, in T:B cocultures, PD-1+CXCR3−CXCR5+CD4+ T cells were vastly superior at inducing memory B cell differentiation into plasma cells and IgG secretion (p < 0.05 and p < 0.0005 [Figure 5D]). The failure of the other T cell populations to provide help to memory B cells was not caused by a lack of expansion or death, given that all T cell subsets expanded and survived similarly (data not shown). The majority of resting CXCR5+CD4+ T cells were PD-1− and were poor helpers of memory B cells (Figure 5D). One interpretation is that those PD-1−CXCR5+ cells represent less polarized or differentiated Tfh cells. That interpretation is consistent with, and supported by, the observation that these cells, along with PD-1+CXCR3−CXCR5+ CD4+ T cells, were competent for helping naive B cells in culture (Figure S5B). Thus, we conclude that the highly functional PD-1+CXCR3−CXCR5+CD4+ T cells are resting memory Tfh cells with the most polarized Tfh cell gene signature and the highest capacity for B cell help.
Blood PD-1+CXCR3−CXCR5+CD4+ T Cells Positively Correlate with bnAb Development in HIV+ Donors
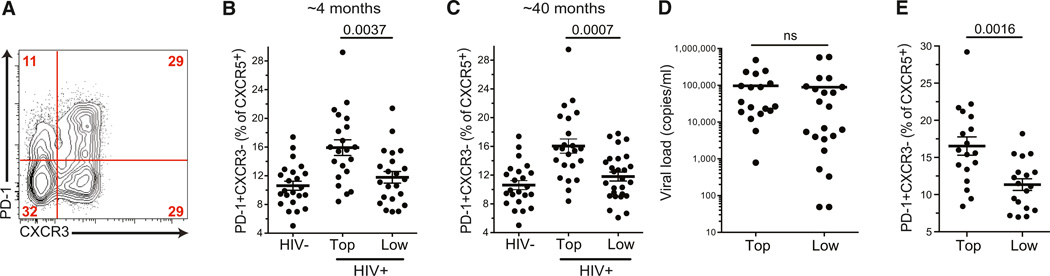
Given that PD-1+CXCR3−CXCR5+CD4+ T cells were the most functional and polarized memory Tfh cells, we then re-evaluated the possible association between blood CXCR5+CD4+ T cells and the development of bnABs against HIV. We hypothesized that memory PD-1+CXCR3−CXCR5+ Tfh cells might serve as a biomarker predicting bnAb development. Strikingly, the frequency of the most functional memory Tfh cells (PD-1+CXCR3−CXCR5+ cells) at the earliest available time point (~4 months postinfection) correlated with the capacity of individuals to subsequently develop bnAbs against HIV many months later (p = 0.0037 [Figures 6A and 6B]). The correlation between PD-1+CXCR3−CXCR5+ Tfh cells and bnAbs was also strong at the time of bnAb development (p = 0.0007 [Figure 6C]). If this population is a memory population, its percentage is expected to be stable over time. PD-1+CXCR3−CXCR5+ Tfh cell frequencies for each individual were correlated at the two time points (Figure S6A), consistent with the proposal that this is a stable memory population. An alternative interpretation would be that PD-1+CXCR3−CXCR5+ Tfh cells represent a stable niche but that the cells constituting this population change over time. Subsequent statistical tests found that PD-1+CXCR3−CXCR5+ cells were the only CXCR5+CD4+ T cell population that correlated with a high-quality HIV antibody response (Figures S1C, S1D, and S6B–S6D). Although blood CXCR3−CXCR5+ cells were previously described as the blood cells possessing the most Tfh-cell-like functional activity (Morita et al., 2011), the proportion of CXCR3− cells among CXCR5+CD4+ T cells did not correlate with bnAb development (Figure S6B). Additionally, PD-1 by itself was not a useful biomarker, given that PD-1+CXCR3+ memory Tfh cell frequencies at either time point failed to correlate with a capacity to develop bnAbs (Figures S6C and S6D).
Figure 6. Highly Functional PD-1+CXCR3−CXCR5+CD4+ T Cells Are Associated with HIV bnAb Development.
(A) A representative flow cytometry plot of an HIV+ donor shows the gates for the PD-1+CXCR3− population (gated on the CXCR5+CD4+ cells).
(B and C) The PD-1+CXCR3− percentage of CXCR5+CD4+ T cells in HIV− and HIV+ groups at the earliest available time point (average of 4 months postinfection) and the time of bnAb development (average of 40 months postinfection). (B) Blood samples were from the earliest available time point after HIV infection, years before the development of bnAbs. “Top” indicates the top neutralizers, and “low” indicates the low neutralizers. Each point represents an individual donor. The PD-1+CXCR3− percentage of CXCR5+ CD4+ T cells (gated on CXCR5+CD4+ T cells) is shown for HIV− and HIV+ groups from the same samples as in Figure 1. Mean and SEM are plotted. (C) Same as in (B), except PBMCs from HIV+ groups were tested at the time of bnAb development.
(D) Viral load (copies/ml) of the individuals in the “top” and “low” neutralizing antibody groups at the ~4-month time point. Bars indicate the mean. “ns” stands for not significant.
(E) To control for any effects of viral load (“VL”), we performed an additional analysis of the ~4-month-time-point samples, excluding individuals with a low viral load (less than 1,000 viral copies/ml, dashed line; individuals dropped for this analysis are highlighted in red in Figure S6E). The PD-1+CXCR3− percentage of CXCR5+CD4+ T cells within the “top” and “low” neutralizing antibody groups at the ~4-month time point are shown for individuals with a viral load > 1,000. Mean and SEM are plotted.
Also see Figure S6.
Although viral loads were not statistically significantly different between top and low neutralizers at the earliest available time point (Figure 6D), which was the primary time point of interest, a statistically significant difference in viral loads did develop over time (Figures S6E and S6G). Therefore, viral load was controlled for as a variable. The statistical significance of the correlation between PD-1+CXCR3−CXCR5+ Tfh cells and bnAbs remained effectively unchanged after correction for viral loads (p = 0.0016, at the ~4-month time point postinfection [Figure 6E]; p = 0.0075 at the time of bnAb development [Figure S6H]), indicating that the association between high PD-1+CXCR3−CXCR5+ cell frequencies and bnAb development is an independent parameter from viral loads. As an alternative approach, multivariate analysis accounting for viral load was also performed, and again the statistical significance of the correlation between PD-1+CXCR3−CXCR5+ cells and bnAb development was independent of viral load (p = 0.00764 at ~4 months postinfection; p = 0.00097 at the time of bnAb development). Thus, the frequency of PD-1+CXCR3−CXCR5+ cells selectively correlates with the ability to develop bnAbs against HIV.
Together, these data provide evidence that PD-1+CXCR3−CXCR5+CD4+ T cells are a human memory CD4+ T cell type with high Tfh cell functionality. Furthermore, the ability of rare HIV-infected individuals to make bnAbs against HIV is associated with the presence of these PD-1+CXCR3−CXCR5+ highly functional memory Tfh cells, supporting the concept that enhanced T cell help to B cells is most likely a critical component of any candidate HIV vaccine designed to elicit potent bnAbs against HIV.
DISCUSSION
Whereas most human vaccines, and many new candidate vaccines, for major infectious killers of humans are dependent on quality antibody responses, our understanding of how Tfh cells support and impact that process in humans is just developing (Crotty, 2011; Streeck et al., 2013). Here, we identified a population of highly functional circulating memory Tfh cells with a strong transcriptional resemblance to GC Tfh cells. Quiescent PD-1+ CXCR3−CXCR5+CD4+ T cells were marked by the expression of a number of Tfh-cell-related molecules, including Maf, TIGIT, POU2AF1, Slamf6, and PD-1. Besides playing a pivotal role in the differentiation and function of Tfh cells, Maf also regulates IL-21 and IL-4. We did not detect elevated Bcl6 in memory PD-1+CXCR3−CXCR5+CD4+ T cells. Bcl6 expression in murine memory Tfh cells was recently found to be downregulated and not expressed higher than in resting non-Tfh memory cells (Choi et al., 2013; Hale et al., 2013; Liu et al., 2012). The lack of elevated Bcl6 suggests that some aspects of Tfh cells are maintained by low Bcl6 levels or are initiated by Bcl6 expression and are sustained by other facets of the Tfh cell program when Bcl6 levels decrease in quiescent memory cells.
Whether circulating CXCR5+ cells are the memory counterpart of Tfh cells has been controversial. We directly identified memory Tfh cells by tetanus-specific MHC class II tetramer staining. PD-1+CXCR3−CXCR5+CD4+ T cells can be defined as TCM cells by virtue of CCR7 expression. Although active Tfh cells are located in secondary lymphoid organs, it is not surprising to find memory Tfh cells in the blood. Indeed, it is reasonable to assume that similarly to other CD4+ TCM cells and memory B cells, memory Tfh cells regularly recirculate to distribute themselves throughout secondary lymphoid tissues. It is intriguing that, although positive, the PD-1+CXCR3−CXCR5+CD4+ T cells expressed lower levels of CCR7 than did PD-1−CXCR5+CD4+ T cells. A coordinated increase in CXCR5 and decrease in CCR7 is important for CD4+ T cell localization to B cell follicles (Haynes et al., 2007), and lower levels of CCR7 might allow for a deeper migration into B cell follicles in vivo.
In our study, the PD-1+CXCR3−CXCR5+CD4+ T cells emerged as the cells with the highest functional ability to help B cells. All of the CXCR5+ cells were capable of upregulating CD40L and producing IL-21 and CXCL13 to some degree. Strikingly, the PD-1+ CXCR3−CXCR5+CD4+ T cells were the only ones capable of supporting efficient antibody responses by memory B cells in vitro. The PD-1+CXCR3−CXCR5+CD4+ T cells appeared to be the most differentiated and highly functional memory Tfh cells, in line with the idea that optimal memory Tfh cells would be able to efficiently provide help to both naive and memory B cells. Nevertheless, we do not exclude that the other CXCR5+ CD4+ T cells are part of the memory Tfh cell family. One possibility is that these cells might be less polarized Tfh cell memory precursors that require further stimuli to complete their differentiation process. The CXCR3+ cells might have been exposed to excessive Th1 stimuli, diverting their Tfh cell differentiation.
A recent study by Bentebibel and colleagues has found that a small population of activated ICOS+CXCR3+CXCR5+ cells transiently appear in human blood after influenza vaccination and that these cells correlate with influenza antibody titers (Bentebibel et al., 2013). That observation was unexpected, given that Morita and colleagues demonstrated that CXCR3+CXCR5+ cells are the worst CXCR5+CD4+ T cells at helping B cells (Morita et al., 2011), and we have confirmed that finding here. The CD4+ T cells arising after influenza vaccination are transient effector cells displaying an activated phenotype, including expression of ICOS and Ki67, and their lineage is unclear, given that many activated human CD4+ T cells can express CXCR5 and other chemokine receptors transiently. In contrast, our study focused on resting cells. Here, we have now demonstrated that quiescent blood CXCR5+CD4+ T cells (lacking ICOS and other activation markers), including the PD-1+CXCR3−CXCR5+ cells that were the focus of this study, are a memory cell population. Notably, the influenza vaccine is notorious for being the least protective vaccine among vaccines commonly used in the United States, even when the influenza strain is well matched (CDC, 2013), and the influenza vaccine also suffers from an unusually short duration of protection after immunization (CDC, 2011). Therefore, the very weak B cell help provided by CXCR3+CXCR5+CD4+ T cells shown both here and by Morita and colleagues (Morita et al., 2011) suggests that the currently available influenza vaccines might generate poor antibody protection precisely because they elicit CXCR3+CXCR5+ cells. We speculate that an influenza vaccine promoting the generation of highly functional PD-1+CXCR3−CXCR5+ memory Tfh cells described herein would result in better antibody responses and efficacious protection against influenza.
Surrogate cell populations in blood are essential as clinical study markers. Although a direct analysis of bona fide active GC Tfh cells from lymph nodes of bnAb+ individuals would have provided valuable information, this was not feasible. Therefore, the study of the relationship between Tfh cells and HIV bnAb development relied upon the definition of highly functional memory Tfh cells in the blood. We identified PD-1+CXCR3−CXCR5+CD4+ T cells as the blood population with the most pronounced phenotypic and functional hallmarks of bona fide Tfh cells. Remarkably, PD-1+CXCR3−CXCR5+CD4+ T cells were the only population of blood CXCR5+ cells in HIV+ individuals to positively correlate with the generation of bnAbs against HIV in this study. We focused our analysis on cell frequency, instead of absolute numbers, in order to minimize unknown confounding factors in HIV+ individuals and to concentrate on biases in the differentiation of different subtypes of memory Tfh cells. However, as with most human immunology studies, we do not know how directly the blood frequencies relate to cell frequencies in the lymph nodes and spleen, the sites of GCs and T:B interactions. We interpret the increased frequency of optimal circulating memory Tfh cells to be a biomarker of the ability to generate strong Tfh cell responses necessary to drive high levels of somatic hypermutation found in HIV bnAbs. Future studies directly interrogating memory Tfh cells in lymphoid tissues would be of great value.
Previously, clinical and viral parameters have been associated with HIV bnAb production (Hessell and Haigwood, 2012). Those previous findings support a model in which antigen load and HIV envelope diversity are important components of the evolution of bnAbs. It is likely that antigen load and a recursive process of antibody mutation and virus escape (Richman et al., 2003) provide the basis for the process of antibody epitope honing to the most conserved HIV envelope epitopes (Liao et al., 2013). However, because most individuals with high viral loads do not develop bnAb responses, the limiting factor (or factors) is clearly something other than viral load, and it is logical to consider host immune factors. Although it is reasonable to assume that multiple factors play important roles in the process of HIV bnAb development, our data show that individuals who make a bnAb response preferentially have higher percentages of the highly functional circulating PD-1+CXCR3−CXCR5+ memory Tfh cells. Consistent with the fact that both antigen and Tfh cells are instrumental parts of the GC response, it is logical that both factors are positively associated with the end product—a highly mutated antibody capable of neutralizing most HIV strains.
Here, we have demonstrated that highly functional memory Tfh cells are present in human blood and that these cells correlate with a clinically important outcome: the development of potent bnAbs against HIV. These findings might facilitate rational vaccine design against HIV and other pathogens. Although it is conceivable that an antibody-dependent HIV vaccine is possible, given the discovery of bnAb+ individuals, the path to eliciting such antibodies by vaccination is unclear (Burton et al., 2012; McMichael and Haynes, 2012). HIV bnAbs are highly affinity maturated, well above the average affinity maturation observed in human memory B cells and well beyond the amount of affinity maturation elicited by current human vaccines (Streeck et al., 2013). Although it was hypothetically possible that Tfh cells are important for bnAb development, the results reported here are evidence of a positive association between a Tfh cell population and bnAb development. These results provide an encouraging indication that generating better Tfh cell responses and Tfh cell memory might be a fruitful vaccine strategy for enhancing the quality of antibody responses against HIV sufficiently to accomplish the generation of HIV bnAbs in the context of a candidate vaccine employing appropriate HIV Env trimer antigens or HIV Env trimer expression. Furthermore, characterization of PD-1−+CXCR3−CXCR5+CD4+ T cells as the most highly functional memory Tfh cells indicates that candidate HIV vaccines should attempt to preferentially elicit these cells for maximal B cell help and affinity maturation after immunization. In addition, PD-1+CXCR3−CXCR5+ memory CD4+ T cells are present in human peripheral blood and therefore can be readily quantified. Finally, the relevance of these results is unlikely to be restricted to HIV vaccine development given that protective immunity to a range of pathogens for which the human immune system has difficulty developing high-quality antibody responses might be limited by the quality of the Tfh cell response and the resultant capacity to generate highly affinity-matured antibody responses.
EXPERIMENTAL PROCEDURES
Human Samples
Fresh blood samples (for Figures 2, 3, 4, and 5) from healthy donors were obtained from the La Jolla Institute for Allergy and Immunology (LJI) blood donor program. Informed consent was obtained from all donors. Peripheral-blood mononuclear cells (PBMCs) were isolated from blood by density-gradient centrifugation using Histopaque-1077 (Sigma-Aldrich). Fresh human tonsils were obtained from the National Disease Resource Interchange. Tonsil preparation was previously described (Kroenke et al., 2012). Details on Protocol C donor recruitment and testing are described in the Supplemental Experimental Procedures. All protocols were approved by LJI and National Disease Resource Interchange institutional review boards.
Flow Cytometry and Cell Sorting
For the intranuclear staining, cells were treated with the FoxP3 Fixation/Permeabilization Kit (eBioscience) and stained in permeabilization buffer (eBioscience). For surface staining, primary staining panels used for phenotypic analysis or for cell sorting are described in Tables S1 and S2. See Supplemental Experimental Procedures for detailed descriptions.
Gene Expression Microarrays
Blood CD4+ T cells were enriched by magnetic-bead isolation (CD4+ T Cell Isolation Kit II, Miltenyi Biotec). After enrichment, CD14−CD16−CD19−CD8−CD4+CD45RA− cells were sorted by fluorescence-activated cell sorting (FACS) into the following five populations: CXCR5−, PD1+CXCR3−CXCR5+, PD1+CXCR3+CXCR5+, PD1−CXCR3−CXCR5+, and PD1−CXCR3+CXCR5+. Cells were directly sorted in buffer RLT for RNA extraction. Sorting of tonsil GC Tfh (CXCR5hi) cells and Tfh (CXCR5int) cells was previously described (Kroenke et al., 2012). RNA was isolated with the RNeasy Micro Kit (QIAGEN). The quantity and quality of the RNA were confirmed with a NanoDrop 2000c (Thermo Fisher Scientific) and an Experion Electrophoresis System (Bio-Rad). Samples (20 ng) were amplified with Illumina MessageAmp II aRNA Amplification Kits (Ambion), hybridized to HumanHT-12_V4 BeadChips (Illumina), and quantified with Genome Studio (Illumina). Normalized data (heatmap of blood cells) or raw data (heatmap of tonsil cells and Volcano plot) were analyzed with the GenePattern software suite.
T:B Coculture Assay
CD4+ T cells were isolated from fresh PBMCs by magnetic-bead negative selection with the CD4+ T Cell Isolation Kit II (Miltenyi Biotec). The CD19−CD14−CD8−CD4+CD45RO+ cells were sorted by FACS into CXCR5−, PD1+ CXCR3−CXCR5+, PD1+CXCR3+CXCR5+, PD1−CXCR3+CXCR5+, and PD1−CXCR3− CXCR5+ subpopulations. B cells were isolated from fresh PBMCs by CD19 magnetic-bead positive selection (Miltenyi Biotec). Memory B cells were then sorted as CD3−CD14−CD4−CD19+IgD−CD27+CD38− cells. Memory B cells (3 × 104 cells/well) were cocultured with the different T cell populations (3 × 104 cells/well) in the presence of staphylococcal enterotoxin B (SEB, 100ng/ml, Toxin Technology) in AIM-V medium (Life Technologies). B cell number, phenotype, and Ig concentrations in the supernatants were measured after 7 days.
Statistical Analysis
A two-tailed Mann-Whitney test was used for evaluating differences in the populations of the HIV clinical cohorts shown in Figures 1 and 6. The Mann-Whitney nonparametric rank-comparison statistical test is optimal for analysis of the clinical cohort because it does not assume a normal distribution of the data, a standard approach with human samples. Corrections for multiple tests were not applied because each test was based on a specific hypothesis specified in advance. Covariance of PD-1+CXCR3−CXCR5+ Tfh cells and viral load was evaluated with the ANCOVA multivariate statistical test (VassarStats). Prism 5.0 (GraphPad) was used for all other data statistical analyses. The statistical analyses of the biological phenotypes and functionality of CXCR5+CD4+ T cell populations from healthy controls (Figures 2, 3, 4, and 5) were done with two-tailed paired t tests. The paired t test is the best statistical test in this case because of the direct comparison of populations within donors. Independent experiments were repeated several times with controls.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank all IAVI Protocol C project study volunteers, clinical team members, and site team members, specifically Melissa Simek and Emmanuel Cormier for sample tracking coordination. We thank Terri Wrin from Monogram Biosciences for generating neutralization data and Khader Ghneim from VGTI for assisting with microarray data. We thank the LIAI flow cytometry core, Anthony Jose, and Kayla Kendric for technical assistance. This work was supported by NIAID grants (AI090970), IAVI, and the CHAVI-ID (UM1-AI10066).
Footnotes
ACCESSION NUMBERS
The microarray data are available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/gds) under the accession numbers GSE50390 (blood CD4+ T cells) and GSE50391 (tonsil CD4+ T cells) and together under the GEO SuperSeries accession number GSE50392.
SUPPLEMENTAL INFORMATION
Supplemental Information includes a full list of IAVI Protocol C principal investigators, Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2013.08.031.
REFERENCES
- Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho I-C, Sharpe AH, Kuchroo VK. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, Flaño E, Mejias A, Albrecht RA, Blankenship D, et al. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci. Transl. Med. 2013;5:76ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettler T, Moeckel F, Cheng Y, Heeg M, Salek-Ardakani S, Crotty S, Croft M, von Herrath MG. OX40 facilitates control of a persistent virus infection. PLoS Pathog. 2012;8:e1002913. doi: 10.1371/journal.ppat.1002913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Förster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Ahmed R, Barouch DH, Butera ST, Crotty S, Godzik A, Kaufmann DE, McElrath MJ, Nussenzweig MC, Pulendran B, et al. A Blueprint for HIV Vaccine Discovery. Cell Host Microbe. 2012;12:396–407. doi: 10.1016/j.chom.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Prevention and control of influenza with vaccines: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2011. MMWR Morb. Mortal. Wkly. Rep. 2011;60:1128–1132. [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Interim adjusted estimates of seasonal influenza vaccine effectiveness - United States, February, 2013. MMWR Morb. Mortal. Wkly. Rep. 2013;62:119–123. [PMC free article] [PubMed] [Google Scholar]
- Chevalier N, Jarrossay D, Ho E, Avery DT, Ma CS, Yu D, Sallusto F, Tangye SG, Mackay CR. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. J. Immunol. 2011;186:5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- Choi YS, Yang JA, Yusuf I, Johnston RJ, Greenbaum J, Peters B, Crotty S. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol. 2013;190:4014–4026. doi: 10.4049/jimmunol.1202963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chtanova T, Tangye SG, Newton R, Frank N, Hodge MR, Rolph MS, Mackay CR. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 2004;173:68–78. doi: 10.4049/jimmunol.173.1.68. [DOI] [PubMed] [Google Scholar]
- Craft JE. Follicular helper T cells in immunity and systemic autoimmunity. Nat Rev Rheumatol. 2012;8:337–347. doi: 10.1038/nrrheum.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annu. Rev. Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J. Exp. Med. 2011;208:987–999. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 2011;334:825–829. doi: 10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes NM, Allen CDC, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J. Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- Hessell AJ, Haigwood NL. Neutralizing antibodies and control of HIV: moves and countermoves. Curr. HIV/AIDS Rep. 2012;9:64–72. doi: 10.1007/s11904-011-0105-5. [DOI] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, Murphy KM. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat. Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 2012;36:986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CH, Lim HW, Kim JR, Rott L, Hillsamer P, Butcher EC. Unique gene expression program of human germinal center T helper cells. Blood. 2004;104:1952–1960. doi: 10.1182/blood-2004-03-1206. [DOI] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu R-B, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J. Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human antibodies that neutralize HIV-1: identification, structures, and B cell ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenkamp A, Nagata K, Murphy K, Wu L, Lanzavecchia A, Sallusto F. Kinetics and expression patterns of chemokine receptors in human CD4+ T lymphocytes primed by myeloid or plasmacytoid dendritic cells. Eur. J. Immunol. 2003;33:474–482. doi: 10.1002/immu.200310023. [DOI] [PubMed] [Google Scholar]
- Liao H-X, Lynch R, Zhou T, Gao F, Alam SM, Boyd SD, Fire AZ, Roskin KM, Schramm CA, Zhang Z, et al. NISC Comparative Sequencing Program. Co-evolution of a broadly neutralizing HIV-1 antibody and founder virus. Nature. 2013;496:469–476. doi: 10.1038/nature12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. J. Exp. Med. 2012;209:1841–1852. S1–S24. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner-Nanan B, Deenick EK, Tangye SG. Early commitment of naïve human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol. Cell Biol. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- Ma CS, Deenick EK, Batten M, Tangye SG. The origins, function, and regulation of T follicular helper cells. J. Exp. Med. 2012;209:1241–1253. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael AJ, Haynes BF. Lessons learned from HIV-1 vaccine trials: new priorities and directions. Nat. Immunol. 2012;13:423–427. doi: 10.1038/ni.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldt B, Rakasz EG, Schultz N, Chan-Hui P-Y, Swiderek K, Weisgrau KL, Piaskowski SM, Bergman Z, Watkins DI, Poignard P, Burton DR. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc. Natl. Acad. Sci. USA. 2012;109:18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Schmitt N, Bentebibel S-E, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell EW, Sigal N, Bendall SC, Nolan GP, Davis MM. Cytometry by time-of-flight shows combinatorial cytokine expression and virus-specific cell niches within a continuum of CD8+ T cell phenotypes. Immunity. 2012;36:142–152. doi: 10.1016/j.immuni.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallikkuth S, Parmigiani A, Silva SY, George VK, Fischl M, Pahwa R, Pahwa S. Impaired peripheral blood T-follicular helper cell function in HIV-infected nonresponders to the 2009 H1N1/09 vaccine. Blood. 2012;120:985–993. doi: 10.1182/blood-2011-12-396648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, et al. CD4 T follicular helper cell dynamics during SIV infection. J. Clin. Invest. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin SA. Correlates of protection induced by vaccination. Clin. Vaccine Immunol. 2010;17:1055–1065. doi: 10.1128/CVI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed A-U, Rahn H-P, Sallusto F, Lipp M, Müller G. Follicular B helper T cell activity is confined to CXCR5(hi)ICOS(hi) CD4 T cells and is independent of CD57 expression. Eur. J. Immunol. 2006;36:1892–1903. doi: 10.1002/eji.200636136. [DOI] [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolf J, Bell SE, Kovesdi D, Janas ML, Soond DR, Webb LMC, Santinelli S, Saunders T, Hebeis B, Killeen N, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J. Immunol. 2010;185:4042–4052. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson N, Gatenby PA, Wilson A, Malik S, Fulcher DA, Tangye SG, Manku H, Vyse TJ, Roncador G, Huttley GA, et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010;62:234–244. doi: 10.1002/art.25032. [DOI] [PubMed] [Google Scholar]
- Streeck H, D’Souza MP, Littman DR, Crotty S. Harnessing CD4+ T cell responses in HIV vaccine development. Nat. Med. 2013;19:143–149. doi: 10.1038/nm.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu. Rev. Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa CG, Cyster JG. How T cells earn the follicular rite of passage. Immunity. 2011;35:671–680. doi: 10.1016/j.immuni.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Protocol G Principal Investigators. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, et al. Protocol G Principal Investigators. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.