Abstract
Strain-promoted azide-alkyne cycloaddition (SPAAC) can be used to generate artificial metalloenzymes (ArMs) from scaffold proteins containing a p-azido-L-phenylalanine (Az) residue and catalytically active bicyclononyne-substituted metal complexes. The high efficiency of this reaction allows rapid ArM formation using Az residues within the scaffold protein in the presence of cysteine residues or various reactive components of cellular lysate. In general, cofactor-based ArM formation allows the use of any desired metal complex to build unique inorganic-protein materials. SPAAC covalent linkage further decouples the native function of the scaffold from the installation process since it is not affected by native amino acid residues; as long as an Az residue can be incorporated, an ArM can be generated. We have demonstrated the scope of this method with respect to both the scaffold and cofactor components and established that the dirhodium ArMs generated can catalyze the decomposition of diazo compounds and both Si-H and olefin insertion reactions involving these carbene precursors.
Keywords: artificial metalloenzyme, biocatalysis, cofactor, click chemistry, dirhodium
Controlling the selectivity of metal catalysts[1] and incorporating metal complexes into biological systems, where they could have a transformative impact on our ability to manipulate life processes, [2] stand as key challenges in synthetic chemistry. A number of researchers have developed artificial metalloenzymes (ArMs) comprised of synthetic metal catalysts and enzyme or protein scaffolds to achieve these goals by combining the reactivity former with the adaptability and efficiency of the latter.[3] Several different approaches to link catalysts and scaffolds have been described. Most of these can be broadly classified as involving dative binding of scaffold residues to metal atoms (as free ions or complexed with other ligands),[4] non-covalent binding of metal complexes, [5] or covalent modification using a functionalized metal complex[6].
Efforts to date have demonstrated the potential of ArMs for a range of applications,[5b,7,8] but they have also highlighted the importance of flexibility with respect to the scaffold proteins,[9] bioconjugation methods,[6] and linker structures[10] used for their preparation. The catalytic activity of ArMs need not depend on the original function of the scaffold protein, so one can instead focus on other properties, including expression level, thermostability, and organic solvent tolerance, that improve biocatalyst utility[11]. Furthermore, because scaffold engineering will likely be required to achieve high selectivity,[12] and because a given scaffold protein may provide a better starting point for such efforts,[9] the ability to select among different scaffolds offers a significant advantage.
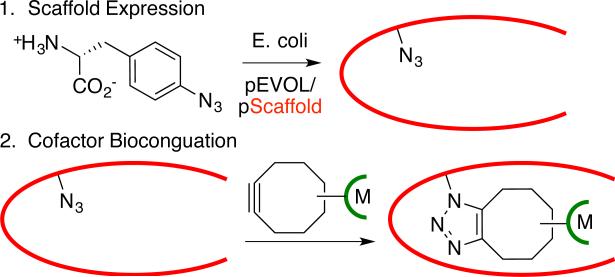
Of the methods outlined above, covalent linkage places the fewest limitations on the nature of the metal catalyst and scaffold protein that can be used for ArM formation.[6] Both metal ion/complex binding and non-covalent linkage require molecular recognition by specific proteins.[4,5] However, selective covalent modification of natural amino acids[6] requires uniquely reactive residues within a protein. Herein, we demonstrate that strain-promoted azide-alkyne cycloaddition (SPAAC)[13] provides a simple means to introduce metal catalysts into proteins (Scheme 1) with the broad scaffold scope of covalent modification while eliminating the constraints of naturally occurring anchor residues (poor reactivity, selectivity, etc.). Several different ArMs were readily constructed using this approach, which should provide an efficient means for generating catalysts for a range of reactions.
Scheme 1.
ArM preparation via SPAAC
SPAAC-based modification of proteins involves expressing a target protein with a genetically encoded azide- or alkyne-bearing amino acid[14] and reacting this protein with an alkyne- or azide-bearing reagent[15]. The high rate, bioorthogonality, and site-specificity of this reaction have led to its wide adoption in chemical biology,[16] but incorporating metal catalysts into proteins has not been described. In general, this requires that the SPAAC reaction occur on the protein interior, rather than its surface,[17] to allow interactions between the protein and the metal secondary coordination sphere (Scheme 1).[18] This is not typically required for the chemical biology applications noted above and demands robust bioconjugation methodology. We envisioned that the central pore of an α,β-barrel protein could provide a suitable environment for azide or alkyne incorporation.[19] Because SPAAC cofactors would be unreactive toward native amino acid functionality, any desired scaffold could be employed for ArM formation.
Reetz and co-workers demonstrated that a cysteine residue in the thermostable α,β-barrel protein tHisF from Thermotoga maritima can be used to introduce a variety of ligands commonly used in transition metal catalysis and even a Pd-complex into this protein, though no activity of the resulting hybrid catalysts was described.[20] Because the bottom of this protein is blocked by a salt-bridge, we hoped that azide or alkyne mutations introduced deep within the 25 Å long central pore of the protein should project SPAAC cofactors up the pore and place the metal complexes within reach of several loops near the pore opening (Scheme 1, Fig. 1A/B, see also supporting information).[21] Extensive biochemical characterization of this protein has revealed that its fold is highly stable, and that it possesses a tryptophan and four tyrosine residues that enable spectroscopic characterization of its folding, both features that make it an ideal test substrate for bioconjugation method development.[22]
Figure 1.
A/B) Structure of wt-tHisF (PDB number 1THF[21a]); colored residues are positions 199 (blue), 50 (orange), and 176 (red). C) HR-ESI-MS of wt-tHisF, tHisF-Az50, and tHisF-Az50-RhBCN. D) Fluorescence spectra (290 nm) of wt-tHisF, tHisF-Az50 (in buffer, 60% ACN, 6M GdmCl), and tHisF-Az50-RhBCN (ACN = acetonitrile; GdmCl = guanidinium chloride).
We used amber stop codon suppression[14] to incorporate p-azido-L-phenylalanine (Az) at representative positions at the top (residue 176), middle (residue 50), and bottom (residue 199) of the central pore of tHisF (A50, A176, and A199)[21]. We observed high levels of scaffold expression and unnatural amino acid incorporation with no apparent azide photolysis based on high resolution ESI mass spectrometry (Fig. 1C), despite A50 and A199 being located on the protein interior[23]. The hexa-histidine tagged scaffold proteins were purified by Ni-affinity chromatography following an initial heat treatment,[20] and both fluorescence (Fig. 1D) and CD spectroscopy (Fig. S5) indicated that little structural perturbation resulted from UAA incorporation.[22] No change in the fluorescence spectrum was observed even in 60% acetonitrile (Fig. 1D), which highlights the organic solvent tolerance of this scaffold protein.[11] A similar approach was used to express variants of a thermostable phytase from Bacillus amyloliquefaciens[24] with Az incorporated at residue 104. This enzyme has an overall cylindrical shape built from six sheets of four-five anti-parallel β-strands arranged around a central pore. The position of the Az residue was approximately 20 Å down this pore, so point mutations N99A, N100A, and D102A, were introduced to facilitate BCN access to the Az residue.
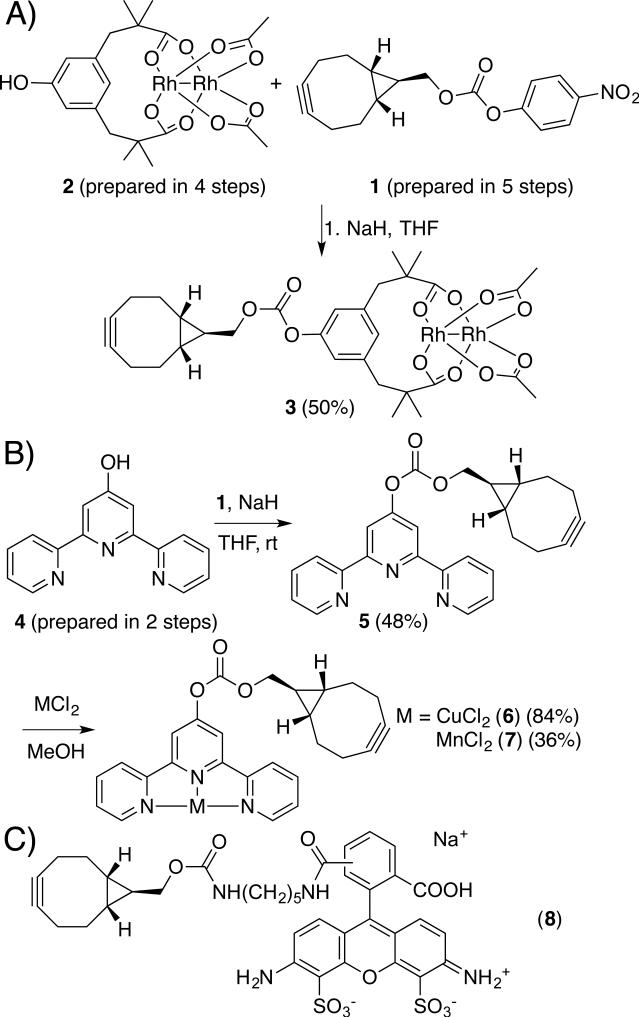
We next developed a modular approach to synthesize alkyne-substituted cofactors. While several alkynes have been developed for SPAAC, we used bicyclo[6,1,0]nonyne (BCN) described by van Delft and co-workers[25] due to its small size, symmetry, and high SPAAC rates[13]. We used carbonate 1[25] as a mild electrophile to which metal complexes bearing a nucleophile could be added (Scheme 2A). We initially targeted dirhodium tetracarboxylate cofactors due to the high activity of these complexes toward a range of carbene insertion reactions[26] that tolerate both air and water[27]. Inspired by the improvements in dirhodium catalysis shown by Du Bois and coworkers using tetramethyl m-benzenedipropionic acid ligands (esp),[28] we prepared hydroxy-esp derivative 2 and reacted this compound with Rh2(TFA)2(OAc)2[29] to form the mixed esp/diacetate complex 3[30] (Scheme 2).
Scheme 2.
Syntheses of cofactors 3, 6, and 7; structure of probe 8.
Two additional BCN cofactors, 6 and 7, containing Cu[31] and Mn[32] terpyridine complexes, were prepared by metallating BCN-terpyridine 5, which was formed from phenol 4 and carbonate 1 (Scheme 2B). Similar metal-terpyridine complexes are known to catalyze a range of C-H insertion reactions.[33] This metallation approach compliments the convergent approach used to prepare 3, and provides additional flexibility for BCN cofactor formation to accommodate the unique reactivity of different metal complexes. Finally, fluorescent probe 8 was prepared in analogy to the approach developed by van Delft (Scheme 2C).[25] The carbonate linkage in all of these cofactors was not hydrolysed even after extended room temperature incubation in various aqueous buffers (e.g. ACN/TRIS or THF/KPi, pH=7.5) based on HPLC analysis.
The reactivity of cofactor 3 toward tHisF-Az50 was then explored. A solution of 3 in acetonitrile (20% v/v ACN/Tris buffer; 5 equiv. 3) was added to a solution of each tHisF mutant (60 M), and the reactions were incubated at 4 °C. ArM formation was monitored by MALDI mass spectrometry, and cofactor consumption was followed by HPLC (the scaffold and ArM could not be resolved). This analysis revealed a depth dependent rate of bioconjugation and final conversions ranging from 50% for Az199 (bottom) to 80% for Az176 (top) (Table 1). While lower temperatures decreased bioconjugation rate, the overall conversion was higher due to decreased ArM denaturation and precipitation over the course of the reaction, so these conditions were utilized for a preparative scale bioconjugation reaction for ArM isolation and characterization. After maximum conversion was observed by MALDI mass spectrometry, the ArM/scaffold mixture was purified by preparative HPLC to remove all traces of unreacted cofactor. While the ArM could not be separated from the scaffold, this has no impact on catalysis since only the ArM contains the cofactor required for catalytic activity. The isolated ArM/scaffold mixture was then characterized by ESI mass spectrometry and HPLC to confirm the composition of the ArM (Fig. 1C) and the absence of free cofactor. Analysis of the ArM/scaffold mixture by fluorescence and circular dichroism spectroscopy also showed no noticeable difference from a sample of pure scaffold, again indicating little if any perturbation to the scaffold structure as a result of bioconjugation (Fig. 1D and Fig. S5).
Table 1.
Mass spectrometry and conversion data for ArMs
| Scaffold (MW)[a] | Cofactor (MW) | MWArM[a] | MWObs[b] | Conv.(%)[b] |
|---|---|---|---|---|
| tHisF-Az50 (28859) | 3 (792) | 29651 | 29614 | 70 |
| tHisF-Az176 (28857) | 3 (792) | 29649 | 29630 | 80 |
| tHisF-Az199 (28857) | 3 (792) | 29649 | 29620 | 50 |
| tHisF-Az176 (28857) | 6 (560) | 29417 | 29435 | 90 |
| tHisF-Az176 (28857) | 7 (550) | 29407 | 29392 | 80 |
| phytase-Az104 (40040) | 3 (792) | 40832 | 40846 | 90 |
Calculated protein MW using tools at www.expasy.org.
Observed MW and approximate conversion from MALDI mass spectrometry.
To demonstrate the generality of our SPAAC ArM approach, we also covalently linked cofactor 3 to sites 176 and 199 of tHisF (Fig. 1) and to Az104 in the central pore of the engineered phytase scaffold. Cofactors 6 and 7 were then linked to tHisF-Az50. In all cases, formation of the desired ArM was confirmed by MALDI mass spectrometry following purification (Table 1). Together, these examples are the first in which the SPAAC reaction has been used to link metal catalysts to proteins. The mild conditions required for this reaction are ideally suited for transition metal cofactors, which may react under other bioconjugation conditions. Also, unlike most SPAAC reactions,[16] which are typically conducted on the surface of proteins, these reactions are conducted within the barrel of the protein, illustrating the high efficiency of this reaction, even when the azide is not exposed to bulk solvent.
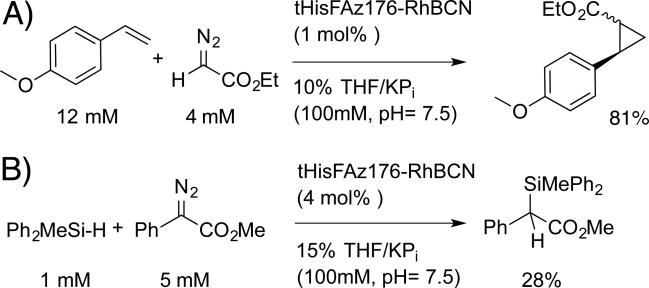
We next evaluated the catalytic activity of purified tHisF-RhBCN ArMs toward a number of dirhodium-catalyzed carbene insertion reactions.[26] Qualitatively, ArM-dependent diazo (e.g. ethyl diazoacetate) decomposition was readily apparent based on visual inspection of the disappearance of the yellow diazo color. Both intermolecular cyclopropanation and Si-H insertion reactions were catalyzed by tHisF-Az50, -Az176, and -Az199 (see Scheme 4 for representative examples), with decreasing conversions observed as the cofactor linkage site was lowered within the scaffold (see Table S3).
Scheme 4.
tHisF-RhBCN catalyzed A) cyclopropanation and B) Si-H insertion
Unfortunately, the ArMs provided lower conversions than cofactor alone (2-OAc provided 99% and 80% yields for cyclopropanation and Si-H insertion, respectively), negligible enantioselectivity, and significant amounts of diazo insertion into the water O-H bond (see Table S3).[27] While a number of selective ArM catalyzed reactions have been reported,[Error! Bookmark not defined.8] achieving such selectivity remains challenging,[12] and improving metal-protein interactions via directed evolution[9,34] will generally be required. The mass spectrometry and spectroscopic data obtained confirm the composition of the ArMs prepared. These data, however, do not provide information on the orientation of cofactors within the scaffolds, and a lack of selectivity could result from cofactors projecting the wrong way out of tHisF and into solution[18] Establishing cofactor orientation is difficult since the site of bioconjugation is distal to the metal catalyst but could provide insight into the poor selectivity observed for the RhBCN ArMs. While detailed structural characterization (i.e. single crystal X-ray crystallography) is underway, we reasoned that surrogate fluorescent probe 8 could also provide quantitative information about cofactor orientation.
Specifically, to establish whether cofactors linked to the central pore of tHisF protrude up through the central pore or down through the bottom of the protein (Fig. 2), we utilized a dual-label FRET approach using surrogate fluorescent probe 8.[35] Cysteine point mutations were introduced at the top (D174C, Fig. 2B) and bottom (D243C, Fig. 2C) of the tHisF-Az50 scaffold exterior. These proteins were reacted with a commercially available Alexa Fluor 594 maleimide probe. A BCN-conjugated Alexa Fluor 488 probe 8 was prepared [25] and reacted with the tHisF-Alexa 488 conjugates. Energy transfer from the Alexa Fluor 488 donor excited at 495 nm to the Alexa 594 acceptor was then measured using both fluorescence intensity and lifetime methods and used to calculate approximate distances between the pore-linked (Az50) donor dye and the exterior-linked (C174 and C243) acceptor dyes (Table 2).[36] Both intensity and lifetime measurements provided similar results consistent with the relative positions and linker lengths used, and both indicated that that the pore linked dye resided substantially closer to the top of the tHisF than to the bottom.[35]
Figure 2.
A) Cartoon schematic of tHisF. B) “Top” (A50/C174) and C) “bottom” (A50/C343) dual-labeled constructs for FRET analysis
Table 2.
FRET data for dual-labeled scaffolds
| Mutant (+Az50) | Energy transfer | Distance (r) in Å | ||
|---|---|---|---|---|
| Intensity | Lifetime | Intensity | Lifetime | |
| D174C (top) | 0.54 | 0.63 | 58 | 55 |
| K243C (bottom) | 0.11 | 0.15 | 88 | 80 |
Given the identical BCN linkage in probe 8 and cofactors 3, 6, and 7, these data provide good evidence for BCN cofactor projection up through the top of the scaffold as intended. We believe the effective length of 3 upon bioconjugation (ca. 20 Å from the α-carbon of the Az residue to either Rh atom), places the metal complex near the mouth of the α,β-barrel (see Fig. S10). The lower yields of reactions catalysed by the dirhodium ArMs relative to those catalysed by 2-OAc suggest that repulsive ArM-substrate interactions occur during catalysis, but these are insufficient to impart selectivity in the reactions studied. Modified cofactor designs and alternate scaffolds that should correct this problem are currently being explored, and the FRET approach outlined above will enable rapid validation of these ArM designs.
We have established that the SPAAC reaction[13] can be used to generate ArMs from scaffold proteins containing a genetically encoded Az residue and catalytically active BCN-linked cofactors. The high efficiency of this reaction allows for rapid ArM formation even when the Az residue is located within, rather than on the surface, of the scaffold protein, which enables the possibility of engineering the scaffold to tune the secondary coordination sphere[18] of the metal catalyst. The bioorthogonality of SPAAC allows for bioconjugation in the presence of cysteine residues[37] in the scaffold, so no additional scaffold modification is necessary for ArM formation. While these properties are widely exploited for various applications in chemical biology, they have not yet been employed for ArM formation and thus provide a number of opportunities for catalysis. We have demonstrated the scope of this method with respect to both the scaffold (tHisF[21] and phytase[24]) and cofactor (Rh2-tetraacetate[26] and Mn- and Cu-terpyridine[33] complexes) components. We also established that the dirhodium ArMs can catalyze the decomposition of diazo compounds and both Si-H and olefin insertion reactions involving these compounds.[26] The simplicity and modularity of our SPAAC approach should facilitate rapid optimization of the ArMs reported herein for selective catalysis and this work is currently underway in our laboratory.
Supplementary Material
Acknowledgements
This work was supported by the Searle Scholars Program, the David and Lucile Packard Foundation, and the NSF under the CCI Center for Selective C–H Functionalization, CHE-1205646. We thank Prof. Peter G. Schultz for providing pEVOL-pAzF and Prof. Dr. Reinhard Sterner for providing pET11c tHisF. We also thank Dr. Justin Jureller at the UC Institute for Biophysical Dynamics NanoBiology Facility (NIH Grant 1S10RR026988-01) and Dr. Elena Solomaha at the UC Biophysics Core Facility for assistance.
Footnotes
Supporting information for this article is available on the WWW
References
- 1.a Mahatthananchai J, Dumas AM, Bode JW. Angew. Chem. Int. Ed. 2012;51:10954–10990. doi: 10.1002/anie.201201787. [DOI] [PubMed] [Google Scholar]; b Strohmeier GA, Pichler H, May O, Gruber-Khadjawi M. Chem. Rev. 2011;111:4141–4164. doi: 10.1021/cr100386u. [DOI] [PubMed] [Google Scholar]; c Zecchina A, Groppo E, Bordiga S. Chem. Eur. J. 2007:2440–2460. doi: 10.1002/chem.200600678. [DOI] [PubMed] [Google Scholar]
- 2.a Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Weeks AM, Chang MCY. Biochemistry. 2011;50:5404–5418. doi: 10.1021/bi200416g. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Sasmal PK, Carregal-Romero S, Parak WJ, Meggers E. Organometallics. 2012;31:5968–5970. [Google Scholar]
- 3.a Lewis JC. ACS Catal. Just Accepted Manuscript, DOI: 10.1021/cs400806a. [Google Scholar]; b Rosati F, Roelfes G. ChemCatChem. 2010;2:916–927. [Google Scholar]; c Heinisch T, Ward TR. Curr. Opin. Chem. Biol. 2010;14:184–199. doi: 10.1016/j.cbpa.2009.11.026. [DOI] [PubMed] [Google Scholar]; d Lu Y, Yeung N, Sieracki N, Marshall NM. Nature. 2009;460:855–862. doi: 10.1038/nature08304. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Abe S, Ueno T, Watanabe Y. Top. Organomet. Chem. 2009;25:25–43. [Google Scholar]
- 4.a Jing Q, Kazlauskus RJ. ChemCatChem. 2010;2:953–957. [Google Scholar]; b Popp B, Ball Z. J. Am. Chem. Soc. 2010;132:6660–6662. doi: 10.1021/ja101456c. [DOI] [PubMed] [Google Scholar]; c Liu X, Yu Y, Hu C, Zhang W, Lu Y, Wang J. Angew. Chem. Int. Ed. 2012;51:4312–4316. doi: 10.1002/anie.201108756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.a Wilson ME, Whitesides GM. J. Am. Chem. Soc. 1978;100:306–307. [Google Scholar]; b Mao J, Ward TR. Chimia. 2008;62:956–961. [Google Scholar]; c Allard M, Dupont C, Robles VM, Doucet N, Lledos A, Maréchal JD, Urvoas A, Mahy JP, Ricoux R. ChemBioChem. 2011;13:240–251. doi: 10.1002/cbic.201100659. [DOI] [PubMed] [Google Scholar]
- 6.a Kaiser ET, Lawrence DS. Science. 1984;226:505–511. doi: 10.1126/science.6238407. [DOI] [PubMed] [Google Scholar]; b Reetz MT, Peyralans JJP, Maichele A, Fu Y, Maywald M. Chem. Commun. 2006;41:4318–4320. doi: 10.1039/b610461d. [DOI] [PubMed] [Google Scholar]; c Deuss PJ, Popa G, Botting C, Laan W, Kamer PCJ. Angew. Chem. Int. Ed. 2010;49:5315–5317. doi: 10.1002/anie.201002174. [DOI] [PubMed] [Google Scholar]; d Heeten R, Muñoz BK, Popa G, Laan W, Kamer PCJ. Dalton Trans. 2010;39:8477–8483. doi: 10.1039/c0dt00239a. [DOI] [PubMed] [Google Scholar]
- 7.a Pierron J, Malan C, Creus M, Gradinaru J, Hafner I, Ivanova A, Sardo A, Ward TR. Angew. Chem. Int. Ed. 2008;47:701–705. doi: 10.1002/anie.200703159. [DOI] [PubMed] [Google Scholar]; b Carey JR, Ma SK, Pfister TD, Garner DK, Kim HK, Abramite JA, Wang ZL, Guo ZJ, Lu Y. J. Am. Chem. Soc. 2004;126:10812–10813. doi: 10.1021/ja046908x. [DOI] [PubMed] [Google Scholar]
- 8.a Lo C, Ringenberg MR, Gnandt D, Wilson Y, Ward TR. Chem. Commun. 2011;47:12065–12067. doi: 10.1039/c1cc15004a. [DOI] [PubMed] [Google Scholar]; b Abe S, Hirata K, Ueno T, Morino K, Shimizu N, Yamamoto M, Takata M, Yashima E, Watanabe Y. J. Am. Chem. Soc. 2009;131:6958–6960. doi: 10.1021/ja901234j. [DOI] [PubMed] [Google Scholar]
- 9.Reetz MT. Top. Organomet. Chem. 2009;25:63–92. [Google Scholar]
- 10.Pordea A, Ward TR. Chem. Commun. 2008;36:4239–4249. doi: 10.1039/b806652c. [DOI] [PubMed] [Google Scholar]
- 11.a Groger H, Asano Y, Bornscheuer UT, Ogawa J. Chem Asian J. 2012;7:1138–1153. doi: 10.1002/asia.201200105. [DOI] [PubMed] [Google Scholar]; b Huisman GW, Collier SJ. Curr. Opin. Chem. Biol. 2013;17:284–292. doi: 10.1016/j.cbpa.2013.01.017. [DOI] [PubMed] [Google Scholar]
- 12.Reetz MT. Tetrahedron. 2002;58:6592–6602. [Google Scholar]
- 13.a Jewett JC, Bertozzi CR. Chem. Soc. Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Debets MF, Van Berkel SS, Dommerholt J, Dirks AJ, Rutjes FPJT, Van FL. Delft, Acc. Chem. Res. 2011;44:805–815. doi: 10.1021/ar200059z. [DOI] [PubMed] [Google Scholar]
- 14.a Xie J, Schultz PG. Methods. 2005;36:227–238. doi: 10.1016/j.ymeth.2005.04.010. [DOI] [PubMed] [Google Scholar]; b Nguyen DP, Lusic H, Neumann DP, Kapadnis PB, Deiters A, Chin JW. J. Am. Chem. Soc. 2009;131:8720–8721. doi: 10.1021/ja900553w. [DOI] [PubMed] [Google Scholar]; c Wang Q, Parrish AR, Wang L. Chem. Biol. 2009;16:323–336. doi: 10.1016/j.chembiol.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Plass T, Milles S, Schultz C, Lemke EA. Angew. Chem. Int. Ed. 2011;50:3878–3881. doi: 10.1002/anie.201008178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a Laughlin ST, Baskin JM, Amacher SL, Bertozzi CR. Science. 2008;320:664–667. doi: 10.1126/science.1155106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Ning XH, Guo J, Wolfert MA, Boons GJ. Angew. Chem., Int. Ed. 2008;47:2253–2255. doi: 10.1002/anie.200705456. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Thomas JD, Cui H, North PJ, Hofer T, Rader C, Burke TR. Bioconjugate Chem. 2012;23:2007–2013. doi: 10.1021/bc300052u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bos J, Fusetti F, Driessen AJM, Roelfes G. Angew. Chem. Int. Ed. 2012;51:7472–7475. doi: 10.1002/anie.201202070. [DOI] [PubMed] [Google Scholar]
- 18.a Davies CL, Dux EL, Duhme-Klair AK. Dalton Trans. 2009;46:10141–10154. doi: 10.1039/b915776j. [DOI] [PubMed] [Google Scholar]; b Hadt RG, Sun N, Marshall NM, Hodgson KO, Hedman B, Lu Y, Solomon EI. J. Am. Chem. Soc. 2012;134:16701–16716. doi: 10.1021/ja306438n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wierenga RK. FEBS Lett. 2001;492:192–198. doi: 10.1016/s0014-5793(01)02236-0. [DOI] [PubMed] [Google Scholar]
- 20.Reetz MT, Rentzsch M, Pletsch A, Taglieber A, Hollmann F, Mondiere RJG, Dickmann N, Hocker B, Cerrone S, Haeger MC, Sterner R. ChemBioChem. 2008;9:552–564. doi: 10.1002/cbic.200700413. [DOI] [PubMed] [Google Scholar]
- 21.a Lang DA, Obmolova G, Thoma R, Kirschner K, Sterner R, Wilmanns M. Science. 2000;289:1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]; b Douangamath A, Walker M, Beismann-Driemeyer S, Vega-Fernandez MC, Sterner R, Wilmanns M. Structure. 2002;10:185–193. doi: 10.1016/s0969-2126(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 22.Carstensen L, Zoldak G, Schmid FX, Sterner R. Biochemistry. 2012;51:3420–3432. doi: 10.1021/bi300189f. [DOI] [PubMed] [Google Scholar]
- 23.Krueger AT, Imperiali B. Chembiochem. 2013;14:788–799. doi: 10.1002/cbic.201300079. [DOI] [PubMed] [Google Scholar]
- 24.Ha NC, Oh BC, Shin S, Kim HJ, Oh TK, Kim YO, Choi KY, Oh BH. Nat. Struct. Biol. 2000;7:147–153. doi: 10.1038/72421. [DOI] [PubMed] [Google Scholar]
- 25.Dommerholt J, Schmidt S, Temming R, Hendriks LJA, Rutjes FPJT, van Hest JCM, Lefeber DJ, Friedl P, van Delft FL. Angew. Chem. Int. Ed. 2010;49:9422–9425. doi: 10.1002/anie.201003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davies HML, Beckwith REJ. Chem. Rev. 2003;103:2861–2904. doi: 10.1021/cr0200217. [DOI] [PubMed] [Google Scholar]
- 27.a Nicolas I, Le Maux P, Simonneaux G. Coord. Chem. Rev. 2008;252:727–735. [Google Scholar]; b Wurz RP, Charette AB. Org. Lett. 2002;4:4531–4533. doi: 10.1021/ol0270879. [DOI] [PubMed] [Google Scholar]; c Antos JM, McFarland JM, Iavarone AT, Francis MB. J. Am. Chem. Soc. 2009;131:6301–6308. doi: 10.1021/ja900094h. [DOI] [PMC free article] [PubMed] [Google Scholar]; d Tishinov K, Schmidt K, Häussinger D, Gillingham DG. Angew. Chem. Int. Ed. 2012;51:12000–12004. doi: 10.1002/anie.201205201. [DOI] [PubMed] [Google Scholar]
- 28.Espino CG, Fiori KW, Kim M, Du Bois J. J. Am. Chem. Soc. 2004;126:15378–15379. doi: 10.1021/ja0446294. [DOI] [PubMed] [Google Scholar]
- 29.Lou Y, Remarchuk TP, Corey EJ. J. Am. Chem. Soc. 2005;127:14223–14230. doi: 10.1021/ja052254w. [DOI] [PubMed] [Google Scholar]
- 30.Sambasivan R, Ball ZT. J. Am. Chem. Soc. 2010;132:9289–9291. doi: 10.1021/ja103747h. [DOI] [PubMed] [Google Scholar]
- 31.Suntharalingam K, White AJP, Vilar R. Inorg. Chem. 2010;49:8371–8380. doi: 10.1021/ic100884p. [DOI] [PubMed] [Google Scholar]
- 32.Chen H, Tagore R, Das S, Incarvito C, Faller JW, Crabtree RH, Brudvig GW. Inorg. Chem. 2005;44:7661–7670. doi: 10.1021/ic0509940. [DOI] [PubMed] [Google Scholar]
- 33.a Das S, Incarvito CD, Crabtree RH, Brudvig GW. Science. 2006;312:1941–1943. doi: 10.1126/science.1127899. [DOI] [PubMed] [Google Scholar]; b Kwong HL, Lee WS. Tetrahedron-Asymmetry. 2000;11:2299–2308. [Google Scholar]
- 34.a Tracewell CA, Arnold FH. Curr. Opin. Chem. Biol. 2009;13:3–9. doi: 10.1016/j.cbpa.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Reetz MT, Rentzsch M, Pletsch A, Maywald M, Maiwald P, Peyralans JJP, Maichele A, Fu Y, Jiao N, Hollmann F, Mondiere R, Taglieber A. Tetrahdron. 2007;63:6404–6414. [Google Scholar]
- 35.Roy R, Hohng S, Ha T. Nat. Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3rd ed. Springer; New York: 2006. pp. 443–472. [Google Scholar]
- 37.Stephanopoulos N, Francis MB. Nat. Chem. Biol. 2011;7:876–884. doi: 10.1038/nchembio.720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.