Abstract
Background
Echinodermata is a diverse Phylum, a sister group to chordates, and contains diverse organisms that may be useful to understand varied mechanisms of germ-line specification.
Results
We tested 23 genes in development of the sea star Patiria miniata that fall into five categories: 1) Conserved germ-line factors; 2) Genes involved in the inductive mechanism of germ-line specification; 3) Germ-line associated genes; 4) Molecules involved in left-right asymmetry; and 5) Genes involved in regulation and maintenance of the genome during early embryogenesis. Overall, our results support the contention that the posterior enterocoel is a source of the germ line in the sea star P. miniata.
Conclusion
The germ line in this organism appears to be specified late in embryogenesis, and in a pattern more consistent with inductive interactions amongst cells. This is distinct from the mechanism seen in sea urchins, a close relative of the sea star clad. We propose that P. miniata may serve as a valuable model to study inductive mechanisms of germ-cell specification and when compared to germ-line formation in the sea urchin S. purpuratus may reveal developmental transitions that occur in the evolution of inherited and inductive mechanisms of germ-line specification.
Introduction
Evolutionary changes have resulted in a diverse series of mechanisms to accomplish the task of germ-line specification. Three extremes in these mechanisms are widely recognized in the animal kingdom and include 1) germ-cell derivation from adult multipotent stem cells (e.g. neoblasts in planaria, I-cells in hydra), 2) inheritance of maternal factors in early embryogenesis (e.g. pole plasm in Drosophila melanogaster, germ plasm in Xenopus laevis), and 3) cell-cell communication resulting in induction of a germ-line lineage (e.g. mouse and axolotl; Extavour and Akam, 2003; Solana, 2013). Because these different mechanisms of germ-line specification are polyphyletic, transitions between these germ-line specification mechanisms appear to have occurred multiple times within animal evolution (Extavour and Akam, 2003).
When an embryo exhibits a germ-line determination mechanism that exceeds one biological threshold or another, the investigator usually classifies that mechanism as either inductive or inherited. While this is important for ease and clarity in communication it does not reflect the biological mechanism(s) effectively. Since germ-line determination is likely a result of multiple parallel pathways and activities leading to determination, the transition from one state to another in an embryo may result from a continuum of changes instead of a series of binary switches. Part of the problem with our current definitions of germ-line specification results from the fact that most of what we know comes from a small set of animals; mostly developing by an inherited mechanism. These animals include D. melanogaster, Caenorhabditis elegans, X. laevis, and Danio rerio, each with rich contributions of genetic, biochemical, and embryological analysis (e.g Seydoux and Braun, 2006; Gao and Arkov, 2013; Voronina, 2012; Lai and King, 2013). Unfortunately, we do not have as rich a data set for understanding the inductive mechanisms of germ-line determination. The mouse is the best understood model of inductive germ line determination and in this embryo, the epigenetic changes essential to prime PGC formation occurs as a result of signaling between cells (e.g. Magnúsdóttir et al., 2012). This epigenetic reconfiguration then diverts the fate of the cells from a somatic direction, and instead leads to a germ line fate. Whether these features seen in mice are conserved in other organisms using inductive mechanisms is not yet clear and having a strong data set of inductive germ-line features from multiple different animals will be important in order to establish a baseline from which comparisons can be made between inherited mechanisms. Reaching this goal may then reveal the features of germ cell specification shared by inductive mechanisms, those that are unique to inherited and inductive mechanisms, and how evolutionary transitions between each mechanism may occur.
Preliminary experiments in sea stars suggest that this animal uses inductive mechanisms to specify their germ cells. The conserved germ-line factor Vasa becomes restricted to the posterior enterocoel (PE; see Figure 1) only by the early larval stage (Juliano and Wessel, 2009). In addition, PE removal experiments show that the cells from this structure contribute to the future germ cell lineage (Inoue et al., 1992). This is very different than what is known in the closely related taxon of sea urchins. Sea urchins specify their germ cells as early as the 32-cell stage when 4 small micromere cells (sMics) arise as the product of two unequal cell divisions (see Figure 1). Multiple germ cell marker RNAs and proteins accumulate in the sea urchin sMic lineage (Juliano et al., 2006; Voronina et al., 2008; Wessel et al., 2013), they express these markers cell autonomously (Yajima et. al., 2012), and removal of these cells results in loss of the germ lineage in adults (Yajima and Wessel, 2011). In comparison to sea urchins, sea stars and all other groups within echinoderms do not have a sMic lineage. This leads us to believe the sMic lineage and the associated mode of early germ-line specification arose independently in the echinoid (sea urchin) lineage from a common ancestor that used inductive processes for germ-line determination.
Figure 1.
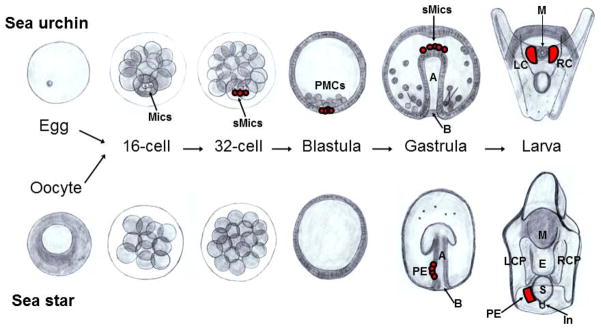
Schematic representation of the developmental stages in sea star and sea urchin. Sea urchins exhibit two asymmetric cleavage events within the cells at the vegetal pole (bottom). These asymmetric divisions result in 4 micromeres forming at the 16-cell stage, followed by 4 large and 4 small micromeres (sMics, in red) forming at the 32-cell stage. The PMC (descendants of the large micromeres) ingress to form the skeleton, whereas descendants of the sMics remain relatively quiescent through embryogenesis, divide only once before gastrulation, and then integrate into the larval coelomic pouches where the adult rudiment forms. Unlike in sea urchins, the sea star has symmetrical cell divisions and does not segregate its germ-line cells during early development. Instead, a PE (in red) projects from the dorsal wall of the archenteron into the blastocoel and then moves to the left side in late gastrula-early larval stages. Left and right anterior (top) coelomic pouches are present on both sides of the esophagus in larvae, and they subsequently extend posteriorly. Later in development, the left coelomic pouch integrates cells of the PE as it extends posteriorly. The blastopore is located at the lower opening of each embryo in the gastrula stage. Mics=Micromeres, sMics=Small micromeres, PMCs=Primary mesenchyme cells, LCP=Left coelomic pouch, RCP=Right coelomic pouch, PE=Posterior enterocoel, LC=Left coelom, RC=Right coelom, M=Mouth, E=Esophagus, S=Stomach, In=Intestine, B=Blastopore, A=Archenteron.
We hypothesize that sea stars use an inductive mechanism of germ cell specification that may represent the ancestral mode of germ cell specification in echinoderms (Extavour and Akam, 2003). To begin to test this hypothesis we undertook the current study with the goal of analyzing the expression of germ cell factors in sea stars based on candidate genes that are important for both inductive and inherited modes of germ cell specification in other organisms. In this way we can compare the expression patterns of germ-line associated markers between sea stars, sea urchins, and other model organisms – especially mice - to test if sea stars specify their germ line in an inductive mode and if so, test if this mode has mechanisms conserved over long periods of evolutionary time.
The genes studied here fall into five categories: 1) Conserved germ-line factors; 2) Genes involved in the inductive mechanism of germ-line specification; 3) Germ-line associated genes; 4) Molecules involved in left-right asymmetry; and 5) Genes involved in regulation and maintenance of the genome during early embryogenesis. Overall, our results support the contention that the PE is a source of the germ line in the sea star P. miniata and that there is no specific accumulation of germ cell markers in any cells prior to PE formation. Our results lead us to conclude that the endomesoderm retains the expression of many pluripotency-associated genes which later give rise to the PE. In addition, we found that PE formation and P. miniata PGC specification is likely determined by inductive interactions amongst cells which simultaneously cause both the accumulation of germ cell determinants and the loss of somatic cell markers in the presumptive PE. We propose the sea star may serve as a valuable model for future study of the inductive mechanisms of germ-line determination, and when compared to the data sets in sea urchins already available, may serve as a useful comparative model for understanding the developmental transitions between an inductive germ-line determination mechanism and an inherited mechanism.
Results and Discussion
We selected the genes used in this study by first identifying genes that are associated with PGC specification in a variety of animals that exhibit diverse mechanisms of germ-line determination. Genes involved in both inherited and inductive mechanisms were chosen as well as those involved in left-right asymmetry. This latter group was selected because the hypothesis being tested is that the PE contributes to the primordial germ cells – the PE structure is on the left side of the midline in the larva. We obtained sequences of D. melanogaster and mouse proteins of all the genes tested here from NCBI (http://www.ncbi.nlm.nih.gov/). Orthologous protein sequences from sea urchins were found by BLAST analysis against the published sea urchin database (Spbase.org). P. miniata orthologous protein sequences were found by BLAST analysis against a nascent P. miniata ovary transcriptome database. The top P. miniata hit was used for reciprocal-BLAST analysis to the non-redundant NCBI database to test orthology. Alignments using these orthologous sequences from Mus musculus, D. melanogaster, S. purpuratus and P. miniata were performed to further test authenticity (Tables 1–5 and Supplementary Figure 1). The lists of primers used for PCR amplification of each gene in sea star and the sizes of predicted and acquired PCR products are shown (Tables 1–5).
Table 1.
Conserved Germ-line Determinants
| Genes | Domains and Function |
Reference | Orthologs (organism) |
%Identities, % Similarities |
NCBI or Spbase Reference number |
Pm transcript number |
Primers sequences | Size of amplification product |
|---|---|---|---|---|---|---|---|---|
| Piwi | PAZ (piwi, argonaute, zwille) domain, required for negative regulation of transposable elements in germ cells | Juliano et al., 2006 | Mm-Piwi1L Dm-Argonaute 3, isoform G Sp-Seawi |
51%, 74% 40%, 59% 50%, 71% |
NP_067286.1 NP_001163498.1 AAG42534.1 |
Pm_33095 | F:CGACGGCAGCCAGATCACCTA R:taatacgactcactatagggCCAGGCAGCAGTACTTCTTGA |
728 bp |
| Nanos | CCHC Zn-finger domain, With partmer, Pumilio, serves as a negative regulator of translation in germ cells |
Juliano et al., 2006 Lai et al., 2013. |
Mm-Nanos2 Dm-Nanos Sp-Nanos2 |
54%, 65% 62%, 73% 39%, 50% |
NP_918953.2 NP_476658.1 NP_001073023.1 |
Pm_4079 | F:GGAGATTGAGAGCGAAGAT R:taatacgactcactatagggTGTTGAATTTCATGAGGCAAA |
971 bp |
| Vasa | CCHC Zn-finger and DEAD-box domains, ATP-dependent RNA helicase involved in germ-line specification | Juliano et al., 2006 | Mm-Vasa Dm-Vasa Sp-Vasa |
61%, 76% 53%, 69% 69%, 82% |
NP_034159.1 NP_723899.1 NP_001139665.1 |
Pm_1519 | F:CGGTCCAGAAGTACGGGATA R:taatacgactcactatagggGTAGAAGCTGGTTGCCTTGC |
992 bp |
| Pumilio | Puf domain, With partner Nanos, RNA-binding protein that negatively regulates translation of target RNA in germ cells | Lai et al., 2013. | Mm-Pum2 Dm-PumF Sp-Pum |
55%, 65% 79%, 88% 65%, 71% |
NP_109648.2 NP_001247002.1 SPU_006847 |
Pm_2787 | F:GGTAGTAACATGGGGGACCAG R:taatacgactcactatagggGGCCTTGTTGTTGACCTTGCT |
805 bp |
| Boule/Dazl | RNA binding protein involved in germ-line determination in mammals | Xu et al., 2001. Shah et al., 2010. |
Mm-Boule Dm-Boule Sp-Boule |
59%, 71% 51%, 62% 37%, 46% |
NP_083543.2 NP_729457.1 SPU_008194.1 |
Pm_22341 | F:TCGGTTCATAACTGCCATCA R:taatacgactcactatagggTTATGGCACCCTGGTGAGAG |
925 bp |
Table 5.
Regulation and genomic maintenance during morphogenesis and early embryogenesis.
| Genes | Domains and Function |
Reference | Orthologs (organism) |
%Identities, % Similarities |
NCBI or Spbase Reference number |
Pm trasncript number |
Primers sequence | Size of amplification product |
|---|---|---|---|---|---|---|---|---|
| Brca1 | N-terminus RING finger domain, two nuclear localization signals and an acidic C-terminus. It is involved in DNA repair, transcriptional regulation, chromatin remodeling, cellular growth control and genome stability | Hoshino et al., 2007 | Mm-Brca1 Sp-Brca1 |
34%, 55% 62%, 75% |
NP_033894.3 SPU_011027.3a |
Pm_3175 | F:GGATCTTCCCAGAGTACGACT R:taatacgactcactatagggGGAGCCAAGAGTTGTCAGAGT |
822 bp |
| Brca2 | N-terminus acidic transcriptional activation domain and a C-terminus DNA binding domain. It is involved in maintenance of genomic stability in response to DNA damaging agents | Rodríguez-Marí et al., 2011 | Mm-Brca2 Sp-Brca2 |
36%, 57% 42%, 62% |
NP_001074470.1 SPU_013435 |
Pm_34269 | F:GGAGAAGCACAGCGAGGGAGG R:taatacgactcactatagggGGCTGGAACCCAGCCTGAAGA |
729 bp |
| Baf250 | It has a DNA binding domain called ARID. It is a component of catalytic cores that regulate the expression of Homeobox genes early in development | Li et al., 2010 | Mm-Arid1B Dm-OsaA Sp-Baf250 |
43%, 69% 38%, 52% 75%, 88% |
NP_001078824.1 NP_732263.1 SPU_023530 |
Pm_47085 | F:CCAGTGTGCATGCCCTCAGTA R:taatacgactcactatagggCCTCTCTCCTTAACTGCATGG |
506 bp |
| Maf | Traffic Jam/Maf is a transcription factor that controls gonad morphogenesis. TJ protein activates piwi expression and tj gene is a piRNA cluster which define the Piwi targets for silencing, in Drosophila | Saito et al., 2009 | Mm-MafB Dm-Tjam Sp-Maf |
51%, 72% 57%, 72% 61%, 80% |
NP_034788.1 NP_609969.2 SPU_025888 |
Pm_71778 | F:CCAAGCCTTGATGAGCTCTAT R:taatacgactcactatagggGGACTACTCGGCAAACTAACG |
629 bp |
Conserved germ-line determinants – Select expression in the Posterior Enterocoel
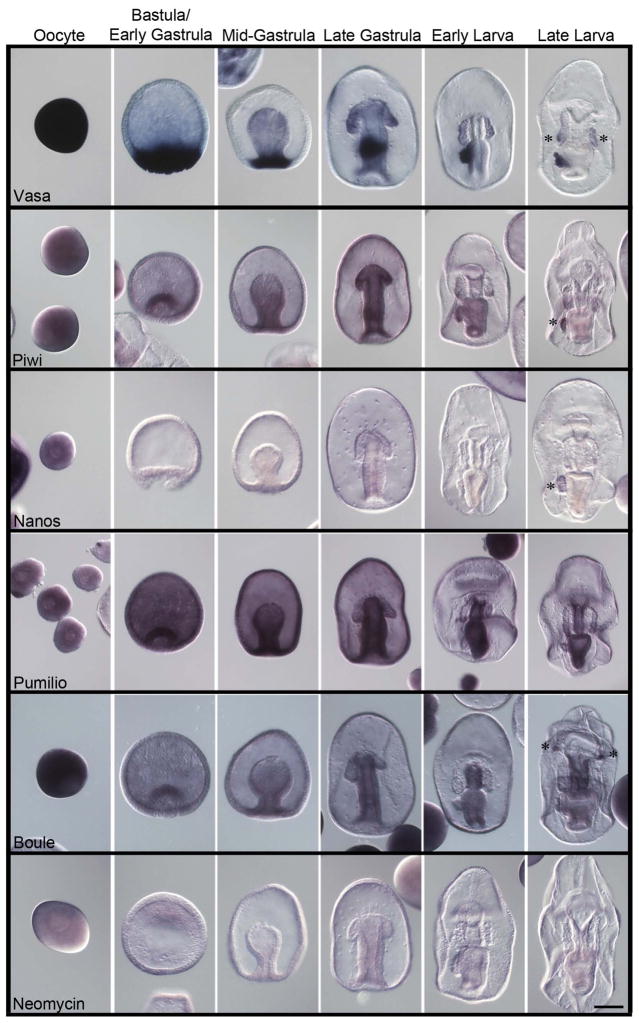
The genes in the first set we explored are those most highly conserved amongst animals as being part of the germ-line determination mechanism (Table 1). Vasa, Nanos, and Piwi are a classical cluster of germ-line factors, found in all animals at some point in the construction or maintenance of a new germ line in both inductive and inherited germ-line formation mechanisms. Vasa is a dead-box helicase involved in regulating the translation of RNAs in the germ-line. We found in the sea star that Vasa gene expression (mRNA accumulation) is ubiquitous in eggs and early embryos and first becomes restricted to the vegetal pole of the blastula. During gastrulation, Vasa mRNA becomes enriched in the middle region of the archenteron and by early larval stages is restricted to the left side of the mid-archenteron where the PE buds. Vasa remains selectively expressed in the PE throughout the development of the larva (Figure 2). We also noted less detectable amounts of Vasa transcripts are present in the perimeter of the left and right coelomic pouches in larval stages (Figure 2; late larva; asterisks). Overall these results support the contention that the PE is at least relevant for consideration of the origin of germ-line determination in this organism. The result also speaks more generally to Vasa function – the transcript is broadly expressed in the egg and early embryo – clearly it is not strictly a germ-line factor. Previous results in sea stars show that Vasa protein accumulates ubiquitously early in development as well. Not until the PE forms does the Vasa protein become restricted (Juliano and Wessel, 2009), much like the expression of its mRNA. This is a particularly important distinction to be made since in sea urchins the protein does not accumulate coincident with the mRNA. Protein translation is widespread in the sea urchin embryo but degradation is selective to the somatic cells. Therefore, the sMics, the PGC lineage in sea urchins, accumulate the Vasa protein selectively even with broad mRNA presence. This selection (degradation of Vasa protein outside of the presumptive PGCs) appears to be a function of at least one E3-ubiquitin ligase activity, Gustavus (Gustafson et al., 2011).
Figure 2.
Expression of conserved germ-line determinants during P. miniata embryonic development. Line 1) Vasa transcripts are widespread in oocytes and become restricted to the vegetal pole in blastula stage embryos. During gastrulation Vasa is then restricted to the center of the archenteron and later to the left side of the archenteron. As soon as the PE is formed Vasa persists in the PE. Line 2) Piwi transcripts are widespread in oocytes, become restricted to the archenteron during gastrulation, then to the center of the archenteron during late gastrulation, and persist in the PE of larva as soon as it is formed. Line 3) Nanos expression is only detected broadly in immature oocytes and in the PE of older larva. Line 4) Pumilio transcripts accumulate broadly throughout the embryo during early development and become enriched in the esophagus and stomach during larval stages. Line 5) Boule transcripts are widespread in oocytes but are not present during development until larval stages. Boule transcripts accumulate in the oral ectoderm/ciliary band and in the esophagus. A sequence for the Neomycin (Neo) resistance gene (Line 6) was used as a negative control for the hybridization procedure. Asterisks (*) represent areas of emphasis for mRNA accumulation. Dorsal views of the larva. Scale bar represents 100 μm.
Piwi is a conserved germ-line factor involved in small RNA-mediated degradation of transposons, and its accumulation is similar to Vasa in P. miniata development. That is, broad early expression with selective accumulation in the PE by the early larval stage (Figure 2). Although the Piwi transcript does not follow as tight an expression domain as Vasa, it is clear that Piwi transcripts accumulate in the archenteron as soon as it is formed and later in gastrulation the Piwi mRNA is enriched in the region where the PE forms - within the mid-region (future mid-gut) of the archenteron (Figure 2, late gastrula, early larva). By the late larval stage Piwi transcripts disappear from the gut, but are retained in the PE (Figure 2, larval stages). In the late larval stage we also note Piwi transcripts in the posterior portions of the left and right anterior coelomic pouches.
In the sea urchin, Piwi expression is much like Vasa, that is, broad early expression with protein accumulation selective to the sMics, and only subsequently during gastrulation does the mRNA become restricted to the sMics (Rodriguez et al., 2005; Juliano et al., 2006). In that respect it is similar in the sea star where the vegetal plate and endodermal tissues are the last to down regulate the Piwi mRNA while being retained in the future germ line, either the sMics in sea urchins or the PE in sea stars (Figure 2, late larva, asterisk). Clearly both Vasa and Piwi gene expression patterns support the contention that the PE contributes to the germ line, and that their expression is not uniquely germ line.
Nanos is another conserved germ-line factor found in all animals studied. Nanos interacts with Pumilio to bind the 3′UTRs of select mRNAs bearing a Pumilio Response Element (PRE, also referred to as Nanos Response Element, NRE). Binding of this complex to the mRNA reduces translation of the encoded protein either through the translational machinery (Cho et al., 2006; Vardy and Orr-Weaver, 2007b) or by degrading the mRNA by recruiting deadenylase activity (Kadyrova et al., 2007; Vardy and Orr-Weaver, 2007a, 2007b). Pumilio is usually expressed more broadly than Nanos and can interact with a variety of regulators of the bound mRNA. Nanos on the other hand is often restricted to the germ line and its expression is associated with decreasing the cell cycle of the PGC through binding of cyclin mRNAs. Mis-expression of Nanos in non-germ line cells will often lead to developmental abnormalities or death in the somatic cells (Lai et al., 2012; Lai and King, 2013). In sea urchins, Nanos is tightly regulated by a variety of means to be specific to the germ line cells (e.g. Oulhen et al., 2013). In the sea star, Pumilio but not Nanos, is broadly expressed throughout much of the embryo early in development. Following gastrulation Pumilio became enriched to the gut but was largely excluded from the coelomic pouches and the PE (Figure 2). Nanos however first appears significantly detectable in the PE when it forms (Figure 2, late larva, asterisk). In this regard, it is like in sea urchins, where Nanos appears only once the sMics form. This is unlike other germ-line marker transcripts that accumulate in larger expression domains prior to restriction (see Vasa and Piwi, Figure 2).
Boule/Dazl expression overlaps that of Pumilio but with distinct characters. Boule gene expression generally is present uniformly in early development with slight enrichment in the endomesoderm. In early larvae, the message is enriched in the esophagus significantly over other regions of the embryo or gut. In later larvae, expression is enriched in the esophagus in addition to a small number of cells (Figure 2, asterisks), which may be the precursors to the dorsal ganglia in late larva (Yankura et al., 2013).
Overall this gene set supports the hypothesis that the PE is a special derivation. In concert, the Piwi, Nanos and Vasa selective mRNA expression, and Vasa protein expression (Juliano and Wessel, 2009) argues that the PE contributes to the germ line. The endomesoderm however, seems to retain many pluripotency-related genes not otherwise present in the ectoderm or future mesodermal cells. Perhaps the endoderm retains broad developmental potency utilizing these various germ cell markers that only later in development become restricted to the PE.
Gene regulatory molecules involved in inductive specification of germ cells in the mouse are conserved in echinoderms
The mechanism of inductive germ-line specification is thought to be the most ancient mechanism used by animals (Extavour and Akam, 2003). Many signaling molecules involved in inductive germ-line specification are not unique to the germ line during animal development and are instead used in a variety of developmental processes. The list of genes involved in the inductive mechanisms of germ-line determination is small – most work thus far has been accomplished only by genetic approaches and only in the mouse. This set of genes is important to test in order to determine if the signaling pathways and downstream effector molecules that are required for inductive germ-line specification in the mouse are conserved in other animals.
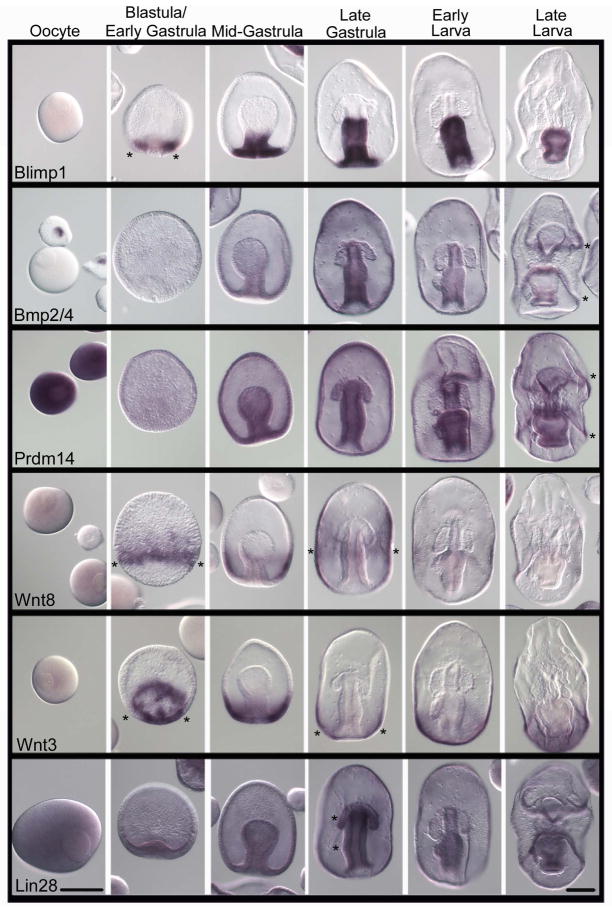
Blimp1 (B lymphocyte-induced maturation protein) also known as Prdm1 (Positive regulatory domain I element of the β-IFN gene promoter, containing Zn-fingers) is a transcriptional regulator. Blimp1 is believed to generally repress its target genes, although some evidence indicates that Blimp1 can also serve as an activator of gene expression in certain contexts (John and Garrett-Sinha, 2009; Magnúsdóttir et al., 2013). Blimp1 was first discovered in the immune system, but it is expressed and functional in many tissues during development (Vincent et al., 2005), and in many animals studied (e.g. de Souza et al., 1999). In mice, Blimp1 is a key factor for germ-line determination (Saitou et al., 2005; Saitou and Yamaji, 2010; Kurimoto et al., 2008; Seervai and Wessel, 2013). Blimp1 appears to repress genes within the presumptive germ cells of mice (including Hox genes, esp. Hoxb1) whereas these same genes remain active in neighboring cells as they begin somatic differentiation. Therefore, Blimp1 in the mouse germ line appears to function in retaining a pluripotent fate by repressing genes important for somatic fates commitments. In contrast, the sea urchin Blimp1 is involved in endoderm gene specification; Blimp1 appears to activate Wnt8 but repress Notch and HesC expression. Knocking down Blimp1 results in a lack of endoderm morphogenesis and perhaps conversion to non-endodermal fates (Livi and Davidson, 2006; Smith and Davidson, 2008; Smith et al., 2008). In the sea star oocyte, Blimp1 transcripts are in low abundance, but in the blastula they accumulate significantly in a torus of cells surrounding the vegetal pole (Figure 3, asterisks; Hinman and Davidson, 2003; Hinman et al., 2007). In mid-gastrulae, expression of Blimp1 extends into the posterior archenteron. In late gastrula and early larval stages, expression is restricted to the midgut and hindgut regions of the archenteron, with a slight clearing in the gut region where the PE will form, followed by a marked absence in the PE. Blimp1 is enriched in the stomach, intestine and anus of the late larva, and enrichment persists later in development to the esophagus. Were Blimp1 to have a conserved function in the sea star as in the mouse, it may be in retaining some potentiality of cell fate in the endoderm. Removing its function may, as in mice, enable cells to differentiate into mesodermal lineages and thereby lose their potential for endodermal and germ-line fates.
Figure 3.
Expression of genes involved in inductive germ-line specification during P. miniata embryonic development. In situ hybridization showing Line 1) Blimp1 transcripts, which first accumulate in the vegetal pole of the blastula, and remain largely endodermal through development. Of note is the rapid loss of Blimp1 in the newly formed PE (see also Figure 7). Line 2) Bmp2/4 transcripts are seen in young oocyte germinal vesicles (oocyte nucleus). During development Bmp2/4 is expressed throughout the embryo except for in the coelomic pouches and PE. Line 3) Prdm14 is present throughout development and largely throughout the embryo with the exception of the coelomic pouches. Lines 4 and 5) Wnt 8 and Wnt 3 (respectively) form a complementary pattern with Wnt3 being more vegetal and Wnt 8 more equatorial in the embryo and larva. No selective accumulation is seen in the coelomic pouches or PE in larval stages. Line 6) Lin28 is expressed broadly during early development until the late gastrula stage when it becomes enriched in the foregut. In larval stages, Lin28 is enriched in the gut and PE and it is distinctly absent from the coelomic pouches. Asterisks (*) show areas of emphasis for transcript detection. Dorsal views of the larva. Scale bar represents 100μm.
Prdm14 is closely related to Blimp1/Prdm1 and is a key regulator of mammalian germ cell development (Yamaji et al, 2008). It plays a critical role in cell fate pluripotency by suppressing the expression of differentiation marker genes. In mice, Prdm14 is expressed early in germ line determination, by 6.5dpc, and its up-regulation is likely in response to Bmp4 signaling. Recently it was shown that Prdm14 functions to ensure pluripotency through two pathways: 1) it antagonizes activation of the fibroblast growth factor receptor (Fgfr) signaling by the core pluripotency transcriptional circuitry, and 2) it represses expression of de novo DNA methyltransferases that would otherwise modify the epigenome to a primed (somatic) epiblast-like state. Prdm14 exerts these effects by recruiting polycomb repressive complex 2 (Prc2) specifically to key genomic sites and repressing nearby gene activity (Yamaji et al, 2013). Prdm14 in the sea star is present widespread and has a very similar expression pattern to Bmp2/4 (below) in embryos and larvae. That is, Prdm14 transcripts accumulate in the archenteron of the gastrula and larvae showed strong Prdm14 staining in the mouth opening, stomach, intestine, anus, and the preoral and postoral ciliary bands, but not in the anterior coeloms nor in the PE (Figure 3, asterisks). In the oocyte, Prdm14 transcripts are ubiquitous (Figure 3). Unlike in mice, Prdm14 in sea stars or sea urchins does not correlate with the accumulation of other PGC markers. Instead, Prdm14 localizes to ciliary bands in urchins and to the embryonic gut and ciliary bands of sea star larva. This leads us to hypothesize that Blimp1 and Prdm14 may be involved in retaining gut pluripotency early in sea star development, which may be an essential role, but this role does not appear to be sequestered to the germ line.
Bmp2/4 is a critical signaling component for germ-line specification in the early mouse embryo (Saitou, et al., 2002; Saitou and Yamaji, 2010). It is expressed by cells in the extra-embryonic tissues and is required for cells in the posterior epiblast to become PGCs. Zhou et al., (2010) also found Bmp4 enhances germ cell derivation in vitro from ES cells, although currently the link is not clear, if any, between the Bmp4 signaling and Blimp1 expression (Toyooka et al., 2003). The sea star, as in sea urchin, appears to have a less diverse family of Bmp signaling molecules and the closest ortholog to the mouse Bmp4 in the sea star is a Bmp2/4 gene (Lapraz et al., 2006; see supplementary Figure 1). Bmp2/4 in sea star is present broadly in development (Figure 3). Remarkably, the Bmp2/4 transcripts are enriched in the nuclei of young oocytes, but not in nuclei of full-grown oocytes or embryonic cells. Some gene expression is regulated by transcript retention in the egg nucleus of the sea urchin (Venezky et al., 1981; Angerer and Angerer, 1981) though this is the first example of selective transcript retention in the germinal vesicle (GV) of a sea star. In gastrula and early larval stages, Bmp2/4 transcripts accumulate very similarly to Prdm14, and show a clear accumulation in the gut of the embryo, but not in the anterior coeloms nor in the PE. In late larvae, Bmp2/4 is expressed largely around the opening of the mouth, anus and the preoral and postoral ciliary bands (Figure 3; asterisks). Recently, another report on Bmp2/4 in P. miniata showed enriched Bmp2/4 expression in the future site of mouth formation in blastula stage embryos (Yankura et al., 2013). We did not see this profile here although it may have been a time point missed in our analysis – we focused on PE formation here and terminated the in situ hybridization analysis to best reveal signal information in later embryos and larvae.
The Wnt signaling pathway is thought to regulate several aspects of germ-line function. These include germ-line stem cell functions (e.g. Golestaneh et al., 2009), germ-cell migration (e.g. Chawengsaksophak et al., 2012), and germ cell-soma interactions in the gonad (e.g. Tanwar et al., 2010). Most importantly, and in regards to mouse inductive PGC specification, Wnt3 signaling is required to prime the cells in the posterior epiblast so they are competent to respond to Bmp4 PGC specification signals (Ohinata et al., 2009). In addition, Wnt8 is a somatic marker in axolotls that is expressed during gastrulation in areas of germ cell precursor formation (Bachvarova et al., 2001, Johnson et al., 2003). We chose to determine the expression patterns of Wnt3 and Wnt8 in the sea star to determine if their expression correlates with PE formation. Moreover, in sea urchins as in many embryos, the Wnt signaling pathway is involved in broad scale axial patterning (e.g. Kumburegama and Wikramanayake, 2009; Stamateris et al., 2010). In sea urchins, the nuclearization of β-catenin (a mark of active canonical Wnt signaling) begins in the micromeres at the 16-cell stage, the precursors to the sMics. β-catenin and Blimp1 further trigger Wnt8 expression (Oliveri et al., 2008; Yamazaki et al., 2012). These expression profiles are common in all indirect-developing sea urchins examined so far (Nakata and Minokawa, 2008; Yamazaki et al., 2010). Furthermore, in sea urchins Wnt8 is the primary component of the early endomesoderm gene regulatory network, and its role is to activate the β-catenin/Tcf signal transduction system (Smith and Davidson, 2008; Oliveri et al., 2008). In sea stars, Wnt8 and Wnt3 expression patterns show a segmented, circumferential pattern with an overlapping border in the posterior third of the ectoderm (Figure 3). As in sea urchins, Wnt8 transcripts are not maternally expressed in the sea star. By the late blastula stage, however, Wnt8 transcript enrichment is seen as a ring around the vegetal half of the embryo (Figure 3; asterisks). This expression pattern continued until mid-gastrula, but its enrichment decreased during early and late larval stages. The Wnt3 expression pattern in P. miniata follows a similar expression pattern as Wnt8, but the circumferential pattern is closer to the vegetal pole of the embryo (Figure 3, asterisks). This pattern continued during the gastrula and larval stages and both genes appeared to have continuous expression domains. Neither of the Wnts is detectable in the invaginated endoderm (where the greatest enrichment occurs for Blimp1, Prdm14, and Bmp2/4 transcripts) nor in the PE (see also Yankura et al., 2013). We conclude from this small sampling that at least Wnt 3 and 8 are not directly linked to PE formation, though indirect instruction remains possible especially if they are conferring competency to respond to later specification signals (e.g. Bmp2/4).
Lin28 plays multiple roles in regulating cellular homeostasis at least in part by binding and regulating miRNAs (West et al., 2009). A well-known target of Lin28 negative regulation is let-7, a miRNA critically involved in developmental regulation in C. elegans (Büssing et al., 2008). Lin28 expression is linked to pluripotency in a variety of animals. For example, Lin28 is essential for proper PGC development in mice (West et al., 2009). Let-7 usually represses Blimp1, therefore, the presence of Lin28 in the germ line suppresses let-7 and enables Blimp1 to accumulate and specify germ-line cells. In sea star oocytes, Lin28 transcripts accumulate ubiquitously (Figure 3) and then become restricted to the vegetal pole of the blastula and then to the archenteron in the early gastrula stage. During late gastrulation, Lin28 transcripts are restricted to the archenteron within two enriched domains. The first one forms a ring in the upper part of the archenteron and the second domain forms a ring closer to the vegetal pole (Figure 3, asterisks). Lin28 transcripts accumulate in the lower part of the forming esophagus of the early larva, and in the stomach, intestine, anus and PE in the late larva, but not in the coelomic pouches. Importantly, a conserved let-7 miRNA was found in sea urchins (Kadri et al., 2011; Song et al., 2012), although its site of accumulation is not known, nor whether it is present in sea stars.
Assuming Lin28 functions similarly in sea stars as it does in other organisms the marked enrichment in the endoderm (along with Blimp1/Prdm1 and Prdm14) supports the hypothesis that it may be involved in retaining pluripotency in this tissue early in sea star development. Since Lin28 transcripts are not present in the PE, either they do not function in germ-line determination, or they have already accomplished their germ-line role (in the endoderm) prior to this time. We hypothesize that the endoderm (invaginated epithelium) may have significant pluripotentiality that is lost in other tissues. We do not see any direct evidence that Bmp2/4, Wnt3, or Wnt8 expression patterns correlate with PE formation. However, it is still possible that these signaling pathways may act on precursor PE cells at an earlier stage to confer the competency to respond to later PGC inducing signals. It is also possible that Bmp signals may act at a distance during PGC specification in this embryo as well.
Germ-line associated genes
We examined the expression patterns of germ-line associated genes to test correlations with PE formation in sea stars. This cohort of genes is associated with sex determination and, potentially, regulation of germ-line factors (Tables 3–5).
Table 3.
Germ-line associated genes.
| Genes | Domains and Function |
Reference | Orthologs (organism) |
%Identities, % Similarities |
NCBI or Spbase Reference number |
Pm transcript number |
Primers sequence | Size of amplification product |
|---|---|---|---|---|---|---|---|---|
| SoxE | HMG box, transcription factor involved in sex determination. | Juliano et al., 2006 | Mm-Sox9 Dm-Sox100B Sp-SoxE |
77%, 90% 61%, 79% 82%, 89% |
NP_035578.3 NP_651839.1 SPU_016881 |
Pm_60849 | F:CGTCTATTCGCGACGCCGTGT R:taatacgactcactatagggGCTTCTTCCTTGGGTGTGGTC |
496 bp |
| Ovo | C2H2 Zn-finger domain, transcription factor involved in Drosophila oogenesis and mouse spermatogenesis | Dai et al., 1998 | Mm-Ovo2L, isoform A Dm-Ovo, isoform A Sp-Ovo |
59%, 71% 69%, 72% 53%, 66% |
NP_081200.2 NP_525077.2 SPU_012448 |
Pm_17822 | F:CCACAGACGACACACATCTCA R:taatacgactcactatagggGTGAAGGCCTTGCTGCAATAC |
503 bp |
| Gustavus | SPRY and SOCS box domains, E3 ubiquitin ligase specific receptor involved in the regulatory balance of Vasa ubiquitylation. |
Styhler et al., 2002 Gustafson et al., 2011 |
Mm-Gustavus Dm-Gustavus isoform G Sp-Gustavus |
62%, 78% 65%, 79% 74%, 90% |
NP_083311.1 NP_001246140.1 SPU_004717 |
Pm_19970 | F:GGAGGATCTTCGGAGCGGTAG TGCC R:taatacgactcactatagggGGCAGTGGTAGCTGGTAGATGTCTT |
804 bp |
| Cnot6 | EEP domain, deadenylase involved in mRNA decay | Swartz et al, 2013. Wahle et al., 2013. |
Mm-Cnot6 Dm-TwinB Sp-Cnot6 |
60%, 77% 59%, 76% 66%, 78% |
NP_997649.1 NP_732967.1 XP_779942.3 |
Pm_19426 | F:CTGGACCTATCGGCGAATAA R:taatacgactcactatagggTGATGGTCTGGATCAGCTTG |
877 bp |
| G-Cadherin | Laminin G, Ca+2-binding EGF-like, and Cadherin tamdem repeat domains, cell– cell adhesion molecule | Yajima and Wessel, 2012. | Mm-Fat tumor suppressor 1 Dm-NCad isoform G Sp-GCad |
26%, 43% 34%, 51% 39%, 54% |
NP_001074755.2 AAN10997.1 SPU_010840 |
Pm_6651 | F:CGACAAGTTCAGGCTAGACTC R:taatacgactcactatagggGTGACGACAATGTCGATGGTG |
690 bp |
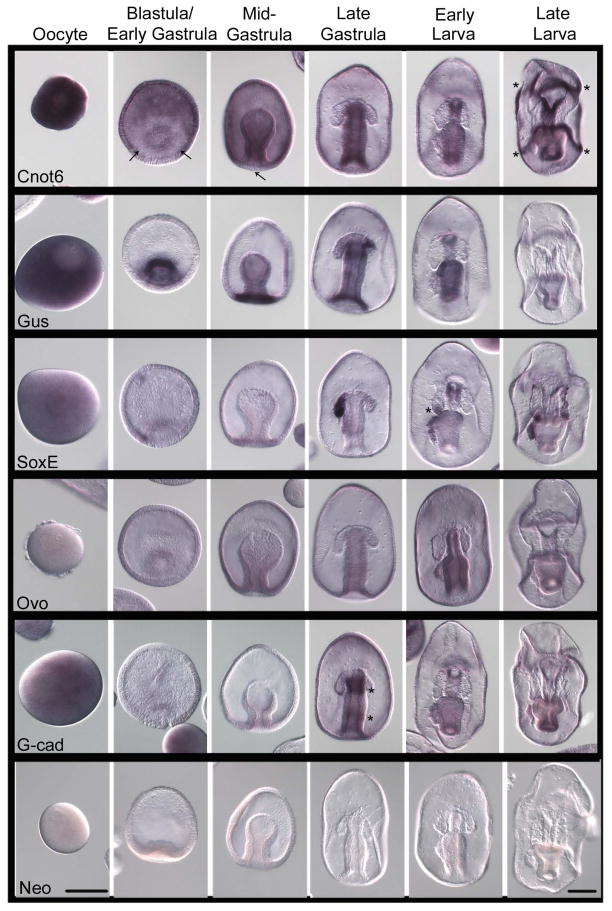
CNOT6 is important for PGC development in sea urchins (Swartz et al., submitted). CNOT6 has deadenylase activity and is recruited to mRNAs for widespread mRNA degradation. CNOT6 is down regulated in the PGCs, and thus these cells retain mRNAs for prolonged times. This is especially important considering the reduced transcriptional activity of the PGCs when compared to their neighboring somatic cells. Indeed, in sea urchins, the sMics retain their maternal (as well as microinjected, exogenously generated) mRNAs for many days whereas their neighboring cells turnover the same transcripts within about one day (Gustafson and Wessel, 2010; Oulhen and Wessel, 2013). CNOT6 is present ubiquitously in the sea urchin embryo except for a marked depletion in the sMics, and this selective depletion of CNOT6 mRNA appears to result from nanos expression selectively in the sMics (Swartz et al., submitted). The CNOT6 mRNA has two nanos/pumilio response elements in its 3′UTR that causes degradation of CNOT6 selectively in the sMics in a nanos-dependent fashion. The CNOT6 present in all other cells of the embryo contributes to the egg – embryo transition e.g. a clearing of the general, maternal, and pluripotent egg mRNAs and “freeing” the somatic cells to differentiate. In contrast, the depletion of CNOT6 in the sMics contributes to the retention of these same mRNAs and their pluripotency (Swartz et al., submitted). It is not clear yet if the sea star has cells that effectively retain maternal mRNAs, so here we determined the expression of CNOT6 to test if such a mechanism may exist. CNOT6 mRNA is present uniformly in the sea star oocytes. By the blastula stage, we detected a depletion of CNOT6 transcripts around the blastopore (Figure 4, arrows). CNOT6 transcripts are enriched in the ectoderm and archenteron of mid-gastrula stage embryos but a clear region remains within the vegetal pole (Figure 4, arrow). In late gastrulae and early larval stages, CNOT6 is still expressed in the ectoderm and in the gut but is depleted in the PE and in the coelomic pouches. In the late larval stage, transcripts are enriched in the gut and both the preoral and postoral ciliary bands (Figure 4, asterisks). We predict that general mRNA retention is greater in the endodermal cells through early larval stages that, once again, emphasizes the endoderm as a tissue retaining its pluripotency.
Figure 4.
Expression of germ-line associated genes during P. miniata embryonic development. Line 1) Cnot6 is nearly uniform throughout development and is absent from the vegetal-most region of the embryo, the coelomic pouches, and the PE as well as the ciliary band in larvae. Line 2) Gustavus (Gus) is enriched in the developing mesoderm in early development. Gus transcripts become enriched in the mouth and stomach in larval stages. Line 3) SoxE (the Sox 9/10 ortholog) is largely absent from the early embryo but accumulates significantly in the left coelomic pouch following gastrulation. SoxE transcripts then accumulate in the PE and the tips of both coelomic pouches during larval stages. Line 4) Ovo is present in the gut of the embryo and larva, with no significant enrichment in the pouches or the PE. Line 5) G-cadherin accumulates in the gut during the late gastrula stage in distinct bands in the foregut and midgut regions. In larval stages G-cadherin becomes enriched in the mouth and stomach. A sequence for the Neomycin (Neo) resistance gene (Line 6) was used as a negative control for the hybridization procedure. Asterisks (*) show areas show areas of emphasis for transcript detection. Arrows emphasize notable areas of transcript depletion. Dorsal views of the larva. Scale bar represents 100 μm.
Gustavus is an E3 ubiquitin ligase that binds Vasa and leads to its degradation. Originally found in Drosophila (Styhler et al., 2002), it is now also known to bind and regulate Vasa protein accumulation in the sea urchin. This function is particularly important in sea urchins since Vasa protein is translated in all cells from ubiquitous maternal mRNA but becomes uniquely retained in sMics as a result of Gustavus-mediated Vasa protein turnover in all somatic cells. Since Vasa protein first appears to be ubiquitous in the sea star embryo, and then clears to become enriched in the PE in late larval stages (Juliano and Wessel, 2009), we tested the location of Gustavus mRNA in the sea star. In sea star oocytes, Gustavus transcripts are present ubiquitously (Figure 4). Gustavus transcripts accumulate in the vegetal pole of the blastula and around the blastopore in early gastrulae (Figure 4). In the late gastrula, Gustavus transcripts are enrich in two rings; one ring forms at the base of the forming anus (blastopore) and another ring forms at the foregut, leaving a less-dense region in the midgut, the site where the PE will form (Figure 4). During early larval stages, Gustavus is express in the gut, but not in the coelomic pouches nor in the PE, the sites of greatest Vasa protein accumulation (Juliano and Wessel, 2009). In late larvae, transcripts remain in the stomach, intestine, anus and ciliary bands (Figure 4). We hypothesize that in the sea star larval stages, Gustavus activity in the midgut may degrade Vasa protein while the lack of Gustavus activity in the PE and anterior coelomic pouches allows Vasa protein to accumulate selectively in these structures.
Echinoderm SoxE is a transcription factor of the HMG family and is the ortholog of the vertebrate member Sox 9 (Howard-Ashby et al., 2006). Sox9 is involved in sex determination in all vertebrates examined; it is regulated positively by SRY in (male) mammals and is enriched in the somatic cells of the presumptive male gonad due to a positive autoregulatory feedback loop (Kashimada and Koopman, 2010; Gilbert, 2010). In other vertebrates, including fish, reptiles, and amphibians, Sox9 may be activated by differing mechanisms, including temperature and other environmental factors and is important for both male and female gene expression leading to sexual dimorphism (Kuroiwa et al., 2002; Maramatsu et al., 2007; Spotila et al., 1998; Uno et al., 2008; Mawaribuchi et al., 2012; Seervai and Wessel, 2013). In sea urchins SoxE is expressed in the left coelomic pouch approximately 50 percent of the time while it has a broader distribution the remaining time (Duboc et al., 2005; Juliano et al., 2006). The SoxE transcript in sea stars is present at low levels in oocytes and is difficult to detect at blastula and mid-gastrula stages. A strong enrichment is then seen in the left anterior coelom and a mild enrichment appears at the midgut in late gastrulae (Figure 4). During early larval development, SoxE transcripts are retained in the most posterior tip of the left anterior coelom (Figure 4, asterisk), then in the same site of both anterior pouches, and in the PE. The expression of SoxE in sea stars is similar to sea urchins in larval stages since it accumulates similarly in the left coelomic pouch and shows a bimodal expression pattern (Juliano et al., 2006).
Ovo is a Zn-finger transcription factor that is important for oogenesis in Drosophila and spermatogenesis in mice (Oliver et al., 1987; Dai et al., 1998). In sea urchins, Ovo is expressed ubiquitously in the blastula stage and is enriched in the vegetal pole at the mesenchyme blastula stage, an expression domain similar to Vasa. However, unlike Vasa, Ovo expression is not detected in gastrula and larval stages (Juliano et al., 2006). In the sea star, Ovo transcripts are not detectable in the oocyte but are detectable ubiquitously at low levels in the blastula stage (Figure 4). During gastrulation Ovo transcripts accumulate throughout the developing gut. In early larvae Ovo transcripts are enriched throughout the gut and esophagus, and in late larva Ovo transcripts are enriched in the gut and pre-oral and post-oral ciliary bands. Although we did not find any obvious correlation between the expression of Ovo and the formation of the PE, its consistent link to the invaginating epithelium may indicate its involvement in the retention of developmental potentiality leading to PE formation.
Changes in cadherin expression are intimately linked to morphogenetic processes that involve the loss of epithelial character and the delamination of cells from an epithelial sheet (Takeichi, 1988; Birchmeier et al., 1993; Gumbiner, 1996), a character often seen in PGCs (McLaren, 2003). In sea urchin embryos, the cellular movements associated with the ingression of primary mesenchyme cells (PMCs) and convergent-extension of the archenteron involve the dynamic regulation of intercellular adhesion, including the loss of cell adhesion molecules (Miller and McClay, 1997; Fink and McClay, 1985). The sMics of the sea urchin appear to retain their cadherin through development until they reach the tip of the archenteron during gastrulation. Premature depletion of the G-cadherin protein appears to disrupt sMic patterning and expression of several cell-specific markers (Yajima and Wessel, 2012) and cadherin orthologs appear important for germ cell function in many animals (e.g. Blaser et al., 2005; Chihara and Nance, 2012). Therefore we were interested to test the profile of the G-cadherin orthology in this sea star.
The sequence of the sea star ortholog of G-cadherin shows significant sequence identity to all classic cadherins, particularly in the cytoplasmic domain, the region predicted to be involved in catenin binding (Supplemental Figure 1). G-cadherin mRNA accumulates ubiquitously in oocytes, but is not detectable in blastula and early gastrula stages (Figure 4). In the late gastrula, G-cadherin transcripts are enriched in two domains; one in the foregut and another in the hindgut (Figure 4, asterisks). This creates a midgut region with less G-cadherin transcripts, the same location in which new morphogenetic movements lead to PE formation. This profile also overlaps the Gustavus mRNA profile and perhaps this region supports the evagination of the epithelium leading to PE formation by decreased cell-cell adhesion. G-cadherin transcripts then decreased overall in the larval stages and became restricted to the mouth opening, stomach, intestine, and anus.
Overall, our analysis of the expression of germ-line associated genes in sea star embryos reveals how mechanisms of PGC biology vary between animals that use different modes of PGC specification. Yet, overall similarities are seen; the RNA degradation machinery such as the deadenylase, CNOT6, may be conserved in its down-regulation in PGCs once they are formed to preserve their inherited transcripts and retain greater developmental plasticity. The ubiquitin pathway and specific E3 ligases (such as Gustavus) may be conservatively involved in down-regulating germ cell determinant protein levels outside of the germ cells once they are specified. Transcription factors that are conserved in somatic sex determination seem to have bi-modal expression patterns and be involved in sex determination regardless of the mode of PGC specification. Finally, changes in adhesion, such as through G-cadherin seem to be linked to germ cell function in many animals.
Left/Right asymmetry molecules
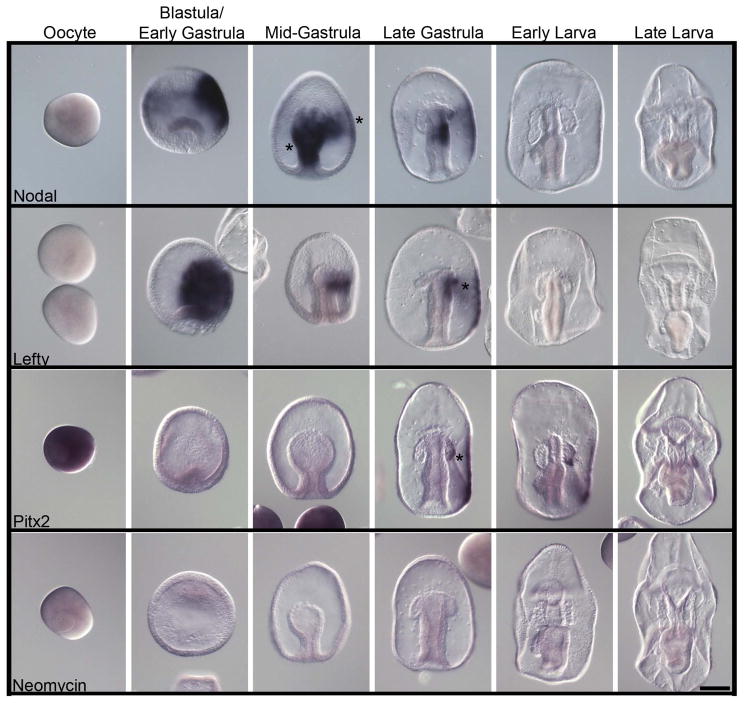
The PE forms only on the left side of the P. miniata gut during development and we hypothesize that conserved left/right signaling pathways may be involved in PE formation (and therefore PGC specification). Consequently, we identified and tested the gene expression patterns of signaling molecules involved in left/right asymmetry. Major mechanisms for establishment of the left/right axis in sea urchin and other organisms include complex epigenetic and genetic cascades (Lin and Xu, 2009; Table 4). The initial symmetry-breaking event is not clearly understood, and is probably different between species (Vandenberg and Levin, 2013), yet this axial organization still requires a specific, and likely conserved, set of gene activities. For example, the Tgf-beta family member Nodal is expressed on the left side of chordate embryos and is widely used to specify structures on the left side differently than on the right. Nodal function activates additional genes involved in development of the left/right asymmetry, including the additional Tgf-beta factor Lefty, and the homeobox transcription factor Pitx2 (Levin et al., 1995; Spéder et al., 2007). In the sea urchin however, this pathway is reversed and is instead expressed on the right side (Duboc et al., 2005). Overexpression of Nodal throughout the sea urchin embryo results in developmental repression of the adult rudiment on the left side, and removal of Nodal in the sea urchin results in duplicated rudiments on both the right and left sides suggesting that a main function of Nodal is repression of developmental derivatives of the right coelomic pouch (Duboc et al., 2005; Bessodes et al., 2012; Luo and Su, 2012; Warner et al., 2012). In the sea star, Nodal is not expressed in oocytes, but accumulates strongly in early blastula stages in the ectoderm (Figure 5). Nodal mRNA remains transiently in the ectoderm until the late gastrula stage and is absent there in larval stages. A second domain of Nodal expression, however, occurs in mid gastrula embryos when Nodal message accumulates in the midgut and on the right side of the invaginated epithelium. We also see Nodal mRNA transiently within the posterior region of the right coelomic pouch, but to a much lesser extent.
Table 4.
Left-Right asymmetry molecules
| Genes | Domains and Function |
Reference | Orthologs (organism) |
%Identities, % Similarities |
NCBI or Spbase Reference number |
Pm transcript number | Primers sequence | Size of amplification product |
|---|---|---|---|---|---|---|---|---|
| Nodal | Tgf-beta ligand Involved in left-right asymmetry | Grande et al., 2009. | Mm-Nodal Dm-Dpp Sp-Nodal |
51%, 74% 42%, 62% 53%, 69% |
NP_038639 NP_477311.1 ABK33664.1 |
gi|307052073|gb|HP126404.1|isotig21602.Pminagast | F:CGGTGGATCGTCTACCCTAA R:taatacgactcactatagggCCCGATCAAATTGTAAAAATGC |
966 bp |
| Lefty | Tgf-beta signaling molecule, involved in left-right asymmetry | Grande et al., 2009. | Mm-Lefty Dm-Dpp Sp-Lefty |
24%, 40% 21%, 36% 28%, 46% |
NP_034224.1 NP_477311.1 NP_001123281.1 |
lcl|scaffold511856 75.8 | F:ATGGAGTCTCGCGTAGCTGT R:taatacgactcactatagggCATGTTTGTTGACGGGTCTG |
537 bp |
| Pitx2 | Homeobox domain, transcription factor involved in left-right asymmetry | Grande et al., 2009. | Mm-Pitx2 Dm-Ptx1 Sp-Pitx2 |
74%, 82% 60%, 65% 73%, 82% |
NP_001035969.1 NP_733410.2 SPU_004599 |
Pm_22862 | F:GCGTCAGGGTGTGGTTTAAG R:taatacgactcactatagggGTTCAAGTTCTGGTGGCTCA |
329 |
Figure 5.
Expression of left/right asymmetry markers during P. miniata embryonic development. Line 1) Nodal is first expressed in the ectoderm at the blastula stage. A second expression domain of Nodal appears in mid-gastrula stage embryos when it is expressed symmetrically in the developing archenteron. Nodal expression becomes restricted to the right side of the archenteron in late-gastrula stage embryos. Line 2) Lefty is first expressed in the ectoderm at the blastula stage. A second expression domain of Lefty appears in late-gastrula stage embryos when it is expressed asymmetrically in the right side of the archenteron. Line 3) Pitx2 is first expressed ubiquitously in oocytes. Pitx2 expression localizes to the right coelomic pouch and right ectoderm in late gastrula staged embryos stage. Pitx2 expression in the right coelomic pouch and right ectoderm persists through late larval stages. A sequence for the Neomycin (Neo) resistance gene (Line 4) was used as a negative control for the hybridization procedure. Asterisks (*) show areas of emphasis for transcriptdetection. Dorsal views of the larva. Scale bar represents 100 μm.
Lefty follows a similar expression pattern as Nodal in the ectoderm. However, Lefty does not accumulate in the archenteron until later in gastrulation when it accumulates in the right side of the archenteron and in the right coelomic pouch (Figure 5, late gastrula, asterisk). Pitx2 transcripts, although ubiquitous in P. miniatao oocytes, do not accumulate significantly in early development until the late gastrula stage. Pitx2 transcripts follow a similar expression domains as Nodal and Lefty, albeit much delayed (Figure 5, late gastrula, asterisk) and this is similar in the closely related sea star Asterina pectinifera (Hibino et al., 2006). Pitx2 is expressed in the posterior portion of the right coelomic pouch which persists from the late gastrula stage until late larval stages, although its accumulation in the right pouch of A. pectinifera is much broader than in P. miniata. We noticed that in the sea star P. miniata, the initial break in left/right asymmetry within the archenteron (as seen with the right-sided expression of Nodal) occurs at the same time that Vasa expression is restricted to the left side of the archenteron.
Therefore, we hypothesize that left/right signaling and Nodal are required for the initial break in left/right asymmetry of Vasa expression and therefore of PE specification. Overall the sea star left/right asymmetry program appears to closely mirror the program in the sea urchin, even with the significant expression in the right side of the archenteron. Thus, the reversal of the left/right program in echinoderms likely occurred prior to the sea urchin-sea star split; phylogenetic analyses suggests Nodal on the right may be ancestral and that reversal may have occurred in the chordate lineage (Duboc et al., 2005; Bessodes et al., 2012; Luo and Su, 2012; Warner et al., 2012). This also may be related to the reversal of the dorsal/ventral program in chordates relative to non-chordates (Grande and Patel 2009).
Genomic maintenance during morphogenesis and early embryogenesis
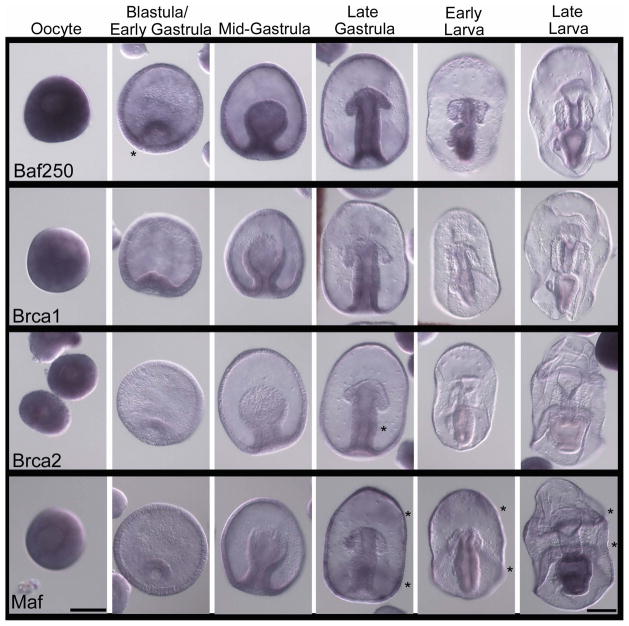
The last group of genes we studied encodes molecules related to genomic and epigenomic regulation during morphogenesis to test if these genes are associated selectively with sea star PE cells (Table 5, Figure 6). Baf250 is an E3 ubiquitin ligase that functions in selective histone (H2B) turnover. Since Baf250 associates with the mammalian SWI/SNF complex it is thought that this histone-turnover machinery regulates epigenetic modifications that lead to selective gene activity (Li et al., 2010). Such modifications are of particular interest in mammalian embryos with the identification of selective epigenetic reprogramming during germ cell formation (Magnúsdóttir et al., 2012). In the sea star P. miniata, Baf250 transcripts accumulate ubiquitously in oocytes. In blastula stage embryos, Baf250 transcripts become enriched in the blastopore (Figure 6, asterisk), and in gastrula embryos transcripts accumulate in the archenteron. During larval stages, Baf250 accumulates in the esophagus, stomach, intestine, anus, and in the PE but not in the coelomic pouches.
Figure 6.
Molecules involved in genomic regulation and maintenance during P. miniata embryonic development. Line 1) Baf250 is expressed ubiquitously in the embryo throughout development with some enrichment in the gut and depletion in the coelomic pouches. Lines 2 and 3) Brca1 and 2 are enriched in the gut of late gastrula embryos and we note no significant accumulation in the PE. Line 4) Maf is most apparent in the stomach of the larva and no specific enrichment in the PE. Asterisks (*) show areas of emphasis for transcript detection. Dorsal views of the larva. Scale bar represents 100 μm.
Recent results in zebrafish suggest that the DNA repair elements Brca1 and Brca2 are involved in germ line development (Shive et al., 2010). We find that Brca1 and 2 in the sea star have overlapping expression profiles: Brca1 transcripts are expressed ubiquitously throughout early development until the late gastrula stage when they become enriched in the midgut (Figure 6). Brca2 transcripts are also present in oocytes and they accumulate in the gut of late gastrula stage embryos (Figure 6, asterisk). The accumulation of both Brca transcripts in oocytes suggests there might be a role for the maternal message in provisioning early blastomeres with Brca proteins that could affect DNA repair during the rapid cleavage divisions that occur.
Traffic jam is an atypical basic leucine zipper transcription factor that regulates somatic-germ cell interactions and its loss results in male and female infertility in Drosophila (Li et al., 2003). Here we found that transcripts of the ortholog of Traffic Jam, called Maf, are ubiquitously distributed in oocytes but are not enriched in blastula and mid-gastrula stage embryos. Maf transcripts are enriched slightly in the ciliary bands of the larva (Figure 6, asterisks but not in the PE).
Conclusions
Our results support the contention that the PE is a source of germ line cells in the sea star P. miniata. Several gene expression patterns reflect a broad initial expression of germ line markers in the embryo followed by a restriction to the PE. These include Vasa, Nanos, and Piwi (see Figure 7). While these results on their own do not prove that the PE contains the germ line, they are complementary to other studies that suggest that these cells give rise to sea star germ cells. The PE is most likely the site of germ cell formation in this animal based on four criteria: 1) PE removal experiments result in larvae with significantly less germ cells (Inoue et al., 1992), 2) Vasa immunolabeling experiments show the PE is the first restricted site of Vasa protein localization (Juliano et al., 2009), 3) conserved germ line factors accumulate selectively in the PE e.g. nanos and piwi in addition to vasa, and 4) the PE exhibits rapid depletion of mRNAs encoding factors involved in somatic cell fates e.g. Blimp1. These observations and the fact that the selective expression in the PE is relatively late in development – following gastrulation - leads us to reason that the germ line in this organism is determined by inductive interactions amongst cells.
Figure 7.
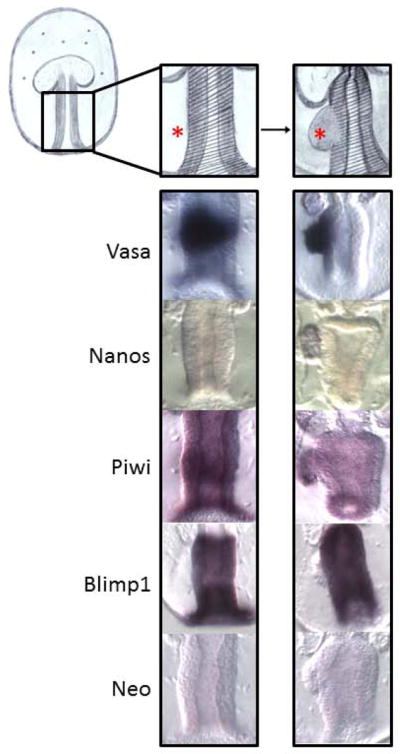
Transcript dynamics during posterior enterocoel formation. The conserved germ-line determinant, Nanos, accumulates specifically in the PE (red asterisk) after formation. During gastrulation, the conserved germ-line determinants Vasa and Piwi, become enriched in the midgut (shadowed area). As soon as the PE is formed Vasa and Piwi transcripts start to become restricted to the PE and to clear from the nearby stomach. This is in stark contrast to the somatic cell marker, Blimp1. During gastrulation, Blimp1 transcripts similarly become enriched in the midgut, however, as soon as the PE is formed, Blimp1 transcripts are restricted to the stomach and clear from the nearby PE.
Blimp1 is a somatic factor in echinoderms (as it is necessary for endomesoderm gene regulatory networks, e.g. Spbase.org) and the selective loss of somatic markers in the PGCs represents a broad theme in the inductive mode of germ cell specification (see also Figure 7). In the mouse inductive mode of germ cell specification, markers of mesoderm, such as Hoxb1 and Hoxa1, are lost from PGCs as soon as they receive PGC inducing signals (Saitou et al., 2002). The loss of Blimp1 transcripts in the PE of sea stars contributes to the hypothesis that when an animal uses the inductive mode for germ cell specification it is conservatively induced from a pluripotential mesodermal lineage.
A consensus revealed in this study was that the gut appears to harbor a gene set making it retain pluripotency and germ line potential. Many factors, including Vasa, Piwi, Blimp1, and Prdm14, are enriched in the endomesoderm of the gut and suggest the gut retains developmental potential whereas the ectoderm is devoid of many of these same factors. This result may simply mean that the endomesoderm forms later in its differentiation program when compared to the ectoderm, or that the cells within this tissue give rise to many more cell types later in development. The additional possibility is that the endomesoderm is the site of germ line formation and retains developmental potency. The gene expression profile on its own is not convincing and will certainly require functional analysis with metrics of germ line success to understand functionality of the genes tested here. However, when the precursor cells to the PGCs are disrupted in sea urchins Vasa is up-regulated throughout the endoderm of the remaining embryo (Voronina et al., 2008). In this experimental case within the sea urchin, Vasa mimics the same broad endodermal expression profile that we see with many pluripotency-related genes in sea stars. Thus, these two echinodermal representatives may have diverged in their germ line determination by transposing the germ line program earlier in sea urchins, and to the sMics. Sea urchins represent a unique model in that this animal retains the ability to use multiple mechanisms for PGC specification upon its disruption (compensatory Vasa upregulation and recovery of germ line cells; Voronina et al., 2008; Yajima and Wessel, 2011). We favor the conclusion that the mechanism in sea urchins is derived and perhaps tending towards an inherited mechanism of germ line determination, especially when compared to the sea star.
Two extreme mechanisms of germ-line determination appear in animal development - inherited vs inductive (Extavour and Akam, 2003; Juliano and Wessel, 2010; Ewen-Campen et al., 2010; Seervai and Wessel, 2013). Embryos using inherited mechanisms usually establish their germ line early in development by acquisition of a specific region of egg cytoplasm - maternally deposited in the oocyte. This is the best known mechanism and is used by many model organisms e.g. fly, worm, frog, and zebrafish. Mice are the best studied organism that uses inductive mechanisms of germ-line determination. In this mechanism, cell interactions are responsible, usually later in development, to establish a germ line lineage. Results presented here support the contention that the sea star PE is a site of germ line formation and the evidence suggests this structure may fits better with the criteria of an inductive mechanism. This means functional studies that determine signaling networks required for PGC specification in this organism may complement the genetic and tissue culture approaches used in mice to reveal inductive germ-line determination mechanisms. Furthermore, comparisons between the differing mechanisms of PGC specification between sea star and sea urchins will be useful to understand transitions that occur in the evolution from one mode to another.
Experimental Procedures
Animals and embryo culture
Patiria miniata were collected from several sites in southern California [www.scbiomarine.com; phalmay@earthlink.net] and embryos were grown basically as described (Foltz et al., 2004). Briefly, sperm were collected from a gonad biopsy and placed into a microfuge tube on ice. Oocytes were collected from a gonad biopsy and matured in vitro with 2 μM 1-Methyl-Adenine. Resultant eggs were fertilized with a dilute sperm suspension and embryos were cultured as previously described (Hinman et al., 2003). Samples from different developmental stages (oocytes; hatched blastula, 18.5 hours post-fertilization (hpf); mid-gastrula, 27.5 hpf; late gastrula, 47 hpf; early larva, 3 days post-fertilization (dpf); late larva, 4–7 dpf) were collected, fixed and stored in 70% ethanol at −20 as described (Arenas-Mena et al., 2000).
RNA analysis
Whole mount in situ RNA hybridizations were performed using digoxigenin-labeled RNA probes as previously described (Arenas-Mena et al., 2000). cDNAs from oocytes and early development stages were used as templates for PCR reactions. Primers designed to amplify each gene of interest included a T7 RNA polymerase sequence in the 5′ end of reverse primers. The resultant PCR products were used as templates for transcription by T7 RNA polymerase to yield an antisense RNA probe with the DIG RNA Labeling Kit (SP6/T7) (Roche Applied Science, IN, USA). Oocytes and embryos were fixed, hybridized, and the signals were detected essentially as described (Arenas-Mena et al., 2000). Negative controls for these experiments included the use of a non-relevant transcript probe (Neomycin). Oocytes and embryos were visualized on a Zeiss Axioplan microscope, and the specimens are oriented with their left side to the left i.e. we position the larva ventral side down, left side of the larva to the left of the image. This is the opposite of the classic human orientation scheme but we think it makes the structures easier to interpret.
Supplementary Material
Table 2.
Genes involved in the inductive mechanism of germ-line specification.
| Genes | Domains and Function |
Reference | Orthologs (organism) |
%Identities, % Similarities |
NCBI or Spbase Reference number |
Pm transcript number |
Primers sequence | Size of amplification product |
|---|---|---|---|---|---|---|---|---|
| Blimp1/Prdm1 | Transcription factor involved in germ line determination in mice |
Saitou et al., 2010. Kurimoto et al., 2008. |
Mm-Prdm1 Dm-Blimp1 Sp-Blimp1/Krox1b |
78%, 87% 67%, 79% 43%, 55% |
AAI29802.1 NP_647982.1 NP_001073021.1 |
Pm_43022 (AAP35029.1) | F:CCATTCTCCGTACTCGTGGT R:taatacgactcactatagggCGGAAGTCTGTGCATGAGAA |
912 bp |
| Prdm14 | PR domain Zn-finger, transcriptional regulator involved in germ line determination in mice |
Saitou et al., 2010. Yamaji et al., 2008 |
Mm-Prdm14 Dm-Prdm14 Sp-Prdm14 |
87%, 94% 37%, 55%** 94%, 97% |
NP_001074678.2 XP_ 794184.3 |
Pm_65577 | F:AACCGCTCTTCCGATCTGT R:taatacgactcactatagggGTGTGACGCGAAGGCTTTT |
402 bp |
| BMP2/4 | TGF-beta ligand signaling molecule. Critical for induction of PGCs in mice |
Saitou et al., 2010. Saitou, 2009. |
Mm-BMP2 Dm-Decapentaplegic, isoformA Sp-BMP 2/4 |
45%, 61% 40%, 55% 50%, 65% |
NP_031579.2 NP_477311.1 SPU_021497 |
Pm_4348 | F:CGTGCCACAGTACATGCTGGA R:taatacgactcactatagggGCTCGCTGACAGACCGAGCTA |
590 bp |
| Lin28 | Cold shock and a cluster of two CCHC Zn-finger domains, RNA binding protein that is required for PGC development |
Bussing et al. 2008. West et al. 2009. |
Mm-Lin28B Dm-Lin28 Sp-Lin28 |
53%, 61% 55%, 75% 64%, 80% |
NP_001026942.1 NP_647983.1 SPU_027195 |
Pm_90489 | F:GGCCGACGAGGGCAAGCTGTG R:taatacgactcactatagggGGCCAGTCACCGACTCCGCCT |
215 bp |
| Wnt3 | Wnt ligand signaling molecule. Involved in germ line competency in mice. |
Saitou et al., 2010. Ohinata et al., 2009. |
Mm-Wnt3 Dm-Wnt2 Sp-Wnt3 |
53%, 69% 43%, 57% 61%, 73% |
NP_033547.1 NP_476810.1 XP_790595.2 |
Combo of: HP125189.1 and sequencing results after cloning | F:TAAATTCATCAGCCCCAAGG R:taatacgactcactatagggATGGCTTCGTTCTTGAATGC |
975 bp |
| Wnt8 | Wnt ligand signaling molecule. Correlates with localization of PGC’s in axolotl. |
Bachvarova et al., 2001. Johnson et al., 2003. |
Mm-Wnt8b Dm-Wingless, isoform B Sp-Wnt8 |
59%, 73% 40%, 58% 48%, 67% |
NP_035850.2 NP_723268.1 NP_999832.1 |
Pm_82262 | F:GCAGCGACAACATCAAATTCG R:taatacgactcactatagggGCTCTTCCGATCTGACGGCTG |
365 bp |
Key findings.
Sea stars have highly conserved germ line factors
These factors are expressed in patterns that suggest the germ line forms from the posterior enterocoel.
The overall data supports the contention that the sea star uses an inductive mechanism of germ line determination, distinct from its near relative, the sea urchin.
Acknowledgments
We gratefully acknowledge grants from the NIH (2R01HD028152), the NSF (IOS-1120972), and CONACyT that made this work possible. We also appreciate the rich intellectual interactions and working environment from members of PRIMO.
Abbreviations
- PE
posterior enterocoel
- sMics
small micromeres
- PRE
pumilio response element
- NRE
nanos response element
- GRN
gene regulatory network
- hpf
hours post-fertilization
- dpf
days post-fertilization
References
- Angerer LM, Angerer RC. Detection of poly A+RNA in sea urchin eggs and embryos by quantitative in situ hybridization. Nucleic Acids Res. 1981;9:2819–2840. doi: 10.1093/nar/9.12.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas-Mena C, Cameron AR, Davidson EH. Spatial expression of Hox cluster genes in the ontogeny of a sea urchin. Development. 2000;127:4631–4643. doi: 10.1242/dev.127.21.4631. [DOI] [PubMed] [Google Scholar]
- Bachvarova RF, Masi T, Hall L, Johnson AD. Expression of AxWnt-8 and Axszl in the urodele, axolotl: comparison with Xenopus. Dev Genes Evol. 2001;211:501–505. doi: 10.1007/s004270100175. [DOI] [PubMed] [Google Scholar]
- Bessodes N, Haillot E, Duboc V, Röttinger E, Lahaye F, Lepage T. Reciprocal signaling between the ectoderm and a mesendodermal left-right organizer directs left-right determination in the sea urchin embryo. PLoS Genet. 2012;8(12):e1003121. doi: 10.1371/journal.pgen.1003121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchmeier W, Weidner KM, Behrens J. Molecular mechanisms leading to loss of differentiation and gain of invasiveness in epithelial cells. J Cell Sci Suppl. 1993;17:159–164. doi: 10.1242/jcs.1993.supplement_17.23. [DOI] [PubMed] [Google Scholar]
- Blaser H, Eisembeiss S, Neumann M, Reichman-Fried M, Thisse B, Thisse C, Raz E. Transition from non-motile behavior to directed migration during early PGC development in zebrafish. J Cell Sci. 2005;118:4027–4038. doi: 10.1242/jcs.02522. [DOI] [PubMed] [Google Scholar]
- Büssing I, Slack FJ, Grosshans H. Let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K, Svingen T, Ng ET, Epp T, Spiller CM, Clark C, Cooper H, Koopman P. Loss of Wnt5a disrupts primordial germ cell migration and male sexual development in mice. Biol Reprod. 2012;86:1–12. doi: 10.1095/biolreprod.111.095232. [DOI] [PubMed] [Google Scholar]
- Chihara D, Nance J. An E-cadherine-mediated hitchhiking mechanism for C. elegans germ cell internañization during gastrulation. Development. 2012;139:2547–2556. doi: 10.1242/dev.079863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho PF, Gamberi C, Cho-Park YA, Cho-Park IB, Lasko P, Sonenberg N. Cap-dependent translational inhibition establishes two opposing morphogen gradients in Drosophila embryos. Curr Biol. 2006;16:2035–2041. doi: 10.1016/j.cub.2006.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Schonbaum C, Degenstein L, Bai W, Mahowald A, Fuchs E. The ovo gene required for cuticle formation and oogenesis in flies is involved in hair formation and spermatogenesis in mice. Genes Dev. 1998;12:3452–63. doi: 10.1101/gad.12.21.3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza FS, Gawantka V, Gómez AP, Delius H, Ang SL, Niehrs C. The zinc finger gene Xblimp1 controls anterior endomesodermal cell fate in Spemann’s organizer. EMBO J. 1999;18:6062–6072. doi: 10.1093/emboj/18.21.6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboc V, Röttinger E, Lapraz F, Besnardeau L, Lepage T. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev Cell. 2005;9:147–158. doi: 10.1016/j.devcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B, Donoughe S, Clarke DN, Extavour CG. Germ cell specification requires zygotic mechanisms rather than germ plasm in a basally branching insect. Curr Biol. 2013;23:835–42. doi: 10.1016/j.cub.2013.03.063. [DOI] [PubMed] [Google Scholar]
- Ewen-Campen B, Schwager EE, Extavour CG. The molecular machinery of germ line specification. Mol Reprod Dev. 2010;77:3–18. doi: 10.1002/mrd.21091. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesist and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Fink RD, McClay DR. Three cell recognition changes accompany the ingression of sea urchin primary mesenchyme cells. Dev Biol. 1985;107:66–74. doi: 10.1016/0012-1606(85)90376-8. [DOI] [PubMed] [Google Scholar]
- Gao M, Arkov AL. Next generation organelles: Structure and role of germ granules in the germline. Mol Reprod Dev. 2013;80(8):610–23. doi: 10.1002/mrd.22115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology. 9. Sinauer Associates; Sunderland, MA: 2010. [Google Scholar]
- Golestaneh N, Beauchamp E, Fallen S, Kokkinaki M, Uren A, Dym M. Wnt signaling promotes proliferation and stemness regulation of spermatogonial stem/progenitor cells. Reproduction. 2009;138:151–62. doi: 10.1530/REP-08-0510. [DOI] [PubMed] [Google Scholar]
- Grande C, Patel NH. Nodal signalling is involved in left-right asymmetry in snails. Nature. 2009;457:1007–1011. doi: 10.1038/nature07603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumbiner BM. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996;84:345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Gustafson EA, Wessel GM. Exogenous RNA is selectively retained in the small micromeres during sea urchin embryos. Mol Reprod Dev. 2010;77:836. doi: 10.1002/mrd.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson EA, Yajima M, Juliano CE, Wessel GM. Post-translational regulation by gustavus contributes to selective Vasa protein accumulation in multipotent cells during embryogenesis. Dev Biol. 2011;349:440–450. doi: 10.1016/j.ydbio.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Nishino A, Amemiya S. Phylogenetic correspondence of the body axes in bilaterians is revealed by the right-sided expression of Pitx genes in echinoderm larvae. Dev Growth Differ. 2006;48:587–595. doi: 10.1111/j.1440-169X.2006.00892.x. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Nguyen A, Davidson EH. Caught in the evolutionary act: precise cis-regulatory basis of difference in the organization of gene networks of sea stars and sea urchins. Dev Biol. 2007;312:584–595. doi: 10.1016/j.ydbio.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Nguyen AT, Davidson EH. Expression and function of a starfish Otx ortholog, AmOtx: a conserved role for Otx proteins in endoderm development that predates divergence of the eleutherozoa. Mech Dev. 2003;120:1165–1176. doi: 10.1016/j.mod.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Davidson EH. Expression of AmKrox, a starfish ortholog of a sea urchin transcription factor essential for endomesodermal specification. Gene Expr Patterns. 2003;3:423–426. doi: 10.1016/s1567-133x(03)00083-8. [DOI] [PubMed] [Google Scholar]
- Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev Biol. 2006;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Inoue C, Kiyomoto M, Shirai H. Germ cell differentiation in starfish The posterior enterocoel as the origin of germ cells in Asterina pectinifera. Dev Growth Differ. 1992;34:413–418. doi: 10.1111/j.1440-169X.1992.00413.x. [DOI] [PubMed] [Google Scholar]
- John SA, Garrett-Sinha LA. Blimp1: a conserved transcriptional repressor critical for differentiation of many tissues. Exp Cell Res. 2009;315:1077–1084. doi: 10.1016/j.yexcr.2008.11.015. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Crother B, White ME, Patient R, Bachvarova RF, Drum M, Masi T. Regulative germ cell specification in axolotl embryos: a primitive trait conserved in the mammalian lineage. Philos Trans R Soc Lond B Biol Sci. 2003;358:1371–1379. doi: 10.1098/rstb.2003.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev Biol. 2006;300:406–415. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Wessel GM. An evolutionary transition of Vasa regulation in echinoderms. Evol Dev. 2009;11:560–573. doi: 10.1111/j.1525-142X.2009.00362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Wessel GM. Developmental biology. Versatile germline genes. Science. 2010;329:640–641. doi: 10.1126/science.1194037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadri S, Hinman VF, Benos PV. RNA deep sequencing reveals differential microRNA expression during development of sea urchin and sea star. PLoS ONE. 2011;6(12):e29217. doi: 10.1371/journal.pone.0029217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrova L, Habara Y, Lee TH, Wharton RP. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 2007;134:1519–1527. doi: 10.1242/dev.002212. [DOI] [PubMed] [Google Scholar]
- Kashimada K, Koopman P. Sry: the master switch in mammalian sex determination. Development. 2010;137:3921–30. doi: 10.1242/dev.048983. [DOI] [PubMed] [Google Scholar]
- Kumburegama S, Wikramanayake AH. Wnt signaling in the early sea urchin embryo. Methods Mol Biol. 2009;469:187–199. doi: 10.1007/978-1-60327-469-2_14. [DOI] [PubMed] [Google Scholar]
- Kurimoto K, Yamaji M, Seki Y, Saitou M. Specification of the germ cell lineage in mice: a process orchestrated by the PR-domain proteins, Blimp1 and Prmd14. Cell cycle. 2008;7:3514–3518. doi: 10.4161/cc.7.22.6979. [DOI] [PubMed] [Google Scholar]
- Kuroiwa A, Uchikawa M, Kamachi Y, Kondoh H, Nishida-Umehara C, Masabanda J, Griffin DK, Matsuda Y. Chromosome assignment of eight SOX family genes in chicken. Cytogenet Genome Res. 2002;98:189–93. doi: 10.1159/000069803. [DOI] [PubMed] [Google Scholar]
- Lai F, Singh A, King ML. Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development. 2012;139:1476–1486. doi: 10.1242/dev.079608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai F, King ML. Repressive translationaly control in germ cells. Mol Reprod Dev. 2013:1–12. doi: 10.1002/mrd.22161. [DOI] [PubMed] [Google Scholar]
- Lapraz F, Röttinger E, Duboc V, Range R, Duloquin L, Walton K, Wu SY, Bradham C, Loza MA, Hibino T, Wilson K, Poustka A, McClay D, Angerer L, Gache C, Lepage T. RTK and TGF-beta signaling pathways genes in the sea urchin genome. Dev Biol. 2006;300:132–52. doi: 10.1016/j.ydbio.2006.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, Johnson RL, Stern CD, Kuehn M, Tabin C. A molecular pathway determining left-right asymmetry in chick embryogenesis. Cell. 1995;82:803–814. doi: 10.1016/0092-8674(95)90477-8. [DOI] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, Godt D. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat Cell Biol. 2003;5:994–1000. doi: 10.1038/ncb1058. [DOI] [PubMed] [Google Scholar]
- Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N. Mammalian SWI/SNF--a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol Cell Biol. 2010;30:1673–88. doi: 10.1128/MCB.00540-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Xu X. Distinct functions of Wnt/beta-catenin signaling in KV development and cardiac asymmetry. Development. 2009;136:207–217. doi: 10.1242/dev.029561. [DOI] [PubMed] [Google Scholar]
- Livi CB, Davidson EH. Expression and function of blimp1/krox, an alternatively transcribed regulatory gene of the sea urchin endomesoderm network. Dev Biol. 2006;293:513–525. doi: 10.1016/j.ydbio.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Luo YJ, Su YH. Opposing nodal and BMP signals regulate left-right asymmetry in the sea urchin larva. PLoS Biol. 2012;10(10):e1001402. doi: 10.1371/journal.pbio.1001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnúsdóttir E, Gillich A, Grabole N, Surani MA. Combinatorial control of cell fate and reprogramming in the mammalian germline. Curr Opin Genet Dev. 2012;22:466–474. doi: 10.1016/j.gde.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Magnúsdóttir E, Dietmann S, Murakami K, Gunesdogan U, Tang F, Bao S, Diamanti E, Lao K, Gottgens B, Surani MA. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nature Cell Biology. 2013;15:905–915. doi: 10.1038/ncb2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maramatsu S, Wakabayashi M, Ohno T, Amano K, Ooishi R, Sugahara T, Shiojiri S, Tashiro K, Suzuki Y, Nishimura R, Kuhara S, Sugano S, Yoneda T, Matsuda A. Functional gene screening system identified TRPV4 as a regulator of chondrogeni differentiation. J Biol Chem. 2007;282:32158–32167. doi: 10.1074/jbc.M706158200. [DOI] [PubMed] [Google Scholar]
- Mawaribuchi S, Yoshimoto S, Ohashi S, Takamatsu N, Ito M. Molecular evolution of vertebrate sex-determining genes. Chromosome Res. 2012;20:139–151. doi: 10.1007/s10577-011-9265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren A. Primordial germ cells in the mouse. Dev Biol. 2003;262:1–15. doi: 10.1016/s0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- Miller JR, McClay DR. Characterization of the role of cadherin in regulating cell adhesion during sea urchin development. Dev Biol. 1997;192:323–339. doi: 10.1006/dbio.1997.8740. [DOI] [PubMed] [Google Scholar]
- Nakata H, Minokawa T. Expression patterns of wnt8 orthologs in two sand dollar species with different developmental modes. Gene Expr Patterns. 2008;9:152–157. doi: 10.1016/j.gep.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Ohinata Y, Ohta H, Shigeta M, Yamanaka K, Wakayama T, Saitou M. A signaling principle for the specification of the germ cell lineage in mice. Cell. 2009;137:571–584. doi: 10.1016/j.cell.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Oliver B, Perrimon N, Mahowald AP. The ovo locus is required for sex-specific germ line maintenance in Drosophila. Genes Dev. 1987;1:913–923. doi: 10.1101/gad.1.9.913. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Tu Q, Davidson EH. Global regulatory logic for specification of an embryonic cell linage. Proc Natl Acad Sci USA. 2008;105:5955–5962. doi: 10.1073/pnas.0711220105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulhen N, Yoshida T, Yajima M, Song J, Sakuma T, Sakamoto N, Yamamoto T, Wessel GM. The 3′UTR of Nanos2 directs enrichment in the germ cell lineage of the sea urchin. Developmental Biology. 2013;377:275–283. doi: 10.1016/j.ydbio.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oulhen N, Wessel GM. Retention of exogenous mRNAs selectively in the germ cells of the sea urchin requires only a 5′-cap and a 3′UTR. Mol Reprod Dev. 2013;80:561–569. doi: 10.1002/mrd.22193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez AJ, Seipel SA, Hamill DR, Romancino DP, Di Carlo M, Suprenant KA, Bonder EM. Seawi--a sea urchin piwi/argonaute family member is a component of MT-RNP complexes. RNA. 2005;11:646–656. doi: 10.1261/rna.7198205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Barton SC, Surani A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418:293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- Saitou M, Payer B, O’Carroll D, Ohinata Y, Surani MA. Blimp1 and the emergence of the germ line during development in the mouse. Cell Cycle. 2005;4:1736–1740. doi: 10.4161/cc.4.12.2209. [DOI] [PubMed] [Google Scholar]
- Saitou M, Yamaji M. Germ cell specification in mice: signaling, transcription regulation, and epigenetic consequences. Reproduction. 2010;139:931–942. doi: 10.1530/REP-10-0043. [DOI] [PubMed] [Google Scholar]
- Seervai RN, Wessel GM. Lessons for inductive germline determination. Mol Reprod Dev. 2013 doi: 10.1002/mrd.22151. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell. 2006;127(5):891–904. doi: 10.1016/j.cell.2006.11.016. [DOI] [PubMed] [Google Scholar]
- Shive HR, West RR, Embree LJ, Azuma M, Sood R, Liu P, Hickstein DD. brca2 in zebrafish ovarian development, spermatogenesis, and tumorigenesis. Proc Natl Acad Sci USA. 2010;107:19350–19355. doi: 10.1073/pnas.1011630107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Davidson EH. Gene regulatory network subcircuit controlling a dynamic spatial pattern of signaling in the sea urchin embryo. Proc Natl Acad Sci USA. 2008;105:20089–20094. doi: 10.1073/pnas.0806442105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J, Kraemer E, Liu H, Theodoris C, Davidson E. A spatially dynamic cohort of regulatory genes in the endomesodermal gene network of the sea urchin embryo. Dev Biol. 2008;313:863–875. doi: 10.1016/j.ydbio.2007.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solana J. Closing the circle of germline and stem cells: the Primordial Stem Cell hypothesis. Evodevo. 2013;4(1):2. doi: 10.1186/2041-9139-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JL, Stoeckius M, Maaskola J, Friedländer M, Stepicheva N, Juliano C, Lebedeva S, Thompson W, Rajewsky N, Wessel GM. Select microRNAs are essential for early development in the sea urchin. Dev Biol. 2012;362:104–113. doi: 10.1016/j.ydbio.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spéder P, Petzoldt A, Suzanne M, Noselli S. Strategies to establish left/right asymmetry in vertebrates and invertebrates. Curr Opin Genet Dev. 2007;17:351–358. doi: 10.1016/j.gde.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Spotila LD, Spotila JR, Hall SE. Sequence and expression analysis of WT1 and Sox9 in the red-eared slider turtle, Trachemys scripta. J Exp Zool. 1998;281:417–427. doi: 10.1002/(sici)1097-010x(19980801)281:5<417::aid-jez7>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- Stamateris RE, Rafiq K, Ettensohn CA. The expression and distribution of Wnt and Wnt receptor in sea urchin development. Gene Expr Patterns. 2010;10:60–64. doi: 10.1016/j.gep.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Lasko P. VASA localization requires the SPRY-domain and SOCS-box containing protein, GUSTAVUS. Dev Cell. 2002;3:865–76. doi: 10.1016/s1534-5807(02)00361-1. [DOI] [PubMed] [Google Scholar]
- Takeichi M. The cadherins: cell–cell adhesion molecules controlling animal morphogenesis. Development. 1988;102:639–655. doi: 10.1242/dev.102.4.639. [DOI] [PubMed] [Google Scholar]
- Tanwar PS, Kaneko-Tarui T, Zhang L, Rani P, Taketo MM, Teixeira J. Constitutive WNT/beta-catenin signaling in murine Sertoli cells disrupts their differentiation and ability to support spermatogenesis. Biol Reprod. 2010;82:422–32. doi: 10.1095/biolreprod.109.079335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Akasu R, Noce T. Embryonic stem cells can form germ cells in vitro. Proc Natl Acad Sci USA. 2003;100:11457–11462. doi: 10.1073/pnas.1932826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Nishida C, Yoshimoto S, Ito M, Oshima Y, Yokoyama S, Nakamura M, Matsuda Y. Diversity in the origins of sex chromosomes in anurans inferred from comparative mapping of sexual differentiation genes for three species of the Raninae and Xenopodinae. Chromosome Res. 2008;16:999–1011. doi: 10.1007/s10577-008-1257-z. [DOI] [PubMed] [Google Scholar]
- Vandenberg LN, Levin M. A unified model for left-right asymmetry? Comparison and synthesis of molecular models of embryonic laterality. Dev Biol. 2013;379:1–15. doi: 10.1016/j.ydbio.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vardy L, Orr-Wraver TL. The Drosophila PNG kinase complex regulates the translation of cyclin B. Dev Cell. 2007a;12:157–66. doi: 10.1016/j.devcel.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Vardy L, Orr-Wraver TL. Regulating translation of maternal messages: multiple repression mechanisms. Trends Cell Biol. 2007b;17:547–554. doi: 10.1016/j.tcb.2007.09.002. [DOI] [PubMed] [Google Scholar]
- Venezky DL, Angerer LM, Angerer RC. Accumulation of histone repeat transcripts in the sea urchin egg pronucleus. Cell. 1981;24(2):385–91. doi: 10.1016/0092-8674(81)90328-7. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Dunn NR, Sciammas R, Shapiro-Shalef M, Davis MM, Calame K, Bikoff EK, Robertson EJ. The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development. 2005;132:1315–1325. doi: 10.1242/dev.01711. [DOI] [PubMed] [Google Scholar]
- Voronina E, Lopez M, Juliano CE, Gustafson E, Song JL, Extavour C, George S, Oliveri P, McClay D, Wessel GM. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol. 2008;314:276–286. doi: 10.1016/j.ydbio.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E. The diverse functions of germline P-Granules in Caenorhabditis elegans. Mool Reprod Dev. 2013 doi: 10.1002/mrd.22136. in press. [DOI] [PubMed] [Google Scholar]
- Warner JF, Lyons DC, McClay DR. Left-Right asymmetry in the sea urchin embryo: BMP and the asymmetrical origins of the adult. PLoS Biol. 2012;10(10):e1001404. doi: 10.1371/journal.pbio.1001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel GM, Brayboy L, Fresques T, Gustafson EA, Oulhen N, Ramos I, Reich A, Swartz SZ, Yajima M, Zazueta V. The biology of the germ line in echinoderms. Mol Reprod Dev. 2013 doi: 10.1002/mrd.22223. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JA, Viswanathan SR, Yabuuchi A, Cunnif K, Takeuchi A, Park IH, Sero JE, Zhu H, Perez-Atayde A, Frazier AL, Surani MA, Daley GQ. A role for Lin28 in primordial germ-cell development and germ-cell malignancy. Nature. 2009;460:909–913. doi: 10.1038/nature08210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Small micromeres contribute to the germline in the sea urchin. Development. 2011;138:237–243. doi: 10.1242/dev.054940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM. Autonomy in specification of primordial germ cells and their passive translocation in the sea urchin. Development. 2012;139:3786–3794. doi: 10.1242/dev.082230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M. Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet. 2008;40:1016–1022. doi: 10.1038/ng.186. [DOI] [PubMed] [Google Scholar]
- Yamaji M, Ueda J, Hayashi K, Ohta H, Yabuta Y, Kurimoto K, Nakato R, Yamada Y, Shirahige K, Saitou M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem Cell. 2013;12:368–382. doi: 10.1016/j.stem.2012.12.012. [DOI] [PubMed] [Google Scholar]
- Yamazaki A, Furuzawa Y, Yamaguchi M. Conserved early expression patterns of micromere specification genes in two echinoid species belonging to the orders clypeasteroida and echinoida. Dev Dyn. 2010;239:3391–3403. doi: 10.1002/dvdy.22476. [DOI] [PubMed] [Google Scholar]
- Yamazaki A, Kidachi Y, Minokawa T. “Micromere” formation and expression of endomesoderm regulatory genes during embryogenesis of the primitive echinoid Prionocidaris baculosa. Dev Growth Differ. 2012;54:566–578. doi: 10.1111/j.1440-169X.2012.01360.x. [DOI] [PubMed] [Google Scholar]
- Yankura KA, Koechlein CS, Cryan AF, Cheatle A, Hinman VF. Gene regulatory network for neurogenesis in a sea star embryo connects broad neural specification and localized patterning. Proc Natl Acad Sci USA. 2013;110:8591–8596. doi: 10.1073/pnas.1220903110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou GB, Meng QG, Li N. In vitro derivation of germ cells from embryonic stem cells in mammals. Mol Reprod Dev. 2010;77:586–594. doi: 10.1002/mrd.21187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.