Abstract
Malaria has been responsible for the highest mortality in most malaria endemic countries. Even after decades of malaria control campaigns, it still persists as a disease of high mortality due to improper diagnosis and rapidly evolving drug resistant malarial parasites. For efficient and economical malaria management, WHO recommends that all malaria suspected patients should receive proper diagnosis before administering drugs. It is thus imperative to develop fast, economical, and accurate techniques for diagnosis of malaria. In this regard an in-depth knowledge on malaria biomarkers is important to identify an appropriate biorecognition element and utilize it prudently to develop a reliable detection technique for diagnosis of the disease. Among the various biomarkers, plasmodial lactate dehydrogenase and histidine-rich protein II (HRP II) have received increasing attention for developing rapid and reliable detection techniques for malaria. The widely used rapid detection tests (RDTs) for malaria succumb to many drawbacks which promotes exploration of more efficient economical detection techniques. This paper provides an overview on the current status of malaria biomarkers, along with their potential utilization for developing different malaria diagnostic techniques and advanced biosensors.
1. Introduction
The World Malaria Report 2012 released by World Health Organization (WHO) summarises the data received from 104 malaria endemic countries [1]. This report estimated around 219 million cases of malaria and a high mortality of about 660,000 people due to the disease in the year 2010. Malaria in humans is transmitted by the bite of more than thirty species of female anopheline mosquitoes. The causative agent is a protozoan parasite of genus Plasmodium. Five species, P. falciparum, P. vivax, P. malariae, P. ovale, and P. knowlesi, are known to affect humans. Characteristic symptoms of the disease include episodes of high fever followed by chills and rigors which are repeated every 48 hrs in falciparum, ovale, or vivax, every 72 hrs in malariae infections, and 24 hrs in knowlesi. P. vivax hypnozoites can lie dormant in the liver and may cause recurrence of the disease [2]. Indiscriminate use of oral artemisinin based monotherapies has been identified as one of the factors that lead to drug resistance, which is a serious problem in malaria management. For example, a sulfadoxine-pyrimethamine combination, which is an effective treatment against multidrug resistant (MDR) malaria, has now been rendered useless in Southeast Asia by its overuse and misuse [1, 3]. WHO recommends that all persons suspected of malaria should receive parasitological confirmation before the drugs being administered, which prevents mismanagement of nonmalarial fevers [4] and lessens overdiagnosis of malaria [5, 6].
In spite of the vast array of tests available today to diagnose malaria, we still await a test that is quantitative and specific to distinguish different Plasmodium species. Although rapid detection tests (RDTs) have facilitated considerably diagnosing malaria, the tests are plagued with many limitations such as variability in results, being nonquantitative, and having poor storage stability in tropical regions. In order to develop an efficient test for diagnosis of malaria, a profound understanding on various malaria-related biomarkers is prerequisite. This review outlines the diagnostic tools for malaria with special focus on the potent biomarkers reported to date and their application for developing rapid and reliable detection techniques including biosensors.
2. Biomarkers for Malaria
Biomarkers are cellular, biochemical, or molecular alterations measurable in biological samples which indicate any biological, pathogenic, or therapeutic responses [7]. There has been no established classification system to categorize biomarkers to date. However, Frank and Hargreaves [8] bring clarity to the biomarkers by classifying them into three types: Type 0, Type 1, and Type 2. Type 0 biomarkers are measures of the natural history of disease and correlate with clinical outcomes; Type 1 biomarkers usually determine the biological effect of a therapeutic intervention; and Type 2 biomarkers are the equivalent of “surrogacy” markers where a surrogate point has been defined as a biomarker intended to substitute for a clinical end point, with the latter being a characteristic or variable that reflects how a patient feels, functions, or survives. Biomarkers are useful for disease management as well as formulating strategies before the onset of disease in case of asymptomatic malaria. In malaria endemic areas like Africa, where transmission is high, it has been found that many P. falciparum infected individuals exhibit asymptomatic malaria [9]. The prevalence of these asymptomatic infections can be as high as 52%.
2.1. Lactate Dehydrogenase
2.1.1. Metabolic Role
The red blood cell (RBC) is a nonproliferating cell with a modest requirement of around 5 μmol glucose/24 hrs/109 RBCs. However, Plasmodium is a voracious scavenger of blood glucose that increases the RBC glucose consumption up to 100-fold. In case of P. falciparum about 60–70% of the glucose is converted to lactic acid and excreted; however, this percentage varies amongst different Plasmodium species and in vitro culture conditions [10]. During the intraerythrocytic stages, the parasite principally relies on anaerobic respiration for ATP generation from glucose, and the NAD+ is regenerated by conversion of pyruvate to lactate while the mitochondria contribute minimally to the ATP pool [11]. This reaction is catalysed by lactate dehydrogenase (LDH), the final enzyme of the glycolytic pathway in Plasmodium. The absence of F0 a and b subunits of the mitochondrial F0-F1 ATP synthase further implies the absence of mitochondrial role in energy generation. However, since parts of the genome sequence are still not known, and the genome being A+T rich that causes difficulty in identifying enzymes, the lack of role of mitochondria in energy generation cannot be ascertained [12]. Nevertheless, due to the dependence on the glycolytic cycle for energy generation, enzymes involved in this pathway are overexpressed [13]. It has been found that P. falciparum LDH (PfLDH) RNA expression level gradually increases, with the peak expression being at 24 to 30 hrs in the intraerythrocytic cycle. This expression declines to zero in the schizont stage. A similar profile that slightly lags behind the RNA expression was observed for the enzyme activity as well [14]. With the help of microarray technology using P. falciparum transcription, it was demonstrated that all glycolytic enzymes are upregulated at the early trophozoite stage during the asexual cycle, coinciding with the time of maximal metabolic activity by the parasite [15].
2.1.2. Structure and Kinetic Parameters of LDH
The parasite LDH (pLDH) is a tetramer where each monomer consists of two domain LDH folds. The larger domain comprises the Rossmann fold that binds the cofactor NADH, while the catalytic residues (His 195, Asp 168, and Arg 171) are located in the other domain. These residues are conserved across all Plasmodium species except P. knowlesi, which lacks His 195. The active site of the enzyme is located between these two domains. Each monomer of the pLDH protein carries one NADH molecule, and each of the four cofactors occupies identical positions in each monomer [16].
Protozoal LDHs display some major structural and kinetic differences compared to their mammalian counterparts that may be exploited to develop selective drugs and detection systems for malaria. PvLDH and PfLDH share 26% and 29% sequence identity with human LDH-A (hLDH-A), respectively. Structurally the parasite enzyme differs in having a five-residue insertion in its active site loop which closes down over the active site during catalysis [16] and also causes a displacement of ~1 A° of the nicotinamide moiety of the NADH cofactor [17, 18]. This insertion “DKEWN” (D-Aspartic acid, E-Glutamic acid, K-Lysin, W-Tryptophan, and N-Asparagine) was used as a common diagnostic epitope by Hurdayal et al. [19] to selectively detect pLDH from human LDH isoforms. The five amino acid insertions initially observed for P. falciparum [20] were later discovered in all five Plasmodium species and in the LDH I and II of T. gondii [21, 22]. However, this feature was not found in all apicomplexan parasites, for instance, Cryptosporidium parvum which causes cryptosporidiosis, a parasitic infection of the mammalian intestinal tract.
Kinetically, the parasite enzyme differs by not being inhibited by excess of the substrate pyruvate, a feature not shared by mammalian enzymes [23]. In the latter, substrate inhibition occurs due to the slow release of the reduced cofactor NAD+ from the active site due to the formation of a covalent adduct with pyruvate. Reduced substrate inhibition is attributed to a single amino acid substitution in the protozoal enzyme (Ser163Leu). The single amino acid mutation has also been exploited as a general method to reduce substrate inhibition in L-lactate dehydrogenase enzymes [24]. The substrate inhibition in plasmodial counterparts is lower by around 175- and 35-fold than that shown by the human heart and muscle isoforms, respectively [25]. The mechanism for the lower substrate inhibition is stated to be the weaker binding of pyruvate to enzyme-cofactor complex rather than slower release of NAD+ [16, 26]. Another kinetic difference is the ability of the pLDH to efficiently use the synthetic cofactor APAD+ (3-acetylpyridine adenine dinucleotide) [17, 27]. Increase in entropy on APAD+ binding altering the rate of active site movement or a higher oxidation potential of APAD+ than NAD+ leading to faster hydride transfer to APAD+ is ascribed as one of the reasons for this cofactor preference [16, 28, 29]. By using lactate and NAD+ as inhibitors in a forward reaction of pyruvate to lactate direction it was revealed that this enzyme exhibits ordered sequential bi-bi mechanism of binding in which the NADH cofactor binds before the pyruvate substrate is attached.
Another striking difference of pLDH from other dehydrogenases is its substrate specificity due to long substrate specificity loop. The enzyme efficiency decreases with the increase in number of methylene groups in the substrate, as is seen when pyruvate was replaced with α-ketobutyrate. Also the enzyme does not show an activity with phenylpyruvate [25], a behaviour that distinguishes the enzyme from the LDH of T. gondii. Although LDHs from P. falciparum and T. gondii have very similar structures, the latter has an additional loop insertion of two residues and several changes in its active site that renders the contrast property [29, 30]. The presence of a positively charged lysine amino acid at position 102 gives the Toxoplasma enzyme a mild malate dehydrogenase activity [25]; this finding, however, is not supported by the previous report [16]. A characteristic feature of this family of enzymes is that the value of K m for substrates is pH dependent, as the substrate pyruvate binds only when the active site histidine is in the protonated state and lactate is in deprotonated state [31].
pLDH is an attractive target for antimalarial drug design because of its three important attributes. (1) It controls the production of plasmodial ATP, (2) it has unique amino acids at the active site compared to its counterparts from other species, and (3) the protein data bank (PDB) contains many X-ray crystallographic structures of pLDH complexed with various compounds which provide ideal targets for modelling of inhibitors (See Table S1 in Supplementary Material available online at http://dx.doi.org/10.1155/2014/852645). From the inhibition studies of various compounds including gossypol and its derivatives against the parasite enzyme pLDH [32–37] (Table S2), it has been revealed that these compounds specifically target pLDH and not human LDH. The findings underline the fact that there are structural differences between the two enzymes that may be exploited for selective detection of pLDH for malaria diagnosis.
2.2. Histidine-Rich Protein II
2.2.1. Occurrence
For the first time in the avian malarial parasite Plasmodium lophurae, numerous cytoplasmic granules were isolated and studied. It was shown to have an unusual polypeptide composition whose amino acid analysis showed 73% histidine, 7.5% proline, 7% alanine, 6% glutamic acid, and 2.1% aspartic acid [38]. This unusual histidine-rich polypeptide raised interest in the structure of the protein and its function. Histidine-rich proteins (HRPs) are also observed in many organisms with different functions [39–42].
P. falciparum synthesizes a unique set of soluble HRPs during the asexual erythrocytic development which are denoted as HRPs I, II, and III in the order of their discovery [43]. HRP I protein also known as knob-associated protein (KAHRP-I) is found in Knob+ strain which means phenotypically that it expresses knob-like protrusion on the cell surface and is suggested to help in the cytoadherence of infected erythrocytes to the venular endothelium [44] and partially contributes to the high parasitemia and hypoxia associated with P. falciparum. HRP II, which is exclusive to P. falciparum, is found in both Knob+ and Knob− strains and is reported to have many functions such as heme binding and heme detoxification by forming hemozoin [45, 46]. It has been projected as a model vaccine against malaria too [47]. HRP III also known as small histidine-alanine-rich protein (SHARP) is a smaller protein and is not found as abundant as the other two proteins. The HRP III sequence of FC27 strain, a Plasmodium isolate FCQ27/PNG from Papua New Guinea, was reported and shown to have polymorphisms in the gene's repeats [48]. It shares high homology with HRP II [49]. Also varying degrees of cross-reactivity between HRP II and HRP III have also been reported for HRP II MAbs: 2G12, MAb87, and 1D6 [50].
2.2.2. Genetic and Structural Organization
The genomic sequence of hrp II contains a hydrophobic signal peptide, an intervening intron and an extensive region of tandem repeats that encodes a 35 kDa polypeptide consisting mostly of histidine, alanine, and aspartic acid. There are 18 tripeptides (Ala-His-His), 3 pentapeptides (Ala-His-His-Ala-Ala), and 33 hexapeptides (Ala-His-His-Ala-Ala-Asp). There is about 85–90% homology between the tandem repeat domains and the regions flanking the repeats of hrp II and hrp III genes which imply that both have originated in the duplication of an ancestral sequence [49]. Another study of the flanking 5′ and 3′ untranslated regions (UTR) of the hrp II and hrp III genes revealed a 5′ UTR intron which has been shown to have high conservation as the coding intron and a 3′ UTR, having a much greater homology than the histidine repeat region of the coding sequences [51]. Figure 1 demonstrates the homology between the two genes. Both hrps II and III share many similarities, but still it is unclear how these genes function and why HRP II is released in large quantities.
Figure 1.

Schematic alignment of hrp II and hrp III genes including 5′ and 3′ UTRs. INT and SEC L stand for intron and secretory leader, respectively. The gene consists of a hydrophobic signal peptide (brown), an intervening intron, and an extensive region of tandem repeats (pink). The high homology (85–90%) between the tandem repeat domains and the regions flanking the repeats of hrp II and hrp III genes is shown [51].
HRP II transports from the parasite, through the host cell cytoplasm, to the culture supernatant in vitro [43] which also accounts for its presence in the serum of plasma, cerebrospinal fluid, and urine of infected patients [52, 53]. It is also present in food vacuole [54], digestive vacuole [55], and membrane surface of the infected RBCs [50]. In short, HRP II is found everywhere and released in abundance which is also a reason for its importance as an antigen biomarker of malaria. The CD spectrum of HRP II in aqueous medium was indicative of random coil and the spectrum changes dramatically from random coil to 310-helix conformation as soon as heme molecules are titrated in the solution [56]. The 310-helix conformation is commonly observed in proteins as short sequences of three to four residues, mostly associated with N- or C-termini of α-helices [57, 58]. However, the length of 310-helix is found to be unusually long in case of HRP II. Along with the changes in the secondary structure of the protein, dimerization of HRP II monomers by formation of intermolecular disulfide bonds also takes place as shown in Figure 2. This process of dimerization depends on heme binding. There is no X-ray crystal structure of HRP II available till now.
Figure 2.

Fe+3 PPIX (protoporphyrin IX) binding to HRP II is able to bring about an interaction between two monomers resulting in the formation of an intermolecular disulphide bond.
2.2.3. Function of HRP II
The actual function of the HRP II is not yet adequately known. However, many reports have demonstrated that its main function is heme binding that may link its role in the heme detoxification in malaria parasites. Spectroscopic studies showed that a single HRP II molecule may bind to multiple heme molecules [59]. By using biuret assay based spectroscopic method, it was determined that 15 heme molecules bind to one HRP II molecule [56]. The K d values for heme binding were calculated to be 0.34 μM. In another report the number of binding sites in HRP II-heme complex is estimated as 18 with K d of 0.94 μM [60], which is in agreement with another report [55]. Hence the estimated binding sites of heme molecules are around 15–18. Further, it was confirmed that the heme molecules in protein heme complex are in low spin, six coordinated, and bis-imidazole ligated. There are also reports on heme-artemisinin adducts. Artemisinin is an antimalarial drug that bears a peroxide function and is triggered only in parasite infected hosts. The drug binds with higher affinity to HRP II as compared to the heme moiety [61].
HRP II is also known to initiate the formation of hemozoin. In vitro heme polymerization assay performed at the optimum pH 4.0 with native and recombinant HRP II promoted the formation of hemozoin. Polymerization increased with time, protein concentration, and initial concentration of heme [55]. A study conducted on biomineralized dendrimeric templates showed potential of heme binding to metal moieties and initiated hemozoin formation [62]. The formation of hemozoin is, however, a complex phenomenon and needs further investigation to be understood adequately.
At an acidic pH electrostatic interaction takes place between the strongly positive charged histidine of HRP II and negative groups on actin and phosphatidylinositol 4,5-biphosphate (PIP2) of host cells. An important role of HRP II as a buffering protein has been suggested as it helps the parasite to stabilize the changes to the cytoskeleton induced by other released parasitic proteins [63]. Another function of HRP II is the neutralization of bacterial toxin LPS [64]. The observed LPS-neutralizing effect of HRPs has probably resulted from the electrostatic interactions between histidines and the negatively charged phosphate groups of LPS.
2.3. Hemozoin
Hemozoin is an insoluble microcrystalline product formed from the digestion of blood by Plasmodium spp. and few other species of blood-feeding parasites [65–67]. Originally, Giovanni Maria Lancisi (1717) had observed that internal organs of malaria victims are decolorized; the biochemical reason of it was, however, not known. Later on the cause was attributed to hemozoin. Hemozoin plays a role as visible marker in identifying malarial parasites and hence, it is popularly termed as malaria pigment. The parasites infect the RBCs and digest hemoglobin resulting in release of amino acids and toxic-free heme (ferriprotoporphyrin IX) which is polymerized to hemozoin. The crystal structure of hemozoin consists of an unusual polymer of hemes linked between the central ferric ion of one heme and a carboxylate side-group oxygen of another heme [68]. Later on its crystal structure was confirmed to be similar to synthetic β-hematin using X-ray diffraction powder pattern [69]. Though there has been extensive study on the structure and characterization of hemozoin crystals, the process of its nucleation is not yet clearly known. There are reports about initiation of hemozoin formation occuring autocatalytically, while other reports suggest that it requires external aids [70]. Most popular belief is that HRP II helps in the initiation of hemozoin formation as it binds firmly to heme molecule in aqueous environment. However, there are other mechanisms that support the view that it is promoted by polar membrane lipids [71] and neutral lipid bodies [72–74]. Since it is a survival tactic of the parasite from the heme toxicity, it has been targeted for malaria drug study for many years [75].
2.4. Other Biomarkers
2.4.1. Aldolase
Aldolase is a key enzyme in the glycolytic pathway of the parasite. It catalyses the cleavage of fructose-1,6-bisphosphate into glyceraldehyde-3-phosphate and dihydroxyacetone phosphate [76]. The genomic sequence of aldolase consists of two exons interrupted by one intron. The first one encodes only one amino acid, the initiation methionine, while the second one encodes the residual 368 amino acids of the protein. The enzyme is homotetrameric protein with each subunit of approximately 40 kDa [77]. Crystal structure of plasmodium aldolase has been studied at a resolution of 3 A° to determine possible structure based inhibitors [78]. The aldolase is localized in the cytoplasm of the parasite as an active and soluble form and is also found to be associated with the membrane fraction as an insoluble form. The enzyme has high degree of sequence diversity from the host and thus has the potential as a drug target. There have been studies on inhibition of recombinant P. falciparum aldolase by several candidates like rabbit antibodies, a 19-residue synthetic peptide, and phosphorothioate antisense oligodeoxynucleotides [77, 79]. In higher vertebrates there are three tissue-specific aldolase isoenzymes whereas only one aldolase enzyme has been identified in blood sucking parasites, for example, Trypanosoma brucei [80], Giardia lamblia [81], and P. falciparum [82], suggesting that multiple isoenzymes are not necessary for the completion of protozoan parasite life cycles. In an experiment examining stage-specific expression of aldolase isoenzymes in P. berghei, two genes of aldolase (aldo-1 and aldo-2) were found where aldo-1 sequence was found to be identical to P. falciparum aldolase, whereas aldo-2 had 13% sequence diversity [83]. In another study the above statement was contradicted and it was reported that only single aldolase gene occurs in the following malaria species: P. berghei, P. chabaudi, P. vinckei, and P. yoelii and from the human malaria parasite P. vivax [84]. Many reports have shown poor sensitivity of aldolase RDTs which encouraged studying more its genetic diversity. Lee et al. [85] studied the diversity in P. falciparum and P. vivax aldolase and showed that aldolase is highly conserved, indicating that antigenic diversity is not a cause of variable RDT sensitivity.
2.4.2. Glutamate Dehydrogenase
Glutamate dehydrogenases (GDHs) are ubiquitous enzymes that occupy an important branch point between carbon and nitrogen metabolism. They are generally involved in ammonium assimilation (NADP-dependent GDHs) or glutamate catabolism (NAD-dependent GDHs). P. falciparum express three GDH isozymes. PfGDH1 is a NADP-dependent glutamate dehydrogenase which is a homohexamer with a subunit of M r 49,500 [86]. It has been postulated to play a role in the parasite's redox metabolism [13]. The crystal structures of PfGDH 1 and 2 have been solved to a resolution of 2.7 A° [87] and 3.1 A° [88], respectively. GDHs possess a unique N terminal residue extension not found in the mature human enzyme and are present throughout the intraerythrocytic cycle of the parasite. Furthermore GDHs are absent in the host RBC making them a potent biomarker [86]. PfGDH1 was used to detect the presence of P. falciparum using western blotting [89]. Monoclonal antibodies in combination with colloidal gold were used in an immunochromatographic assay for diagnosis of P. falciparum. This assay showed a sensitivity and specificity of 86.6% and 96.4%, respectively [90].
2.4.3. Serological Biomarkers for Cerebral Malaria
Cerebral malaria (CM) is a life-threatening complication of malaria and is defined as an unarousable coma with a P. falciparum infection in the absence of other causes of encephalopathy. If left untreated, it is fatal within 24–72 hrs [91]. The advantage of an early immunologically relevant serological biomarker for CM would be stratifying febrile patients into groups: those who are likely to develop CM and those who are likely to develop severe malaria (SM) or mild malaria (MM). Although no clear biomarkers have yet been identified for CM, studies show a conspicuous relationship between chemokine interferon inducible protein (CXCL10 and CXCL4) and severity of CM [12, 92]. It was observed that patients with CM have significantly elevated levels of CXCL-10 and CXCL-4.
Several other serological biomarkers for CM have also been identified. For example, elevated levels of soluble tumor necrosis factor receptor (sTNF-R) and soluble Fas ligand (sFas), a 26 kDa glycoprotein generated from its membrane bound form by a metalloproteinase-like protease, may indicate CM neuropathy [92]. Cerebrospinal fluid (CSF) and serum levels of twelve cytokines and chemokines were monitored and it was found that elevated levels of interleukin 8 (IL-8), interleukin 1 receptor antagonist (IL-1ra), and TNFα were present in children infected with CM as compared to healthy controls [93]. The levels of endothelial regulators like angiopoietin I (ANG I) and ANG II are also affected during CM. It was found that ANG II and ratio of ANG II: ANG I could be used to accurately distinguish between CM and MM patients. This study was done with serum [94] as well as whole blood [95] angiopoietin levels. Potency of a neuroprotective factor, erythropoietin (EPO), was assessed to predict the risk of developing neurological sequelae after recovery to CM [96]. EPO has also been used for adjunctive therapy to prevent neuronal damage [12] in addition to its use as a prognostic biomarker. Serum levels of soluble adhesion molecules, like intracellular adhesion molecule I (ICAM-I), vascular cell adhesion molecule I (VCAM-I), and so forth, were studied in Gambian children and were found to be elevated in children with SM compared to MM [97]. Kassa et al. [98] have exploited the amphipathic nature of hemozoin to capture serum proteins that differ between malaria-infected individuals and healthy individuals. LC-MS/MS analysis of the captured proteins showed that serum amyloid A (SAA), apolipoprotein E (ApoE), and lipopolysaccharide (LPS) binding protein may have potential to act as prognostic biomarkers. Complement system components like C1q, C3, C4, and C5a are affected in malaria patients. Depressed levels of C3 were found in severe and uncomplicated malaria as compared to healthy controls [12, 99].
Microparticles (MPs), also known as microvesicles, are fragments of plasma membrane shed by various cell types under physiological stress conditions and have also been linked with pathophysiology [100]. MPs have been reported in the serum of patients suffering from malaria and diabetes [101, 102], systemic lupus erythematosus [103], and acute coronary syndromes [104] and in conditions of severe trauma [105]. There is a dramatic difference in the plasma levels of MP of endothelial origin among Malawian children suffering from CM, severe malarial anaemia, and uncomplicated malaria caused by P. falciparum [106]. Mfonkeu et al. [107] demonstrated by using fluorescence activated cell sorting (FACS) that MP of platelet origin may be a relevant marker in the routine followup since the levels of MP dramatically increased during CM and decreased when the patient was cured.
The serological biomarkers, however, are difficult to use reliably in diagnosing malaria. For instance, CXCR3 and its ligands have been implicated in case of several neurological diseases like West Nile virus [108, 109], Toxoplasma gondii [12], and HIV infections [110]. Serological biomarkers may be used to indicate CM only after the malaria diagnosis is confirmed using an antigenic biomarker. Potential diagnostic antigenic biomarkers have been summarised in Table 1 for better understanding.
Table 1.
A brief summary of diagnostic antigenic markers of malaria.
| Name of biomarker | Chemical nature | Localization | Salient features | References |
|---|---|---|---|---|
| pLDH | Homotetrameric protein with each monomer of 34 kDa | Inside infected RBCs | Presence of five amino acid residue insertions (DKEWN) in active site loop. Ability to actively utilize synthetic cofactor APAD+. Reduced pyruvate substrate inhibition. |
[16, 17, 25] |
|
| ||||
| HRP II | A 35 kDa protein | Secreted in serum of infected patient | Unique tandem repeats (Ala-His-His-Ala-Ala-Asp). 310-Helix conformation when bound to heme. Secreted in abundance in serum, CSF, and urine of infected patients. |
[49, 56] |
|
| ||||
| Hemozoin | β-Hematin | Inside digestive vacuole of parasite | Consists of Fe(III)PPIX centrosymmetric dimmers linked by hydrogen bonds. Accumulates in the digestive vacuole of parasite and appears as cluster as observed under electron microscope. |
[69, 75] |
|
| ||||
| Aldolase | Homotetrameric protein with each subunit of 40 kDa | Inside infected RBCs | High sequence diversity from host and has potential as a drug target. Used for following response to therapy as it is detected only at high parasitemia. |
[77, 111, 112] |
|
| ||||
| pGDH | Homohexameric protein, with each monomer being 49.5 kDa | Inside infected RBCs | Plays a role in parasite's redox metabolism. Not found in host RBC making it a potent biomarker. Appears throughout the erythrocytic cycle. |
[13, 86] |
3. Detection of Malaria
3.1. Microscopic and Conventional Analytical Techniques
Microscopy for malaria diagnosis has been used since 1904 when Gustav Giemsa introduced a mixture of methylene blue and eosin to stain the parasite [113]. This method is, however, time consuming and labour intensive and requires expert skills. To enhance the sensitivity of microscopic detection, fluorescent microscopy using fluorophores such as acridine orange and benzothiocarboxypurine has frequently been used in addition to quantitative buffy coat technique. Although microscopy was considered as “gold standard” for malaria diagnosis, recent reports suggest otherwise. Bayesian latent class models were used to estimate sensitivities and specificities of various diagnostic tests and revealed that microscopy is a poor reference test [114].
Apart from microscopy, nucleic acid techniques such as PCR and nested PCR [120, 121] and LAMP have also been used for malaria detection. LAMP can be conducted under isothermal conditions and does not need expensive thermocyclers [120]. However, this method may pose danger of cross-contamination during addition of dye for visualization of results. Also this method is not suitable for target DNA greater than 500 bp, as this causes a hindrance to strand displacement [122]. Most of these conventional methods have been summarised in Table 2.
Table 2.
Comparison of conventional detection techniques with advanced biosensors for malaria detection.
| Types of test | Principle of the method | Instrument used | Sensitivity and specificity | Detection limit (parasites/μL) | Response time (min) | Instrument cost |
|---|---|---|---|---|---|---|
| Peripheral blood smears (PBS) | Morphological changes in the stages of parasite by thick and thin blood smears | Optical microscope | Depends on the instrument quality and the skill of the handler | 5–10 | 30–60 | Low |
| Quantitative buffy coat (QBC) | Blood staining by acridine orange | Epifluorescent microscope | Higher than PBS test | <15 | >5 | Moderate |
| RDTs | Detection based on antigen-antibody interactions and enzyme assay | Disposable dipsticks | Moderate at higher parasitemia (>100 parasite/μL) | 50–100 | 10–15 | low |
| PCR | Specific amplification of malaria DNA | Thermocycler | High | ≥1 | 45–360 depending on the methods | High |
| Serological tests | Detection of malaria antigen or antiparasite antibodies in blood | Elisa, WB | Relatively high | 30–60 | Not mentioned | Moderate |
| LAMP | Detection of turbidity after amplifying DNA sequences | Turbidity meter | High | <60 | >5 | Moderate |
| Microarrays | Hybridization of DNA and quantification by fluorescent based detection | DNA chip | High | <60 | Not mentioned | High |
| Flow cytometry | Detection of hemozoin | Flow cytometer | Variable sensitivity, high specificity | <1/sample | Poor correlation with parasitemia | High |
| Automated blood cell counters | Detection of hemozoin in activated monocyte | Hematology analyzers | Variable sensitivity and specificity | <1/sample | 5–20 | High |
| Mass spectrometry | Identification of heme | Laser desorption mass spectrometry | Undetermined | <1/sample | 100 for whole blood | High |
| Amperometric immunosensor | Based on detecting target (HRP II) with the help of antibodies and modified electrodes that generate electricity on interaction | Cyclic voltammetry | Sensitivity is >95% and specificity >90% |
8 ng HRP II/mL | Not mentioned d | Moderate |
| Piezoelectric immunosensor | Based on antibody interactions with target HRP II on the quartz crystal | Cyclic voltammetry | Less sensitive than amperometric immunosensor | 12 ng HRP II/mL | Not mentioned | Moderate |
| Magneto- immunosensor | Based on antibodies targeted to HRP II which was successfully performed in a sandwich assay on magnetic micro- and nanoparticles |
Amperometric controller and Microplate reader for optical measurements | High sensitivity | 0.36 ng HRP II/mL | Not mentioned | Moderate |
| Aptasensor | Based on detecting LDH | Electrochemical impedance spectroscopy | High sensitivity | 108.5 fM for PvLDH and 120.1 fM for PfLDH | Not mentioned | Moderate |
Flow cytometry (FCM) and automated blood cell counting techniques can be used to detect hemozoin or Plasmodium dsDNA in infected erythrocytes; however, the use of Plasmodium dsDNA as a marker in flow cytometry can result in false positives because pathological conditions are characterized by an efflux of normoblasts and erythroblasts in blood [123]. LDMS is used for detection of heme in hemozoin. Hemozoin strongly absorbs UV light resulting in vaporization of individual heme molecules. LDMS only detects small molecules (<1.5 kDa) and favours phospholipids and porphyrins; hence, it is ideally suited for heme detection; however, the detection is only semiquantitative and cannot differentiate between species [124].
A unified sandwich ELISA targeting PfLDH and HRP II was reported that allowed concurrent measurement of the biomolecules [125]. ELISA is an efficient method to detect malaria in a short time frame. Despite the advantages such as high sensitivity, the application of ELISA remains restricted to research settings and blood bank screening where large number of samples have to be screened every day, since there are no commercial species-specific ELISA tests kits for all species [126].
3.2. Rapid Detection Approaches
3.2.1. Rapid Diagnostic Tests (RDTs)
RDTs are quick and portable immunochromatographic dipsticks whose sensitivity generally reaches >95% at P. falciparum density of 1000–2000 parasites/μL [127]. They rely on capture of parasite antigen from peripheral blood using monoclonal antibodies conjugated to either a liposome containing selenium dye or gold particles. A second monoclonal antibody applied to a strip of nitrocellulose acts as the immobile phase. The antigen-antibody complex in the mobile phase migrates along the strip and is captured by the monoclonal antibody of the immobile phase, thus producing a visible coloured line [128].
Dipsticks can be divided into two classes based on their target antigenic biomarkers, namely, HRP II (only for P. falciparum) and pLDH. Most commercial RDTs that detect P. falciparum target HRP II and few others target pLDH. To enhance the efficiency of malaria detection, RDTs targeting many biomarkers at the same time are prepared. The combined immunochromographic-malaria dipstick (ICT) targets HRP II and aldolase for the diagnosis of P. falciparum and P. vivax [111, 112]. The authors suggest using aldolase based RDT for monitoring response to therapy as it is sensitive only at higher parasitemia in sample. CareStart (Access Bio, Princeton, NJ, USA) Malaria HRP II/pLDH Combo Test performed well for detection of P. falciparum (based on HRP II) and pan LDH for non-falciparum infections, but sensitivities for P. ovale and P. malariae were poor [129].
Dipstick operation is varied and may include dipping a nitrocellulose test strip into a blood specimen, placing a blood drop directly on the test strip, or onto a sample pad impregnated with labelled monoclonal antibody (mAb). Results are usually obtained in 5 to 15 minutes [130]. In many ways ICTs appear ideal: they are rapid (<30 mins) and can be easily used by healthcare workers or semiskilled volunteers.
Variations in RDT performance have been seen with season, year, age of patient, and presence or absence of fever during consultation [131]. The most common cause of poor performance of RDTs on the tropics is exposure to high temperature/humidity [132]. The loss in activity is attributed to damage to monoclonal antibodies or the nitrocellulose membrane. This was further confirmed by Chiodini et al. [133] who tested five RDTs from the same lot but stored at different temperatures of 35°C, 45°C, and 60°C. Although dipsticks are widely used, they have certain limitations such as poor species and stage differentiation and quantification, false positive results due to persistent HRP II antigenemia [134, 135] and rheumatoid factors in some patients [136, 137], and false negative results due to excess antigen in case of high parasitemia [138] or HRP II gene deletion [139–141]. The sensitivity of Parascreen (Zephyr Biomedical Systems) was found low for P. falciparum infections. This RDT was not acceptable for malaria diagnosis under the field conditions in the Peruvian Amazon due to HRP II and HRP III gene deletions in the malaria parasite genome [142–144]. Since HRP II persists after parasite clearance, the presence or absence of pLDH appears to be a more reliable diagnostic target, as it is produced by live parasites [118, 139, 145]. HRP II antigenemia is suspected to be a result of gametocytemia [146].
Genetic polymorphism may lead to false negative results as was observed when a countrywide assessment of polymorphism was carried out for HRP II, pLDH, and aldolase genes in Madagascar. Higher levels of polymorphism were observed for HRP II and HRP III genes predicting that about 9% of Malagasy isolates could not be detected at parasite densities <250 parasites/μL [147]. This genetic variation was seen both within and among different countries. Logistic regression analysis predicted that due to this variation only 84% of the P. falciparum infections in the Asia-Pacific region are likely to be detected at densities <250 parasites/μL. HRP III is also suspected to play a role in the performance of HRP II based tests [148]. However, a study on the assessment of plasma concentration, and hence the disease severity, has indicated that sequence polymorphism is not a significant cause of variation in HRP II concentration in plasma samples from African children [149]. In African children it is a major challenge to distinguish severe falciparum malaria from other severe febrile illnesses with P. falciparum infection. Plasma HRP II has prognostic significance and provides a tool to assess the risk of “true” severe malaria as compared to other severe illnesses in parasitemic African children [150]. This justifies the development of plasma HRP II concentration as a method for assessing severe falciparum malaria in African children. Several reviews outline various RDTs [151] and their evaluation reports [130]. In spite of over 100 such reports their comparative assessment is, however, difficult due to difference in population sizes, their clinical and epidemiological characteristics, different trial guidelines, and so forth [152]. A comparison of sensitivity and specificity of few of the field evaluations done during the last decade is shown in Table 3. It can be observed from the table that the sensitivity and selectivity of RDTs significantly vary over different studies and thus there is a need for more sensitive, selective, and reliable methods for rapid detection of malaria. One such effort was made by Peng et al. [153] by developing the Wondfo rapid diagnostic test which is a nanogold based immunochromatography assay that uses monoclonal antibodies. It is a more rapid and sensitive parasite detection method and results showed very good concordance with microscopy examination with a sensitivity and specificity of 95.49% and 99.53%, respectively.
Table 3.
Comparison of evaluation reports of various RDTs done in the recent years.
| Dipstick | Standard | Population | Sensitivity (%) |
Specificity (%) |
Reference |
|---|---|---|---|---|---|
| CareStart (Access Bio, Princeton, NJ, USA) | GTTS* | Southwestern Uganda | 95.6 | 91.5 | [154] |
| Vistapan (Mitra, New Delhi, India) | 91.9 | 89.6 | |||
| Parabank ( Orchid/Zephyr, Goa, India) | 84.7 | 94.3 | |||
| Paracheck pf (Orchid/Zephyr, Goa, India) | 94 | 87.3 | |||
| Optimal-IT (DiaMed, Cressier, Switzerland) | GTS** | Gabon Children under 11 years |
94 | 97 | [155] |
| Acon (Acon Labs, San Diego, CA, USA) | 94 | 90 | |||
| PALUTOP+4 (ALL.DIAG, Strasbourg, France) | GTTS* and PCR | Madagascar | 95.4 | 97.1 | [156] |
| Optimal-IT (DiaMed, Cressier, Switzerland) | 75.8 | 99.0 | |||
| ParaHIT f test (Span Diagnostic Ltd., Surat, India) | GTTS* and PCR | Tanzania | 69.2 | 100 | [157] |
| Malaria Pf | GTTS* | Uganda | 98 | 72 | [135] |
| Paracheck Pf (Orchid Biomedical Systems, Goa, India) | GTTS* | Kenya | 91.7 | 96.7 | [158] |
| Malar-Check_Pf test | GTS** | Brazil | 97.4% | 88.5% | [159] |
| Makromed Dipstick Assay | PCR | Canada | 97.0% | 96.0% | [160] |
| ParaSight-F (Becton Dickinson, USA) | Thin blood smears and Quantitative Buffy Coat malaria test |
France | 94% | 99% | [161] |
| ParaSight-F (Becton Dickinson, USA) | Microscopy | Iquitos, Peru, and Maesod, Thailand | 95% | 86% | [162] |
| Paracheck Pf (Orchid Biomedical Systems) | Microscopy | India | 93% | 84% | [163] |
| ParaHIT-f (Span Diagnostics) | GTS** | Tanzania | 90.7% | 73.5% | [164] |
| ParaHIT-f (Span Diagnostics) | Microscopy | India | 87.5% | 97% | [163] |
**Giemsa thick smear.
*Giemsa thick and thin smear.
3.2.2. Advanced Techniques: Biosensors
A biosensor is a self-contained integrated device that is capable of providing specific quantitative or semiquantitative analytical information using a biological recognition element that is retained in direct spatial contact with a transduction element [165]. The biorecognition elements commonly reported for developing biosensors are enzyme, antibody, aptamer, DNA, and cells. The transduction principles widely used to develop biosensors are electrochemistry, piezoelectricity, and optical spectroscopy. Biosensor research holds promise for developing stable portable devices for rapid, sensitive, selective, reproducible, and economical detection of many analytes of clinical importance. It has the advantage of being used by semiskilled operators in point of care testing and by patients themselves. However, research on biosensors for detecting malaria is in nascent stage as evident from the limited number of the publications in this area. The biorecognition systems reported so far for developing malaria biosensors are mostly confined to aptamer and antibody. Aptamers are single-stranded nucleic acids (DNA, RNA, and modified RNA or DNA) that can uniquely bind to a ligand with high affinity and specificity. Unique aptamer candidates are generally picked up from 1012 to 1015 combinatorial oligonucleotide libraries over multiple rounds of in vitro selection, the process commonly known as SELEX [166]. Aptamers offer several advantages over the conventional antibodies; for instance, SELEX allows greater control over aptamer binding conditions, robustness of phosphodiester backbone renders them more stability as compared to protein based antibody, and they can be easily amplified using PCR and are highly specific [167]. It is due to these advantages that aptamer is gradually replacing antibody as the biorecognition element. The biomarkers against which aptamer or antibody has been raised to develop malaria biosensors till now are limited to pLDH and HRP II. The prominent malaria biosensors reported so far are briefly described below.
(1) Electrochemical Biosensors. Lee et al. [115] had developed an aptasensor targeting pLDH, the schematic of which is shown in Figure 3. Electrochemical impedance spectroscopy was the detection platform because of its sensitivity [168] and label-free attributes [169]. The detection limit reported with the sensor was 108.5 fM and 120.1 fM for PvLDH and PfLDH, respectively [115].
Figure 3.
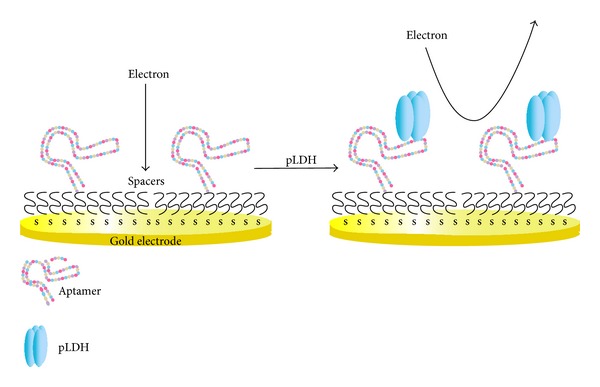
Detection of pLDH by electrochemical impedance spectroscopy. The aptamer is shown as a chain of different coloured circles, representing the four bases A, T, G, and C. Capture of pLDH by aptamer results in a decrease in electron transfer to electrode. The pLDH aptasensor can distinguish between malaria positive blood samples of two major species (P. vivax and P. falciparum) and has a detection limit of 1 pM. [115].
Different electrochemical immunosensors have also been developed using antibody raised against HRP II. The first disposable amperometric HRP II based immunosensor was developed for the detection of P. falciparum malaria. To construct the immunosensor, disposable screen-printed electrodes (SPEs) were modified with multiwall carbon nanotubes (MWCNTs) and Au nanoparticles as shown in Figure 4. Nano-Au/MWCNT/SPEs yielded the highest-level immunosensing with a detection limit of 8.0 ng/mL. The amperometric immunosensor was compared to a commercial RDT Paracheck Pf kit (Orchid Biomedical Systems) keeping microscopy as gold standard. The Paracheck Pf kit exhibited a sensitivity of 79% and a specificity of 81% whereas the developed amperometric immunosensor showed a sensitivity of 96% and a specificity of 94% [116].
Figure 4.

Fabrication of a sandwich immunoassay using screen-printed electrodes (SPEs) modified with gold nanoparticles and carbon nanotubes. This approach used ALP-conjugated antibodies which produce 1-naphthol as the hydrolyzed product of 1-naphthylphosphate (the enzymatic substrate) for the amperometric detection of HRP II [116]. BSA: bovine serum albumin; ALP: alkaline phosphatase.
An immunosensor based on magnetic micro- and nanoparticles were also developed for targeting HRP II [117], the configuration of which is shown in Figure 5. A sandwich assay was performed on both kinds of particles by using a secondary monoclonal antibody labeled with the horseradish peroxidase (HRP) enzyme. Both optical and electrochemical detection methods were studied and compared. The electrochemical magneto-immunosensor coupled with magnetic nanoparticles had shown better analytical performance in terms of detection limit (0.36 ng mL−1).
Figure 5.

Detection of HRP II based on a magnetic sandwich immunoassay performed on magnetic beads or nanoparticles modified with monoclonal anti-HRP II IgM antibody. Detection is done using a monoclonal IgG antibody labelled with horse radish peroxidase to obtain an optical or electrochemical signal [117].
(2) Optical Biosensors. An immunocapture spectrophotometric diagnostic assay based on PfLDH, which showed a detection limit of 50 parasites/μL and sensitivity of 88%, has been developed by Piper et al. [118]. A schematic of this assay is shown in Figure 6. This assay could be further recreated to develop an advanced biosensor with enhanced selectivity. Since it incorporates the functional activity of PfLDH in a secondary enzymatic reaction for generating signal, this assay considerably reduces the number of false positives, which may occur on nonspecific binding of serum proteins to antibodies.
Figure 6.
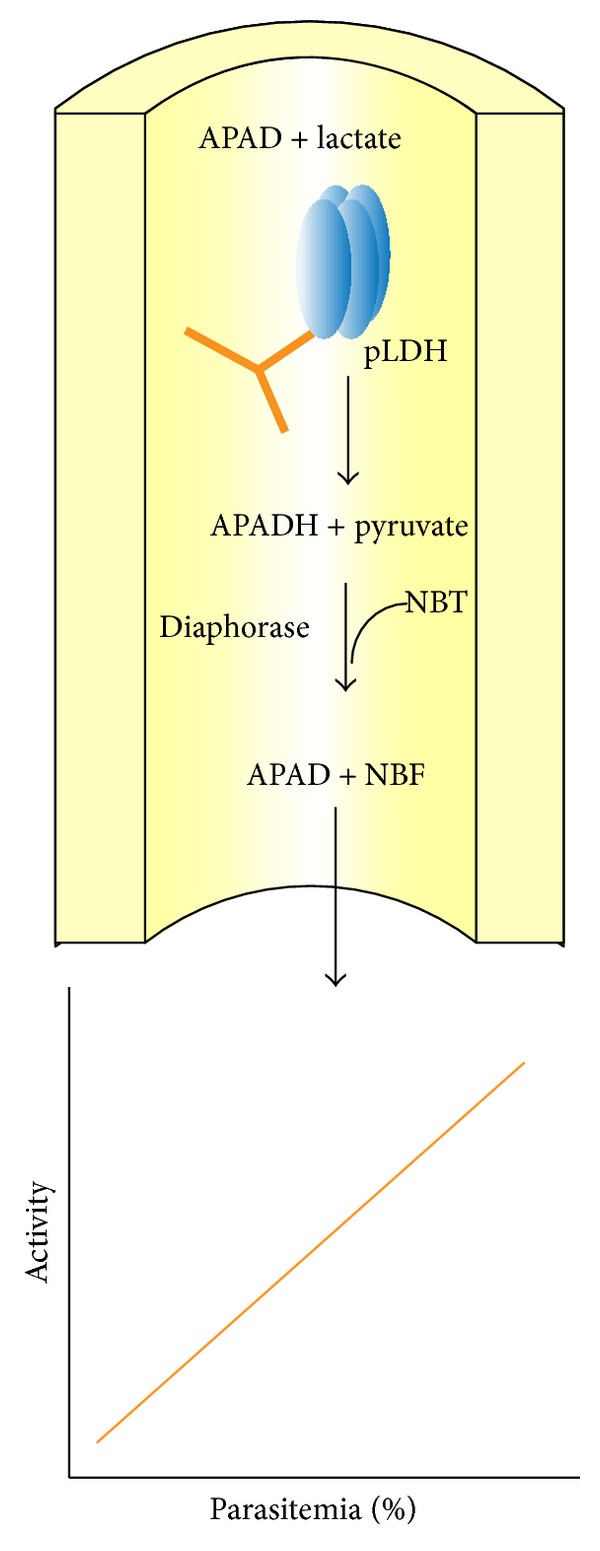
The immunocapture Plasmodium lactate dehydrogenase (ICpLDH) assay. A schematic of the reaction is shown in which the pLDH is immobilized using a monoclonal antibody. The enzyme activity can be measured using a coupled enzyme assay that generates APADH. The latter reduces nitro blue tetrazolium, a chromogenic substrate, using an enzyme diaphorase. Activity is quantified spectrophotometrically at 650 nm and plotted as a function of percentage parasitemia [118].
Besides PfLDH and HRP II, hemozoin has also been used as a biomarker for detecting malaria microscopically. In fact, the characteristic morphology of the crystals can also be considered to distinguish different species of parasites [170]. Hemozoin crystals are paramagnetic in nature and hence show cotton-mouton effect in the presence of an external magnetic field which corresponds linearly to the hemozoin concentration [171]. Based on this concept an in vivo diagnostic method using a magnetooptic fingertip probe was developed and the method was confirmed by a small preliminary clinical trial [171, 172]. Following this method the hemozoin concentrations of less than 0.02 μg/mL could be detected.
(3) Piezoelectric Biosensors. A piezoelectric immunosensor was developed using mixed self-assembled monolayers (SAMs) of thioctic acid and 1-dodecanethiol, the schematic configuration of which is shown in Figure 7. The characteristics of the immunosensor applied to detect HRP II are linear range of 15–60 ng/mL, minimum detection limit of 12 ng/mL, and storage stability (t 1/2) of 14 days. The sensor was tested on clinical human serum samples and the results were compared with the commercially available ICT kit (NOW) which showed good agreement [119].
Figure 7.
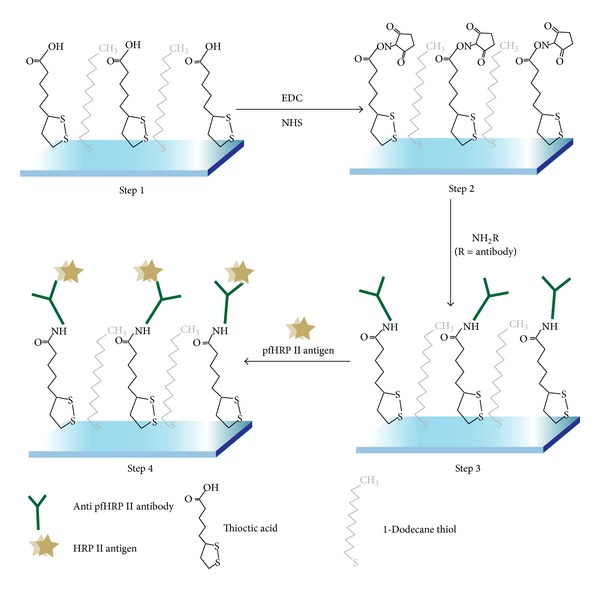
Schematic diagram for preparation of piezoelectric immunosensor for HRP II. The mixed self-assembled monolayers (SAMs) of thioctic acid and 1-dodecanethiol were formed on gold surface of quartz crystal. The rabbit anti-PfHRP II antibodies were coupled on mixed SAM modified gold surface of quartz crystal via NHS/EDC activation method. The amount of HRP II molecules bound on the sensitive area of the electrodes is quantitatively measured as a decrease in resonant frequency [119].
The malarial biosensor research, as evident from the above references, is more inclined towards electrochemical based biosensors and the reason may be attributed to some advantages of this biosensing platform; for example, label-free, portable, and small sample volume is needed for the analysis. From the sensitivity point of view electrochemical biosensors targeting HRP II and pLDH showed sensitivity even in the femtomolar range as compared to the other biosensors (Table 2). Comparing the two biorecognition elements exploited for developing malaria biosensor, namely, antibody and aptamer, the latter showed better performance in terms of sensitivity while the specificity of the aptamer developed by Lee et al. [115] is yet to reach the desired level as it could not discriminate between PfLDH and PvLDH.
4. Conclusion and Future Perspective
In the last two decades, biomarker research on malarial pathogenicity has taken a leap as witnessed from the discovery of many interesting molecules and functional entities which are significantly distinct from the host counterpart and other host pathogens. Among the biomarkers, pLDH, HRP II, aldolase, and hemozoin formation have received increasing attention for their possible utilizations as drug target and for developing malaria detection techniques. The increase in drug resistance of malaria parasites at alarming rates, caused primarily by the indiscriminate use and overprescription of drugs, has made the current treatment protocol inefficient. Therefore, a rapid, selective, sensitive, and less expensive detection technique is a need of the hour for quick parasitological confirmation in the patients before the administration of drugs. Though RDTs are a milestone in malaria detection due to their rapid detection capability and portable nature, these cannot be exclusively relied upon for reproducible and unambiguous results. In this regard, biosensors have emerged as a reliable diagnostic technique for malaria due to many inherent advantages set by their tenet including reusability, quantitative nature, enhanced signals at low sample volume, and better storage and operational stability. As revealed from the studies, every biomarker has some strengths and limitations. In order to develop an efficient biosensor, the strength of a biomarker needs to be exploited for which a profound understanding on the physiology and metabolism is essential, on the basis of which the biomarker is being instituted. Biosensor development and its confluence with biomarker discovery are therefore important to reach the desired goal in diagnosis and treatment of malaria.
Supplementary Material
Table S1 : List of PDB entries for Plasmodium lactate dehydrogenase.
Table S2: List of inhibitors developed against lactate dehydrogenase from P.falciparum.
Acknowledgment
The financial assistance of the Department of Biotechnology, India, was used to carry out the work.
Abbreviations
- ACT:
Artemisinin based combination therapies
- ApoE:
Apolipoprotein E
- WHO:
World Health Organization
- RDT:
Rapid detection tests
- MDR:
Multidrug resistant
- FACS:
Fluorescence activated cell sorting
- RBC:
Red blood cell
- ATP:
Adenosine triphosphate
- NAD+:
Nicotinamide adenine dinucleotide (oxidized)
- LDH:
Lactate dehydrogenase
- PfLDH:
Plasmodium falciparum lactate dehydrogenase
- PvLDH:
Plasmodium vivax lactate dehydrogenase
- pLDH:
Parasite lactate dehydrogenase
- RNA:
Ribonucleic acid
- NADH:
Nicotinamide adenine dinucleotide (reduced)
- APAD:
3-Acetylpyridine adenine dinucleotide
- PDB:
Protein data bank
- HRP:
Histidine-rich proteins
- mcfp-4:
Mytilus californianus foot protein 4
- HRG:
Histidine-rich glycoprotein
- MA:
Monoclonal antibodies
- UTR:
Untranslated regions
- CD:
Circular dichroism
- pGDH:
Parasite glutamate dehydrogenase
- PfGDH:
Plasmodium falciparum glutamate dehydrogenase
- CM:
Cerebral malaria
- SM:
Severe malaria
- MM:
Mild malaria
- IL:
Interleukin
- TNF:
Tissue necrosis factor
- ANG:
Angiopoietin
- EPO:
Erythropoietin
- ICAM:
Intracellular adhesion molecule
- LC-MS/MS:
Liquid chromatography-mass spectrometry/mass spectrometry
- SAA:
Serum amyloid A
- LPS:
Lipopolysaccharide
- PM:
Placental malaria
- MP:
Microparticles
- CXCR3:
Chemokine (C-X-C motif) receptor 3
- PCR:
Polymerase chain reaction
- LAMP:
Loop-mediated isothermal amplification
- IFA:
Immunofluorescence antibody
- FCM:
Flow cytometry
- LDMS:
Laser desorption mass spectrometry
- TDR:
Tropical disease research
- FIND:
Foundation for Innovative New Diagnostics
- CDC:
Center for Disease Control and Prevention
- ICT:
Immunochromographic-malaria dipstick
- ELISA:
Enzyme-linked immunosorbent assay
- SELEX:
Systematic evolution of ligands by exponential enrichment
- SPE:
Screen printed electrodes
- MWCNTs:
Multiwall carbon nanotubes
- SAM:
Self-assembled monolayers
- GNI:
Gross national income.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Priyamvada Jain and Babina Chakma contributed equally to this work.
References
- 1.WHO world malaria report 2012. http://www.who.int/malaria/publications/world_malaria_report_2012/wmr2012_full_report.pdf.
- 2.Centres for Disease Control and Prevention. http://www.cdc.gov/malaria/about/biology/parasites.html.
- 3.Choi S-R, Beeler AB, Pradhan A, et al. Generation of oxamic acid libraries: antimalarials and inhibitors of plasmodium falciparum lactate dehydrogenase. Journal of Combinatorial Chemistry. 2007;9(2):292–300. doi: 10.1021/cc060110n. [DOI] [PubMed] [Google Scholar]
- 4.Chanda P, Hamainza B, Mulenga S, Chalwe V, Msiska C, Chizema-Kawesha E. Early results of integrated malaria control and implications for the management of fever in under-five children at a peripheral health facility: a case study of Chongwe rural health centre in Zambia. Malaria Journal. 2009;8(1, article 49) doi: 10.1186/1475-2875-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hume JCC, Barnish G, Mangal T, Armázio L, Streat E, Bates I. Household cost of malaria overdiagnosis in rural Mozambique. Malaria Journal. 2008;7, article 33 doi: 10.1186/1475-2875-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Msellem MI, Mårtensson A, Rotllant G, et al. Influence of rapid malaria diagnostic tests on treatment and health outcome in fever patients, Zanzibar—a crossover validation study. PLoS Medicine. 2009;6(4) doi: 10.1371/journal.pmed.1000070.e1000070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hulka BS. Epidemiological studies using biological markers: issues for epidemiologists. Cancer Epidemiology Biomarkers and Prevention. 1991;1(1):13–19. [PubMed] [Google Scholar]
- 8.Frank R, Hargreaves R. Clinical biomarkers in drug discovery and development. Nature Reviews Drug Discovery. 2003;2(7):566–580. doi: 10.1038/nrd1130. [DOI] [PubMed] [Google Scholar]
- 9.Dal-Bianco MP, Köster KB, Kombila UD, et al. High prevalence of asymptomatic Plasmodium falciparum infection in Gabonese adults. American Journal of Tropical Medicine and Hygiene. 2007;77(5):939–942. [PubMed] [Google Scholar]
- 10.Jensen MD, Conley M, Helstowski LD. Culture of plasmodium falciparum: the role of pH, glucose, and lactate. Journal of Parasitology. 1983;69(6):1060–1067. [PubMed] [Google Scholar]
- 11.Fry M, Webb E, Pudney M. Effect of mitochondrial inhibitors on adenosinetriphosphate levels in Plasmodium falciparum. Comparative Biochemistry and Physiology B. 1990;96(4):775–782. doi: 10.1016/0305-0491(90)90230-q. [DOI] [PubMed] [Google Scholar]
- 12.Olszewski KL, Llinás M. Central carbon metabolism of Plasmodium parasites. Molecular and Biochemical Parasitology. 2011;175(2):95–103. doi: 10.1016/j.molbiopara.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth E., Jr. Plasmodium falciparum carbohydrate metabolism: a connection between host cell and parasite. Blood Cells. 1990;16(2-3):453–460. [PubMed] [Google Scholar]
- 14.Pfaller MA, Krogstad DJ, Parquette AR, Nguyen Dinh P. Plasmodium falciparum: stage-specific lactate production in synchronized cultures. Experimental Parasitology. 1982;54(3):391–396. doi: 10.1016/0014-4894(82)90048-0. [DOI] [PubMed] [Google Scholar]
- 15.Bozdech Z, Llinás M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biology. 2003;1(1, article e5) doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown WM, Yowell CA, Hoard A, et al. Comparative structural analysis and kinetic properties of lactate dehydrogenases from the four species of human malarial parasites. Biochemistry. 2004;43(20):6219–6229. doi: 10.1021/bi049892w. [DOI] [PubMed] [Google Scholar]
- 17.Dunn CR, Banfield MJ, Barker JJ, et al. The structure of lactate dehydrogenase from Plasmodium falciparum reveals a new target for anti-malarial design. Nature Structural Biology. 1996;3(11):910–915. doi: 10.1038/nsb1196-912. [DOI] [PubMed] [Google Scholar]
- 18.Winter VJ, Cameron A, Tranter R, Sessions RB, Brady RL. Crystal structure of Plasmodium berghei lactate dehydrogenase indicates the unique structural differences of these enzymes are shared across the Plasmodium genus. Molecular and Biochemical Parasitology. 2003;131(1):1–10. doi: 10.1016/s0166-6851(03)00170-1. [DOI] [PubMed] [Google Scholar]
- 19.Hurdayal R, Achilonu I, Choveaux D, Coetzer THT, Dean Goldring JP. Anti-peptide antibodies differentiate between plasmodial lactate dehydrogenases. Peptides. 2010;31(4):525–532. doi: 10.1016/j.peptides.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Bzik DJ, Fox BA, Gonyer K. Expression of Plasmodium falciparum lactate dehydrogenase in Escherichia coli. Molecular and Biochemical Parasitology. 1993;59(1):155–166. doi: 10.1016/0166-6851(93)90016-q. [DOI] [PubMed] [Google Scholar]
- 21.Yang S, Parmley SF. A bradyzoite stage-specifically expressed gene of Toxoplasma gondii encodes a polypeptide homologous to lactate dehydrogenase. Molecular and Biochemical Parasitology. 1995;73(1-2):291–294. doi: 10.1016/0166-6851(95)00124-j. [DOI] [PubMed] [Google Scholar]
- 22.Yang S, Parmley SF. Toxoplasma gondii expresses two distinct lactate dehydrogenase homologous genes during its life cycle in intermediate hosts. Gene. 1997;184(1):1–12. doi: 10.1016/s0378-1119(96)00566-5. [DOI] [PubMed] [Google Scholar]
- 23.Sessions RB, Dewar V, Clarke AR, Holbrook JJ. A model of Plasmodium falciparum lactate dehydrogenase and its implications for the design of improved antimalarials and the enhanced detection of parasitaemia. Protein Engineering. 1997;10(4):301–306. doi: 10.1093/protein/10.4.301. [DOI] [PubMed] [Google Scholar]
- 24.Hewitt CO, Eszes CM, Sessions RB, et al. A general method for relieving substrate inhibition in lactate dehydrogenases. Protein Engineering. 1999;12(6):491–496. doi: 10.1093/protein/12.6.491. [DOI] [PubMed] [Google Scholar]
- 25.Shoemark DK, Cliff MJ, Sessions RB, Clarke AR. Enzymatic properties of the lactate dehydrogenase enzyme from Plasmodium falciparum. FEBS Journal. 2007;274(11):2738–2748. doi: 10.1111/j.1742-4658.2007.05808.x. [DOI] [PubMed] [Google Scholar]
- 26.Gomez MS, Piper RC, Hunsaker LA, et al. Substrate and cofactor specificity and selective inhibition of lactate dehydrogenase from the malarial parasite P. falciparum. Molecular and Biochemical Parasitology. 1997;90(1):235–246. doi: 10.1016/s0166-6851(97)00140-0. [DOI] [PubMed] [Google Scholar]
- 27.Chaikuad A, Fairweather V, Conners R, Joseph-Horne T, Turgut-Balik D, Brady RL. Structure of lactate dehydrogenase from Plasmodium vivax: complexes with NADH and APADH. Biochemistry. 2005;44(49):16221–16228. doi: 10.1021/bi051416y. [DOI] [PubMed] [Google Scholar]
- 28.Reddy SG, Scapin G, Blanchard JS. Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes. Biochemistry. 1996;35(41):13294–13302. doi: 10.1021/bi9615809. [DOI] [PubMed] [Google Scholar]
- 29.Kavanagh KL, Elling RA, Wilson DK. Structure of toxoplasma gondii LDH1: active-site differences from human lactate dehydrogenases and the structural basis for efficient APAD+ use. Biochemistry. 2004;43(4):879–889. doi: 10.1021/bi035108g. [DOI] [PubMed] [Google Scholar]
- 30.Dando C, Schroeder ER, Hunsaker LA, et al. The kinetic properties and sensitivities to inhibitors of lactate dehydrogenases (LDH1 and LDH2) from Toxoplasma gondii: comparisons with pLDH from Plasmodium falciparum. Molecular and Biochemical Parasitology. 2001;118(1):23–32. doi: 10.1016/s0166-6851(01)00360-7. [DOI] [PubMed] [Google Scholar]
- 31.Baldwin RL. Structure and mechanism in protein science. A guide to enzyme catalysis and protein folding. Protein Science. 2000;9:p. 207. [Google Scholar]
- 32.Royer RE, Deck LM, Campos NM, Hunsaker LA, Vender Jagt DL. Biologically active derivatives of gossypol: synthesis and antimalarial activities of peri-acylated gossylic nitriles. Journal of Medicinal Chemistry. 1986;29(9):1799–1801. doi: 10.1021/jm00159a043. [DOI] [PubMed] [Google Scholar]
- 33.Deck LM, Royer RE, Chamblee BB, et al. Selective inhibitors of human lactate dehydrogenases and lactate dehydrogenase from the malarial parasite Plasmodium falciparum. Journal of Medicinal Chemistry. 1998;41(20):3879–3887. doi: 10.1021/jm980334n. [DOI] [PubMed] [Google Scholar]
- 34.Cameron A, Read J, Tranter R, et al. Identification and activity of a series of azole-based compounds with lactate dehydrogenase-directed antimalarial activity. The Journal of Biological Chemistry. 2004;279(30):31429–31439. doi: 10.1074/jbc.M402433200. [DOI] [PubMed] [Google Scholar]
- 35.Conners R, Schambach F, Read J, et al. Mapping the binding site for gossypol-like inhibitors of Plasmodium falciparum lactate dehydrogenase. Molecular and Biochemical Parasitology. 2005;142(2):137–148. doi: 10.1016/j.molbiopara.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Menting JGT, Tilley L, Deady LW, et al. The antimalarial drug, chloroquine, interacts with lactate dehydrogenase from Plasmodium falciparum. Molecular and Biochemical Parasitology. 1997;88(1-2):215–224. doi: 10.1016/s0166-6851(97)00095-9. [DOI] [PubMed] [Google Scholar]
- 37.Read JA, Wilkinson KW, Tranter R, Sessions RB, Brady RL. Chloroquine binds in the cofactor binding site of Plasmodium falciparum lactate dehydrogenase. Journal of Biological Chemistry. 1999;274(15):10213–10218. [PubMed] [Google Scholar]
- 38.Kilejian A. A unique histidine rich polypeptide from the malaria parasite, Plasmodium lophurae. Journal of Biological Chemistry. 1974;249(14):4650–4655. [PubMed] [Google Scholar]
- 39.Zhao H, Waite JH. Proteins in load-bearing junctions: the Histidine-rich metal-binding protein of mussel byssus. Biochemistry. 2006;45(47):14223–14231. doi: 10.1021/bi061677n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burch MK, Blackburn MN, Morgan WT. Further characterization of the interaction of histidine-rich glycoprotein with heparin: evidence for the binding of two molecules of histidine-rich glycoprotein by high molecular weight heparin and for the involvement of histidine residues in heparin binding. Biochemistry. 1987;26(23):7477–7482. doi: 10.1021/bi00397a042. [DOI] [PubMed] [Google Scholar]
- 41.Ge R, Watt RM, Sun X, et al. Expression and characterization of a histidine-rich protein, Hpn: potential for Ni2+ storage in Helicobacter pylori. Biochemical Journal. 2006;393(1):285–293. doi: 10.1042/BJ20051160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheel J, Ziegelbauer K, Kupke T, et al. Hisactophilin, a histidine-rich actin-binding protein from Dictyostelium discoideum. Journal of Biological Chemistry. 1989;264(5):2832–2839. [PubMed] [Google Scholar]
- 43.Howard RJ, Uni S, Aikawa M. Secretion of a malarial histidine-rich protein (Pf HRP II) from Plasmodium falciparum-infected erythrocytes. Journal of Cell Biology. 1986;103(4):1269–1277. doi: 10.1083/jcb.103.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pologe LG, Pavlovec A, Shio H, Ravetch JV. Primary structure and subcellular localization of the knob-associated histidine-rich protein of Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 1987;84(20):7139–7143. doi: 10.1073/pnas.84.20.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leech JH, Barnwell JW, Aikawa M. Plasmodium falciparum malaria: association of knobs on the surface of infected erythrocytes with a histidine-rich protein and the erythrocyte skeleton. Journal of Cell Biology. 1984;98(4):1256–1264. doi: 10.1083/jcb.98.4.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lynn A, Chandra S, Malhotra P, Chauhan VS. Heme binding and polymerization by Plasmodium falciparum histidine rich protein II: influence of pH on activity and conformation. FEBS Letters. 1999;459(2):267–271. doi: 10.1016/s0014-5793(99)01260-0. [DOI] [PubMed] [Google Scholar]
- 47.Kilejian A. Histidine rich protein as a model malaria vaccine. Science. 1978;201(4359):922–924. doi: 10.1126/science.567375. [DOI] [PubMed] [Google Scholar]
- 48.Stahl HD, Kemp DJ, Crewther PE, et al. Sequence of a cDNA encoding a small polymorphic histidine- and alanine-rich protein from Ptasmodium falciparum. Nucleic Acids Research. 1985;13(21):7837–7846. doi: 10.1093/nar/13.21.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wellems TE, Howard RJ. Homologous genes encode two distinct histidine-rich proteins in a cloned isolate of Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(16):6065–6069. doi: 10.1073/pnas.83.16.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rock EP, Marsh K, Saul AJ, et al. Comparative analysis of the Plasmodium falciparum histidine-rich proteins HRP-I, HRP-II and HRP-III in malaria parasites of diverse origin. Parasitology. 1987;95, part 2:209–227. doi: 10.1017/s0031182000057681. [DOI] [PubMed] [Google Scholar]
- 51.Sullivan DJ, Jr., Ayala YM, Goldberg DE. An unexpected 5° untranslated intron in the P. falciparum genes for histidine-rich proteins II and III. Molecular and Biochemical Parasitology. 1996;83(2):247–251. doi: 10.1016/s0166-6851(96)02768-5. [DOI] [PubMed] [Google Scholar]
- 52.Parra ME, Evans CB, Taylor DW. Identification of Plasmodium falciparum histidine-rich protein 2 in the plasma of humans with malaria. Journal of Clinical Microbiology. 1991;29(8):1629–1634. doi: 10.1128/jcm.29.8.1629-1634.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez-del Valle M, Quakyi IA, Amuesi J, Quaye JT, Nkrumah FK, Taylor DW. Detection of antigens and antibodies in the urine of humans with Plasmodium falciparum malaria. Journal of Clinical Microbiology. 1991;29(6):1236–1242. doi: 10.1128/jcm.29.6.1236-1242.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Desakorn V, Dondorp AM, Silamut K, et al. Stage-dependent production and release of histidine-rich protein 2 by Plasmodium falciparum. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2005;99(7):517–524. doi: 10.1016/j.trstmh.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 55.Sullivan DJ, Jr., Gluzman IY, Goldberg DE. Plasmodium hemozoin formation mediated by histidine-rich proteins. Science. 1996;271(5246):219–222. doi: 10.1126/science.271.5246.219. [DOI] [PubMed] [Google Scholar]
- 56.Schneider EL, Marletta MA. Heme binding to the histidine-rich protein II from Plasmodium falciparum. Biochemistry. 2005;44(3):979–986. doi: 10.1021/bi048570p. [DOI] [PubMed] [Google Scholar]
- 57.Barlow DJ, Thornton JM. Helix geometry in proteins. Journal of Molecular Biology. 1988;201(3):601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 58.Karpen ME, de Haseth PL, Neet KE. Differences in the amino acid distributions of 310-helices and α- helices. Protein Science. 1992;1(10):1333–1342. doi: 10.1002/pro.5560011013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Choi CYH, Cerda JF, Hsiu-An C, Babcock GT, Marletta MA. Spectroscopic characterization of the heme-binding sites in Plasmodium falciparum histidine-rich protein 2. Biochemistry. 1999;38(51):16916–16924. doi: 10.1021/bi991665k. [DOI] [PubMed] [Google Scholar]
- 60.Pandey AV, Bisht H, Babbarwal VK, Srivastava J, Pandey KC, Chauhan VS. Mechanism of malarial haem detoxification inhibition by chloroquine. Biochemical Journal. 2001;355(2):333–338. doi: 10.1042/0264-6021:3550333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Accardo A, Laurent SA-L, Mazarguil H, Meyer M, Robert A, Meunier B. Interaction of iron(II)-heme and artemisinin with a peptide mimic of Plasmodium falciparum HRP-II. Journal of Inorganic Biochemistry. 2007;101(11-12):1739–1747. doi: 10.1016/j.jinorgbio.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 62.Ziegler J, Chang RT, Wright DW. Multiple-antigenic peptides of histidine-rich protein II of Plasmodium falciparum: dendrimeric biomineralization templates. Journal of the American Chemical Society. 1999;121(11):2395–2400. [Google Scholar]
- 63.Benedetti CE, Kobarg J, Pertinhez TA, et al. Plasmodium falciparum histidine-rich protein II binds to actin, phosphatidylinositol 4,5-bisphosphate and erythrocyte ghosts in a pH-dependent manner and undergoes coil-to-helix transitions in anionic micelles. Molecular and Biochemical Parasitology. 2003;128(2):157–166. doi: 10.1016/s0166-6851(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 64.Bosshart H, Heinzelmann M. Endotoxin-neutralizing effects of histidine-rich peptides. FEBS Letters. 2003;553(1-2):135–140. doi: 10.1016/s0014-5793(03)00997-9. [DOI] [PubMed] [Google Scholar]
- 65.Oliveira MF, Silva JR, Dansa-Petretski M, et al. Haem detoxification by an insect. Nature. 1999;400(6744):517–518. doi: 10.1038/22910. [DOI] [PubMed] [Google Scholar]
- 66.Chen MM, Shi L, Sullivan DJ., Jr. Haemoproteus and Schistosoma synthesize heme polymers similar to Plasmodium hemozoin and β-hematin. Molecular and Biochemical Parasitology. 2001;113(1):1–8. doi: 10.1016/s0166-6851(00)00365-0. [DOI] [PubMed] [Google Scholar]
- 67.Pisciotta JM, Ponder EL, Fried B, Sullivan D. Hemozoin formation in Echinostoma trivolvis rediae. International Journal for Parasitology. 2005;35(10):1037–1042. doi: 10.1016/j.ijpara.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 68.Slater AFG, Swiggard WJ, Orton BR, et al. An iron-carboxylate bond links the heme units of malaria pigment. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(2):325–329. doi: 10.1073/pnas.88.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pagola S, Stephens PW, Bohle DS, Kosar AD, Madsen SK. The structure of malaria pigment β-haematin. Nature. 2000;404(6775):307–310. doi: 10.1038/35005132. [DOI] [PubMed] [Google Scholar]
- 70.Adams PA, Egan TJ, Ross DC, Silver J, Marsh PJ. The chemical mechanism of β-haematin formation studied by Mössbauer spectroscopy. Biochemical Journal. 1996;318(1):25–27. doi: 10.1042/bj3180025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bendrat K, Berger BJ, Cerami A. Haem polymerization in malaria. Nature. 1995;378(6553):138–139. doi: 10.1038/378138a0. [DOI] [PubMed] [Google Scholar]
- 72.Fitch CD, Cai G-Z, Chen Y-F, Shoemaker JD. Involvement of lipids in ferriprotoporphyrin IX polymerization in malaria. Biochimica et Biophysica Acta. 1999;1454(1):31–37. doi: 10.1016/s0925-4439(99)00017-4. [DOI] [PubMed] [Google Scholar]
- 73.Jackson KE, Klonis N, Ferguson DJP, Adisa A, Dogovski C, Tilley L. Food vacuole-associated lipid bodies and heterogeneous lipid environments in the malaria parasite, Plasmodium falciparum. Molecular Microbiology. 2004;54(1):109–122. doi: 10.1111/j.1365-2958.2004.04284.x. [DOI] [PubMed] [Google Scholar]
- 74.Pisciotta JM, Coppens I, Tripathi AK, et al. The role of eutral lipid nanospheres in Plasmodium falciparum haem crystallization. Biochemical Journal. 2007;402(1):197–204. doi: 10.1042/BJ20060986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Solomonov I, Osipova M, Feldman Y, et al. Crystal nucleation, growth, and morphology of the synthetic malaria pigment β-hematin and the effect thereon by quinoline additives: the malaria pigment as a target of various antimalarial drugs. Journal of the American Chemical Society. 2007;129(9):2615–2627. doi: 10.1021/ja0674183. [DOI] [PubMed] [Google Scholar]
- 76.Srivastava IK, Schmidt M, Certa U, Döbeli H, Perrin LH. Specificity and inhibitory activity of antibodies to Plasmodium falciparum aldolase. Journal of Immunology. 1990;144(4):1497–1503. [PubMed] [Google Scholar]
- 77.Döbeli H, Trzeciak A, Gillessen D, et al. Expression, purification, biochemical characterization and inhibition of recombinant Plasmodium falciparum aldolase. Molecular and Biochemical Parasitology. 1990;41(2):259–268. doi: 10.1016/0166-6851(90)90189-s. [DOI] [PubMed] [Google Scholar]
- 78.Kim H, Certa U, Döbeli H, Jakob P, Hol WGJ. Crystal structure of fructose-1,6-bisphosphate aldolase from the human malaria parasite plasmodium falciparum. Biochemistry. 1998;37(13):4388–4396. doi: 10.1021/bi972233h. [DOI] [PubMed] [Google Scholar]
- 79.Shear HL, Wanidworanun CC, Nagel RL. Antisense oligonucleotides targeting malarial aldolase inhibit the asexual erythrocytic stages of Plasmodium falciparum. Molecular and Biochemical Parasitology. 1999;102(1):91–101. doi: 10.1016/s0166-6851(99)00087-0. [DOI] [PubMed] [Google Scholar]
- 80.Clayton CE. Structure and regulated expression of genes encoding fructose biphosphate aldolase in Trypanosoma brucei. The EMBO Journal. 1985;4(11):2997–3003. doi: 10.1002/j.1460-2075.1985.tb04035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Henze K, Morrison HG, Sogin ML, Miklós Müller MM. Sequence and phylogenetic position of a class II aldolase gene in the amitochondriate protist, Giardia lamblia. Gene. 1998;222(2):163–168. doi: 10.1016/s0378-1119(98)00499-5. [DOI] [PubMed] [Google Scholar]
- 82.Knapp B, Hundt E, Kupper HA. Plasmodium falciparum aldolase: gene structure and localization. Molecular and Biochemical Parasitology. 1990;40(1):1–12. doi: 10.1016/0166-6851(90)90074-v. [DOI] [PubMed] [Google Scholar]
- 83.Meier B, Döbeli H, Certa U. Stage-specific expression of aldolase isoenzymes in the rodent malaria parasite Plasmodium berghei. Molecular and Biochemical Parasitology. 1992;52(1):15–27. doi: 10.1016/0166-6851(92)90032-f. [DOI] [PubMed] [Google Scholar]
- 84.Cloonan N, Fischer K, Cheng Q, Saul A. Aldolase genes of Plasmodium species. Molecular and Biochemical Parasitology. 2001;113(2):327–330. doi: 10.1016/s0166-6851(01)00226-2. [DOI] [PubMed] [Google Scholar]
- 85.Lee N, Baker J, Andrews KT, et al. Effect of sequence variation in Plasmodium falciparum histidine-rich protein 2 on binding of specific monoclonal antibodies: implications for rapid diagnostic tests for malaria. Journal of Clinical Microbiology. 2006;44(8):2773–2778. doi: 10.1128/JCM.02557-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wagner JT, Lüdemann H, Färber PM, Lottspeich F, Krauth-Siegel RL. Glutamate dehydrogenase, the marker protein of Plasmodium falciparum cloning, expression and characterization of the malarial enzyme. European Journal of Biochemistry. 1998;258(2):813–819. doi: 10.1046/j.1432-1327.1998.2580813.x. [DOI] [PubMed] [Google Scholar]
- 87.Werner C, Stubbs MT, Krauth-Siegel RL, Klebe G. The crystal structure of Plasmodium falciparum glutamate dehydrogenase, a putative target for novel antimalarial drugs. Journal of Molecular Biology. 2005;349(3):597–607. doi: 10.1016/j.jmb.2005.03.077. [DOI] [PubMed] [Google Scholar]
- 88.Zocher K, Fritz-Wolf K, Kehr S, Fischer M, Rahlfs S, Becker K. Biochemical and structural characterization of Plasmodium falciparum glutamate dehydrogenase 2. Molecular and Biochemical Parasitology. 2012;183(1):52–62. doi: 10.1016/j.molbiopara.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 89.Rodríguez-Acosta A, Domínguez NG, Aguilar I, Girón ME. Characterization of Plasmodium falciparum glutamate dehydrogenase-soluble antigen. Brazilian Journal of Medical and Biological Research. 1998;31:1149–1155. doi: 10.1590/s0100-879x1998000900008. [DOI] [PubMed] [Google Scholar]
- 90.Li Y, Ning Y-S, Li L, Peng D-D, Dong W-Q, Li M. Preparation of a monoclonal antibodies against Plasmodium falciparum glutamate dehydrogenase and establishment of colloidal gold-immunochromatographic assay. Di Yi Jun Yi Da Xue Xue Bao. 2005;25(4):435–438. [PubMed] [Google Scholar]
- 91.Lucchi NW, Jain V, Wilson NO, Singh N, Udhayakumar V, Stiles JK. Potential serological biomarkers of cerebral malaria. Disease Markers. 2011;31(6):327–335. doi: 10.3233/DMA-2011-0854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Armah HB, Wilson NO, Sarfo BY, et al. Cerebrospinal fluid and serum biomarkers of cerebral malaria mortality in Ghanaian children. Malaria Journal. 2007;6, article 147 doi: 10.1186/1475-2875-6-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.John CC, Panoskaltsis-Mortari A, Opoka RO, et al. Cerebrospinal fluid cytokine levels and cognitive impairment in cerebral malaria. American Journal of Tropical Medicine and Hygiene. 2008;78(2):198–205. [PMC free article] [PubMed] [Google Scholar]
- 94.Lovegrove FE, Tangpukdee N, Opoka RO, et al. Serum angiopoietin-1 and -2 levels discriminate cerebral malaria from uncomplicated malaria and predict clinical outcome in African children. PLoS ONE. 2009;4(3) doi: 10.1371/journal.pone.0004912.e4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Conroy AL, Lafferty EI, Lovegrove FE, et al. Whole blood angiopoietin-1 and-2 levels discriminate cerebral and severe (non-cerebral) malaria from uncomplicated malaria. Malaria Journal. 2009;8(1, article 295) doi: 10.1186/1475-2875-8-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Casals-Pascual C, Idro R, Gicheru N, et al. High levels of erythropoietin are associated with protection against neurological sequelae in African children with cerebral malaria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(7):2634–2639. doi: 10.1073/pnas.0709715105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jakobsen PH, Morris-Jones S, Ronn A, et al. Increased plasma concentrations of sICAM-1, sVCAM-1 and sELAM-1 in patients with Plasmodium falciparum or P. vivax malaria and association with disease severity. Immunology. 1994;83(4):665–669. [PMC free article] [PubMed] [Google Scholar]
- 98.Kassa FA, Shio MT, Bellemare M-J, Faye B, Ndao M, Olivier M. New inflammation-related biomarkers during malaria infection. PLoS ONE. 2011;6(10) doi: 10.1371/journal.pone.0026495.e26495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wenisch C, Spitzauer S, Florris-Linau K, et al. Complement activation in severe Plasmodium falciparum malaria. Clinical Immunology and Immunopathology. 1997;85(2):166–171. doi: 10.1006/clin.1997.4417. [DOI] [PubMed] [Google Scholar]
- 100.Freyssinet J-M. Cellular microparticles: what are they bad or good for? Journal of Thrombosis and Haemostasis. 2003;1(7):1655–1662. doi: 10.1046/j.1538-7836.2003.00309.x. [DOI] [PubMed] [Google Scholar]
- 101.Haest CWM. Interactions between membrane skeleton proteins and the intrinsic domain of the erythrocyte membrane. Biochimica et Biophysica Acta. 1982;694(4):331–352. doi: 10.1016/0304-4157(82)90001-6. [DOI] [PubMed] [Google Scholar]
- 102.Frasch SC, Henson PM, Nagaosa K, Fessler MB, Borregaard N, Bratton DL. Phospholipid flip-flop and phospholipid scramblase 1 (PLSCR1) co-localize to uropod rafts in formylated met-leu-phe-stimulated neutrophils. Journal of Biological Chemistry. 2004;279(17):17625–17633. doi: 10.1074/jbc.M313414200. [DOI] [PubMed] [Google Scholar]
- 103.Bernal-Mizrachi L, Jy W, Jimenez JJ, et al. High levels of circulating endothelial microparticles in patients with acute coronary syndromes. American Heart Journal. 2003;145(6):962–970. doi: 10.1016/S0002-8703(03)00103-0. [DOI] [PubMed] [Google Scholar]
- 104.Andree HAM, Nemerson Y. Tissue factor: regulation of activity by flow and phospholipid surfaces. Blood Coagulation and Fibrinolysis. 1995;6(3):189–197. [PubMed] [Google Scholar]
- 105.Jacoby RC, Owings JT, Holmes J, Battistella FD, Gosselin RC, Paglieroni TG. Platelet activation and function after trauma. Journal of Trauma—Injury, Infection and Critical Care. 2001;51(4):639–647. doi: 10.1097/00005373-200110000-00003. [DOI] [PubMed] [Google Scholar]
- 106.Combes V, Taylor TE, Juhan-Vague I, et al. Circulating endothelial microparticles in Malawian children with severe falciparum malaria complicated with coma. Journal of the American Medical Association. 2004;291(21):2542–2544. doi: 10.1001/jama.291.21.2542-b. [DOI] [PubMed] [Google Scholar]
- 107.Mfonkeu JBP, Gouado I, Kuaté HF, et al. Elevated cell-specific microparticles are a biological marker for cerebral dysfunctions in human severe malaria. PLoS ONE. 2010;5(10) doi: 10.1371/journal.pone.0013415.e13415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klein RS, Lin E, Zhang B, et al. Neuronal CXCL10 directs CD8+ T-cell recruitment and control of West Nile virus encephalitis. Journal of Virology. 2005;79(17):11457–11466. doi: 10.1128/JVI.79.17.11457-11466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhang B, Ying KC, Lu B, Diamond MS, Klein RS. CXCR3 mediates region-specific antiviral T cell trafficking within the central nervous system during west nile virus encephalitis. Journal of Immunology. 2008;180(4):2641–2649. doi: 10.4049/jimmunol.180.4.2641. [DOI] [PubMed] [Google Scholar]
- 110.Sui Y, Potula R, Dhillon N, et al. Neuronal apoptosis is mediated by CXCL10 overexpression in simian human immunodeficiency virus encephalitis. American Journal of Pathology. 2004;164(5):1557–1566. doi: 10.1016/S0002-9440(10)63714-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tjitra E, Suprianto S, Dyer M, Currie BJ, Anstey NM. Field evaluation of the ICT Malaria P.f/P.v immunochromatographic test for detection of Plasmodium falciparum and Plasmodium vivax in patients with a presumptive clinical diagnosis of malaria in eastern Indonesia. Journal of Clinical Microbiology. 1999;37(8):2412–2417. doi: 10.1128/jcm.37.8.2412-2417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eisen DP, Saul A. Disappearance of pan-malarial antigen reactivity using the ICT Malaria P.f/P.v(TM) kit parallels decline of patent parasitaemia as shown by microscopy. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2000;94(2):169–170. doi: 10.1016/s0035-9203(00)90262-1. [DOI] [PubMed] [Google Scholar]
- 113.Fleischer B. Editorial: 100 Years ago: Giemsa’s solution for staining of plasmodia. Tropical Medicine and International Health. 2004;9(7):755–756. doi: 10.1111/j.1365-3156.2004.01278.x. [DOI] [PubMed] [Google Scholar]
- 114.Goncalves L, Subtil A, de Oliveira MR, Rosario V, Lee PW, Shaio MF. Bayesian latent class models in malaria diagnosis. Plos ONE. 2012;7 doi: 10.1371/journal.pone.0040633.e40633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee S, Song K-M, Jeon W, Jo H, Shim Y-B, Ban C. A highly sensitive aptasensor towards Plasmodium lactate dehydrogenase for the diagnosis of malaria. Biosensors and Bioelectronics. 2012;35(1):291–296. doi: 10.1016/j.bios.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 116.Sharma MK, Rao VK, Agarwal GS, et al. Highly sensitive amperometric immunosensor for detection of plasmodium falciparum histidine-rich protein 2 in serum of humans with malaria: comparison with a commercial kit. Journal of Clinical Microbiology. 2008;46(11):3759–3765. doi: 10.1128/JCM.01022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.de Souza Castilho M, Laube T, Yamanaka H, Alegret S, Pividori MI. Magneto immunoassays for plasmodium falciparum histidine-rich protein 2 related to malaria based on magnetic nanoparticles. Analytical Chemistry. 2011;83(14):5570–5577. doi: 10.1021/ac200573s. [DOI] [PubMed] [Google Scholar]
- 118.Piper R, LeBras J, Wentworth L, et al. Immunocapture diagnostic assays for malaria using Plasmodium lactate dehydrogenase (pLDH) American Journal of Tropical Medicine and Hygiene. 1999;60(1):109–118. doi: 10.4269/ajtmh.1999.60.109. [DOI] [PubMed] [Google Scholar]
- 119.Sharma MK, Rao VK, Merwyn S, Agarwal GS, Upadhyay S, Vijayaraghavan R. A novel piezoelectric immunosensor for the detection of malarial Plasmodium falciparum histidine rich protein-2 antigen. Talanta. 2011;85(4):1812–1817. doi: 10.1016/j.talanta.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 120.Lee PW, Ji DD, Liu CT, Rampao HS. Application of loop-mediated isothermal amplification for malaria diagnosis during a follow up study in Sao Tome. Malaria Journal. 2012;11, article 408 doi: 10.1186/1475-2875-11-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Morassin B, Fabre R, Berry A, Magnaval J-F. One year’s experience with the polymerase chain reaction as a routine method for the diagnosis of imported malaria. American Journal of Tropical Medicine and Hygiene. 2002;66(5):503–508. doi: 10.4269/ajtmh.2002.66.503. [DOI] [PubMed] [Google Scholar]
- 122.Lau Y-L, Fong M-Y, Mahmud R, et al. Specific, sensitive and rapid detection of human plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malaria Journal. 2011;10, article 197 doi: 10.1186/1475-2875-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Malleret B, Claser C, Ong ASM, et al. A rapid and robust tri-color flow cytometry assay for monitoring malaria parasite development. Scientific Reports. 2011;1, article 118 doi: 10.1038/srep00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Feldman AB. Progress toward rapid malaria screening based on mass spectrometry. Lab Tech, 2006, http://www.cli-online.com/fileadmin/artimg/progress-toward-rapid-malaria-screening-based-on-mass-spectrometry.pdf.
- 125.Martin SK, Rajasekariah G-H, Awinda G, Waitumbi J, Kifude C. Unified parasite lactate dehydrogenase and histidine-rich protein ELISA for quantification of Plasmodium falciparum. American Journal of Tropical Medicine and Hygiene. 2009;80(4):516–522. [PubMed] [Google Scholar]
- 126.Noedl H, Yingyuen K, Laoboonchai A, Fukuda M, Sirichaisinthop J, Miller RS. Sensitivity and specificity of an antigen detection ELISA for malaria diagnosis. American Journal of Tropical Medicine and Hygiene. 2006;75(6):1205–1208. [PubMed] [Google Scholar]
- 127.Björkman A, Mårtensson A. Risks and benefits of targeted malaria treatment based on rapid diagnostic test results. Clinical Infectious Diseases. 2010;51(5):512–514. doi: 10.1086/655689. [DOI] [PubMed] [Google Scholar]
- 128.Moody A. Rapid diagnostic tests for malaria parasites. Clinical Microbiology Reviews. 2002;15(1):66–78. doi: 10.1128/CMR.15.1.66-78.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Maltha J, Gillet P, Bottieau E, Cnops L, van Esbroeck M, Jacobs J. Evaluation of a rapid diagnostic test (CareStart Malaria HRP-2/pLDH (Pf/pan) Combo Test) for the diagnosis of malaria in a reference setting. Malaria Journal. 2010;9(1, article 171) doi: 10.1186/1475-2875-9-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wongsrichanalai C. Rapid diagnostic techniques for malaria control. Trends in Parasitology. 2001;17(7):307–309. doi: 10.1016/s1471-4922(01)01925-0. [DOI] [PubMed] [Google Scholar]
- 131.Abeku TA, Kristan M, Jones C, et al. Determinants of the accuracy of rapid diagnostic tests in malaria case management: evidence from low and moderate transmission settings in the East African highlands. Malaria Journal. 2008;7, article 202 doi: 10.1186/1475-2875-7-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jorgensen P, Chanthap L, Rebueno A, Tsuyuoka R, Bell D. Malaria rapid diagnostic tests in tropical climates: the need for a cool chain. American Journal of Tropical Medicine and Hygiene. 2006;74(5):750–754. [PubMed] [Google Scholar]
- 133.Chiodini PL, Bowers K, Jorgensen P, et al. The heat stability of Plasmodium lactate dehydrogenase-based and histidine-rich protein 2-based malaria rapid diagnostic tests. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2007;101(4):331–337. doi: 10.1016/j.trstmh.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 134.Mayxay M, Pukrittayakamee S, Chotivanich K, Looareesuwan S, White NJ. Persistence of Plasmodium falciparum HRP-2 in successfully treated acute falciparum malaria. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2001;95(2):179–182. doi: 10.1016/s0035-9203(01)90156-7. [DOI] [PubMed] [Google Scholar]
- 135.Kyabayinze DJ, Tibenderana JK, Odong GW, Rwakimari JB, Counihan H. Operational accuracy and comparative persistent antigenicity of HRP2 rapid diagnostic tests for Plasmodium falciparum malaria in a hyperendemic region of Uganda. Malaria Journal. 2008;7, article 221 doi: 10.1186/1475-2875-7-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Laferi H, Kandel K, Pichler H. False positive dipstick test for malaria. The New England Journal of Medicine. 1997;337(22):1635–1636. doi: 10.1056/NEJM199711273372219. [DOI] [PubMed] [Google Scholar]
- 137.Iqbal J, Sher A, Rab A. Plasmodium falciparum histidine-rich protein 2-based immunocapture diagnostic assay for malaria: cross-reactivity with rheumatoid factors. Journal of Clinical Microbiology. 2000;38(3):1184–1186. doi: 10.1128/jcm.38.3.1184-1186.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Jelinek T, Amsler L, Grobusch MP, Nothdurft HD. Self-use of rapid tests for malaria diagnosis by tourists. The Lancet. 1999;354(9190):p. 1609. doi: 10.1016/s0140-6736(99)01969-8. [DOI] [PubMed] [Google Scholar]
- 139.Palmer CJ, Lindo JF, Klaskala WI, et al. Evaluation of the optimal test for rapid diagnosis of Plasmodium vivax and Plasmodium falciparurn malaria. Journal of Clinical Microbiology. 1998;36(1):203–206. doi: 10.1128/jcm.36.1.203-206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Iqbal J, Siddique A, Jameel M, Hira PR. Persistent histidine-rich protein 2, parasite lactate dehydrogenase, and panmalarial antigen reactivity after clearance of Plasmodium falciparum monoinfection. Journal of Clinical Microbiology. 2004;42(9):4237–4241. doi: 10.1128/JCM.42.9.4237-4241.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lee N, Baker J, Bell D, McCarthy J, Cheng Q. Assessing the genetic diversity of the aldolase genes of Plasmodium falciparum and Plasmodium vivax and its potential effect on performance of aldolase-detecting rapid diagnostic tests. Journal of Clinical Microbiology. 2006;44(12):4547–4549. doi: 10.1128/JCM.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gamboa D, Ho M-F, Bendezu J, et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS ONE. 2010;5(1) doi: 10.1371/journal.pone.0008091.e8091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Maltha J, Gamboa D, Bendezu J, et al. Rapid diagnostic tests for malaria diagnosis in the Peruvian Amazon: impact of pfhrp2 gene deletions and cross reactions. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0043094.e43094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bendezu J, Rosas A, Grande T, et al. Field evaluation of a rapid diagnostic test (Parascreen™) for malaria diagnosis in the Peruvian Amazon. Malaria Journal. 2010;9(1, article 154) doi: 10.1186/1475-2875-9-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Makler MT, Palmer CJ, Ager AL. A review of practical techniques for the diagnosis of malaria. Annals of Tropical Medicine and Parasitology. 1998;92(4):419–433. doi: 10.1080/00034989859401. [DOI] [PubMed] [Google Scholar]
- 146.Tjitra E, Suprianto S, McBroom J, Currie BJ, Anstey NM. Persistent ICT malaria P.f/P.v Panmalarial and HRP2 antigen reactivity after treatment of Plasmodium falciparum malaria is associated with gametocytemia and results in false-positive diagnoses of Plasmodium vivax in convalescence. Journal of Clinical Microbiology. 2001;39(3):1025–1031. doi: 10.1128/JCM.39.3.1025-1031.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mariette N, Barnadas C, Bouchier C, Tichit M, Ménard D. Country-wide assessment of the genetic polymorphism in Plasmodium falciparum and Plasmodium vivax antigens detected with rapid diagnostic tests for malaria. Malaria Journal. 2008;7, article 219 doi: 10.1186/1475-2875-7-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Baker J, McCarthy J, Gatton M, et al. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 (PfHRP2) and its effect on the performance of PfHRP2-based rapid diagnostic tests. Journal of Infectious Diseases. 2005;192(5):870–877. doi: 10.1086/432010. [DOI] [PubMed] [Google Scholar]
- 149.Ramutton T, Hendriksen I, Mwanga-Amumpaire J, et al. Sequence variation does not confound the measurement of plasma PfHRP2 concentration in African children presenting with severe malaria. Malaria Journal. 2012;11:p. 276. doi: 10.1186/1475-2875-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hendriksen ICE, Mwanga-Amumpaire J, von Seidlein L, et al. Diagnosing severe falciparum malaria in parasitaemic African children: a prospective evaluation of plasma PfHRP2 measurement. PLOS Medicine. 2012;9 doi: 10.1371/journal.pmed.1001297.e1001297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Murray CK, Bennett JW. Rapid diagnosis of malaria. Interdisciplinary Perspectives on Infectious Diseases. 2009;2009:7 pages. doi: 10.1155/2009/415953.415953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wongsrichanalai C, Barcus MJ, Muth S, Sutamihardja A, Wernsdorfer WH. A review of malaria diagnostic tools: microscopy and rapid diagnostic test (RDT) The American Journal of Tropical Medicine and Hygiene. 2007;77(6):119–127. [PubMed] [Google Scholar]
- 153.Peng Y, Wu J, Wang J, Li W, Yu S. Study and evaluation of Wondfo rapid diagnostic kit based on nano-gold immunochromatography assay for diagnosis of Plasmodium falciparum. Parasitology Research. 2012;110:1421–1425. doi: 10.1007/s00436-011-2643-6. [DOI] [PubMed] [Google Scholar]
- 154.Fogg C, Twesigye R, Batwala V, et al. Assessment of three new parasite lactate dehydrogenase (pan-pLDH) tests for diagnosis of uncomplicated malaria. Transactions of the Royal Society of Tropical Medicine and Hygiene. 2008;102(1):25–31. doi: 10.1016/j.trstmh.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 155.Mawili-Mboumba DP, Akotet MKB, Ngoungou EB, Kombila M. Evaluation of rapid diagnostic tests for malaria case management in Gabon. Diagnostic Microbiology and Infectious Disease. 2010;66(2):162–168. doi: 10.1016/j.diagmicrobio.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 156.Rakotonirina H, Barnadas C, Raherijafy R, et al. Accuracy and reliability of malaria diagnostic techniques for guiding febrile outpatient treatment in malaria-endemic countries. American Journal of Tropical Medicine and Hygiene. 2008;78(2):217–221. [PubMed] [Google Scholar]
- 157.Nicastri E, Bevilacqua N, Schepisi MS, et al. Accuracy of malaria diagnosis by microscopy, rapid diagnostic test, and PCR methods and evidence of antimalarial Overprescription in non-severe febrile patients in two Tanzanian hospitals. American Journal of Tropical Medicine and Hygiene. 2009;80(5):712–717. [PubMed] [Google Scholar]
- 158.de Oliveira AM, Skarbinski J, Ouma PO, et al. Performance of malaria rapid diagnostic tests as part of routine malaria case management in Kenya. American Journal of Tropical Medicine and Hygiene. 2009;80(3):470–474. [PubMed] [Google Scholar]
- 159.Avila PE, Kirchgatter K, Brunialti KCS, Oliveira AM, Siciliano RF, Di Santi SM. Evaluation of a rapid dipstick test, Malar-Check™, for the diagnosis of Plasmodium falciparum malaria in Brazil. Revista do Instituto de Medicina Tropical de Sao Paulo. 2002;44(5):293–296. doi: 10.1590/s0036-46652002000500012. [DOI] [PubMed] [Google Scholar]
- 160.Richardson DC, Ciach M, Zhong KJY, Crandall I, Kain KC. Evaluation of the Makromed dipstick assay versus PCR for diagnosis of Plasmodium falciparum malaria in returned travelers. Journal of Clinical Microbiology. 2002;40(12):4528–4530. doi: 10.1128/JCM.40.12.4528-4530.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Uguen C, Rabodonirina M, de Pina J-J, et al. ParaSight-F rapid manual diagnostic test of Plasmodium falciparum infection. Bulletin of the World Health Organization. 1995;73(5):643–649. [PMC free article] [PubMed] [Google Scholar]
- 162.Forney JR, Magill AJ, Wongsrichanalai C, et al. Malaria rapid diagnostic devices: performance characteristics of the ParaSight F device determined in a multisite field study. Journal of Clinical Microbiology. 2001;39(8):2884–2890. doi: 10.1128/JCM.39.8.2884-2890.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Singh N, Saxena A, Awadhia SB, Shrivastava R, Singh MP. Evaluation of a rapid diagnostic test for assessing the burden of malaria at delivery in India. American Journal of Tropical Medicine and Hygiene. 2005;73(5):855–858. [PubMed] [Google Scholar]
- 164.McMorrow ML, Masanja MI, Kahigwa E, Abdulla SMK, Kachur SP. Quality assurance of rapid diagnostic tests for malaria in routine patient care in rural Tanzania. American Journal of Tropical Medicine and Hygiene. 2010;82(1):151–155. doi: 10.4269/ajtmh.2010.09-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Schmidt MT, Schaechter M. Topics and Ecological and Environmental Microbiology. New York, NY, USA: Academic Press; 2011. [Google Scholar]
- 166.Cho EJ, Lee J-W, Ellington AD. Applications of aptamers as sensors. Annual Review of Analytical Chemistry. 2009;2:241–264. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 167.McKeague M, DeRosa CM. Challenges and opportunities for small molecule aptamer development. Journal of Nucleic Acids. 2012;2012:20 pages. doi: 10.1155/2012/748913.748913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ohnoa R, Ohnukia H, Wanga H, et al. Electrochemical impedance spectroscopy biosensor with interdigitated electrode for detection of human immunoglobulin A. Biosensors and Bioelectronics. 2013;40:422–426. doi: 10.1016/j.bios.2012.07.052. [DOI] [PubMed] [Google Scholar]
- 169.Min K, Cho M, Han S-Y, Shim Y-B, Ku J, Ban C. A simple and direct electrochemical detection of interferon-γ using its RNA and DNA aptamers. Biosensors and Bioelectronics. 2008;23(12):1819–1824. doi: 10.1016/j.bios.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 170.Noland GS, Briones N, Sullivan DJ., Jr. The shape and size of hemozoin crystals distinguishes diverse Plasmodium species. Molecular and Biochemical Parasitology. 2003;130(2):91–99. doi: 10.1016/s0166-6851(03)00163-4. [DOI] [PubMed] [Google Scholar]
- 171.Newman DM, Heptinstall J, Matelon RJ, et al. A magneto-optic route toward the in vivo diagnosis of malaria: preliminary results and preclinical trial data. Biophysical Journal. 2008;95(2):994–1000. doi: 10.1529/biophysj.107.128140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Newman DM, Matelon RJ, Wears ML, Savage LB. The in vivo diagnosis of malaria: feasibility study into a magneto-optic fingertip probe. IEEE Journal on Selected Topics in Quantum Electronics. 2010;16(3):573–580. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 : List of PDB entries for Plasmodium lactate dehydrogenase.
Table S2: List of inhibitors developed against lactate dehydrogenase from P.falciparum.


