Abstract
Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is an important virulence factor encoded by a family of 59 var genes, including 56 var genes plus 3 small var3 genes. The var genes are among the most diverse sequences in the P. falciparum genome, but the var3 genes are found conserved in most P. falciparum strains. Previous studies have been mainly focused on the typical var genes, while the biological characteristics of the var3 genes remain unknown. In this study, the three var3 genes, PF3D7_0100300, PF3D7_0600400, and PF3D7_0937600, were found to be transcribed in the erythrocytic stages of P. falciparum, with a peak in the transcription level at 16 h post-invasion, but terminated immediately after 16 h post-invasion. The encoded protein of PF3D7_0600400 could be detected in both the late trophozoite stage and schizont stage, while the encoded proteins of PF3D7_0100300 and PF3D7_0937600 could only be detected in the late trophozoite stage and schizont stage, respectively. Thus, the var3 genes of the P. falciparum 3D7 strain were differentially expressed during the erythrocytic development of the parasite.
Keywords: Plasmodium falciparum, var gene, PfEMP1, Expression
Abstract
La protéine 1 de membrane de l’érythrocyte de Plasmodium falciparum (PfEMP1) est un important facteur de virulence, codé par une famille de 59 gènes var, qui comprend cinquante-six gènes var et trois petits gènes var3. Les gènes var sont parmi les séquences les plus diverses dans le génome de P. falciparum, mais sont sont conservés dans la plupart des souches de P. falciparum. Des études antérieures ont été principalement axées sur les gènes var typiques, tandis que les caractéristiques biologiques des gènes var3 restent inconnues. Dans cette étude, il a été trouvé que les trois gènes var3, PF3D7_0100300, PF3D7_0600400 et PF3D7_0937600, sont transcrit dans les stades érythrocytaires de P. falciparum, avec un pic du niveau de la transcription à 16h post- invasion, qui s’arrête immédiatement après 16h post- invasion. La protéine codée par PF3D7_0600400 a pu être détectée à la fois dans le stade trophozoïte tardif et le stade schizonte, tandis que les protéines codées par PF3D7_0100300 et PF3D7_0937600 ne pouvait être détectées, respectivement, qu’au stade trophozoïte final et au stade schizonte. Les gènes var3 de la souche 3D7 de P. falciparum sont donc exprimés de façon différentielle au cours du développement érythrocytaire du parasite.
Introduction
Plasmodium falciparum malaria is still a major threat to human life and health, especially in sub-Saharan Africa [1]. An estimated 300–500 million clinical cases occur each year, with 660,000 deaths [1]. It is a disease that mainly affects immunologically naive individuals, especially children under 5 years old, and women who are in their first or second pregnancy [5, 9]. Malaria vaccine development has been hampered by the frequent antigenic variation of the P. falciparum parasites. Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP1) is one of the most extensively studied variant surface antigens. Members of the PfEMP1 family mediate the cyto-adherence of infected erythrocytes to host receptors, allowing the parasites to avoid splenic clearance, and the tremendous sequence variation within the variant family has enabled the parasites to escape host immune responses [2, 17].
The PfEMP1 family is encoded by var genes which include 56 var genes plus 3 small var-like genes (collectively called var3) [10, 27]. It has previously been shown that var genes can be subgrouped into three major groups (groups A, B, and C) and two intermediate groups B/A and B/C based on the conserved upstream sequence and genomic locations [16, 19]. The var3 genes were also classified into group A based on the N-terminus sequences [19]. However, they are very different from the other var genes in sequence length and secondary structures of the encoded proteins. Earlier studies have found that the expression of group A PfEMP1s was associated with severe malaria with phenotypes of rosetting and adherence to endothelial receptors [15, 18, 32]. The DBLα (α-type Duffy binding-like) domains have been regarded as the molecular markers for classification of the sequence types associated with clinical diseases [13, 23, 31]. Further, previous studies have mainly been focused on the “typical” var genes, while the biological characteristics of the var3 genes have not been extensively explored. Several studies suggested that var3 was indeed expressed by clinical P. falciparum isolates as well as the laboratory-adapted parasite lines, which could be recognized by IgG from children from malaria-endemic regions [3, 30]. The three var3 genes (PF3D7_0100300, PF3D7_0600400, and PF3D7_0937600) are located in subtelomeric regions of chromosome 1, 6, and 9, respectively. Var3 genes are around 4 kb, and the secondary structures of the extracellular regions of the encoded proteins contain only one DBLα and one DBLε domain, whereas the other PfEMP1 proteins are composed of multiple DBL domains and a cystine-rich interdomain region (CIDR). Thus, var3 genes encoded “mini-PfEMP1s” contain a very compressed architecture and an atypical PfEMP1 structure (Fig. 1A). Further, the orthologs of the three var3 genes are detected in nearly all parasite isolates [29], which are as conserved as the var2csa gene that encodes VAR2CSA associated with pregnancy-associated malaria. However, the function of the “mini-PfEMP1s” encoded by the var3 genes is still not known. Further, the atypical intron activity of the var3 genes suggested their expression might be under the control of a different mechanism from other var genes [7]. In this study, the transcription and expression of the individual var3 genes were analyzed by quantitative RT-PCR, Western blot, and immunofluorescence.
Figure 1.

Schematic structures of the three var3 genes and the encoded proteins. A. The domain architecture of the “min-PfEMP1s” encoded by the three var3 genes. NTS: N-Terminal Segment; DBL: Duffy Binding-Like; ATS: Acidic Terminal Segment; TM: Transmembrane region. B. The variable (A1, B1, and C1) and conserved (A2, B2, and C2) regions and the identity in percentage between two sequences.
Methods
Parasite culture
The Plasmodium falciparum 3D7 clone was cultured in human O+ RBC according to standard procedures [28]. The parasites were synchronized by three rounds of treatment at 4 h post-invasion with 5% sorbitol and parasites were harvested every 8 h, including 8, 16, 24, 32, 40, and 48 h post-infection.
Transcription analysis of the var3 genes with Quantitative RT-PCR
RNA at six time points after invasion was extracted using Trizol (Invitrogen, CA, USA) according to the manufacturer’s instructions. The extracted RNAs were treated with DNase I (TaKaRa, Dalian, China) to completely remove the genomic DNA. The RNA was reverse transcribed from Oligo(dT). The cDNAs were used as templates for quantitative RT-PCR with the specific primers; the information regarding the primer sequences is shown in Table 1. All primer sets had amplification efficiency >90%, which was determined automatically by the software. Quantitative RT-PCR was performed on an ABI PRISM® 7500 Real-Time PCR System (Applied Biosystems) applying SYBR® Premix Ex Taq TM (TaKaRa), and the internal control gene seryl-tRNA synthetase (PF3D7_1205100), which is stably expressed during the erythrocytic stage of the parasite, was used for normalization [21]. Further, two genes (PF3D7_0811600 and PF3D7_1361800) mainly expressed at 40–48 h p.i. were also included as external controls. Transcript levels relative to the average level of the internal control gene were calculated as 2−ΔCt (var genes) [20]. The experiment was repeated three times and the transcription levels were represented by the mean values of the three experiments.
Table 1.
The primer sequences of quantitative RT-PCR.
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| PF3D7_0937600 | ATGGCACCGAAAA | GCCGCTTCCAATA |
| PF3D7_0100300 | GGGATCATTATGGGAAGCAC | CGTTCTTGATTTCTACCATCGCA |
| PF3D7_0600400 | ACGCCCAATTTCATGAT | TTCGGCATTTTCGTCAA |
Generation of recombinant protein and variant-specific antibodies
The gene fragments, named PF3D7_0100300 (amino acids 1-962), PF3D7_0600400 (amino acids 1-972), and PF3D7_0937600 (amino acids 1-1044) respectively, encoding the N-terminal regions (about 110 amino acids) of the three “mini-PfEMP1s” were selected as the target sequences. The three target sequences plus an ATS (acidic C-terminal segment) region were amplified from genomic DNA of the 3D7 P. falciparum clone. The ATS region was selected based on the complete conservation of the amino acid sequence (more than 90% identity between different PfEMP1 variants). The information regarding the primer sets is shown in Table 2. The amplified PCR products were cloned into the plasmid pET-28a (Qiagen, Düsseldorf, Germany) and pGEX-4T-1 (GE Healthsystems, Uppsala, Sweden) to construct recombinant plasmids. Expression and purification of His-tagged (ATS-His, PF3D7_0100300-His, PF3D7_0600400-His, and PF3D7_0937600-His) and GST-tagged (ATS-GST, PF3D7_0100300-GST, PF3D7_0600400-GST, and PF3D7_0937600-GST) recombinant proteins were carried out as described [20, 21, 26].
Table 2.
The primer sequences for cloning the gene fragments encoding the recombinant proteins expressed in E. coli.
| Gene name | Forward primer | Reverse primer |
|---|---|---|
| ATS | GTAGATATGATACGGATATT | TTTTGATGGCTCCAGAACAA |
| PF3D7_0937600 | ATGGCACCGAAAAATGGA | TATTTTATCATTGTTACAA |
| PF3D7_0100300 | ATGGCACTAAAAAAAG | CGTTCTTGATTTCTACCATCGCA |
| PF3D7_0600400 | ATGGGATCAGATTATTCG | ACTATTACAATACGCTTC |
Four female Wister rats received four immunizations with His-tagged recombinant proteins on days 0, 15, 30, 45, and 60 (100 μg/rat) emulsified in Freund’s complete (first immunization) and incomplete adjuvant (second to fourth immunizations). Simultaneously, three female New Zealand rabbits were immunized with ATS-His recombinant proteins. The experiments with laboratory animals were approved by the ethical committee of the Department of Agricultural Sciences, Jilin University. Specific IgG fractions in the immunized rats and rabbits were affinity-purified using the Protein G SepharoseTM 4 Fast Flow (GE Healthcare) and the nProtein A SepharoseTM 4 Fast Flow (GE Healthcare), respectively.
Expression characterization of the native proteins
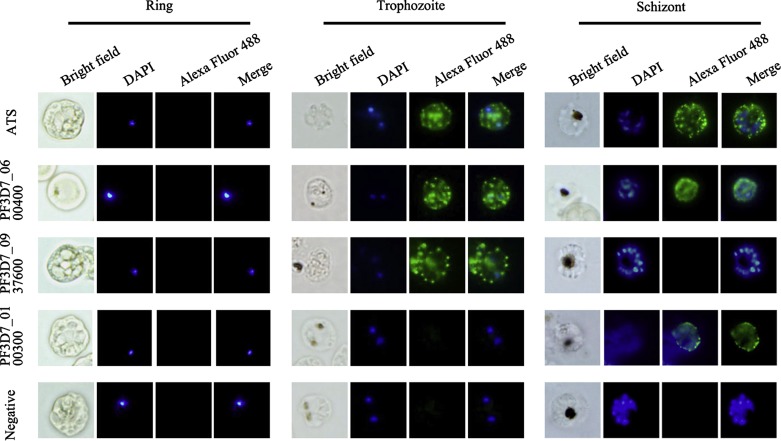
The infected erythrocytes at three developmental stages, including the ring, late trophozoite, and schizont stages, were purified by centrifugation on a gradient Percoll (GE Healthcare) as described (http://www.mr4.org/Portals/3/Methods_In_Malaria_Research-5theditionv5-2.pdf). Western blots and immunofluorescence assays were performed as described [11]. Briefly, the infected erythrocytes with parasites at three developmental stages, including the ring stage, late trophozoite stage, and schizont stage, were purified by gradient Percoll (GE Healthcare). For Western blotting, the total proteins of the infected erythrocytes were denatured in SDS-PAGE loading buffer (250 mM Tris, 1.92 M glycine, and 1% SDS) and subjected to electrophoresis under reducing conditions on an 8% polyacrylamide gel, and the proteins were transferred onto 0.2-μm nitrocellulose membranes (Bio-Rad, CA, USA). Membranes were blocked with 5% milk (Sigma, St. Louis, USA) for 1 h followed by incubation with the anti-ATS-His, anti-PF3D7_0100300-His, anti-PF3D7_0600400-His, and anti-PF3D7_0937600-His IgG (1:1000), respectively, as primary antibodies overnight at 4 °C. The membranes were further incubated with alkaline phosphatase conjugated mouse anti-rat IgG (Sigma, 1:20,000) for 1 h at 37 °C, BCIP/NBT substrate (Sigma) was added to the membranes and the reaction was terminated by rinsing in water after sufficient color development. To further confirm the expression of the three var3 genes, indirect immunofluorescence assays were performed. Thin smears of ring, late trophozoite, and schizont stage P. falciparum-infected erythrocytes were fixed with methanol at −80 °C for 5 min and blocked with PBS containing 5% nonfat milk at 37 °C for 1 h. The smears were then incubated with anti-ATS-His, anti-PF3D7_0937600-His, anti-PF3D7_0600400-His, and anti-PF3D7_0100300-His antibodies (1:50), followed by incubation with Alexa Fluor 488-conjugated goat anti-rat IgG (Invitrogen) (1:1000) at 37 °C for 1 h. Healthy rat IgG was used as a negative control antibody. The parasite nuclei were stained with DAPI (Roche, Basel, Switzerland) at room temperature for 5 min. High-resolution images were captured with a fluorescence microscope (Olympus, BX 53).
Results and discussion
Sequence analyses of the var3 genes
The encoded proteins by the three var3 genes were divided into two regions based on sequence similarity. The most N-terminal regions of around 110 amino acids were variable, with 60%–67% identity between two sequences, while the internal regions were more similar with an identity of 88%–96% (Figs. 1A and 1B). The high similarity of this structurally distinct group of var genes indicated they shared a common origin and were generated by gene recombination [8, 22].
Quantitative RT-PCR analysis of the var3 genes in the 3D7 clone
The transcriptions of the three var3 genes in the P. falciparum 3D7 clone were determined by quantitative RT-PCR. The transcription of the three genes started early after erythrocyte invasion, which was in a similar way to other var genes [6], but with a relatively short time window. The transcription peaked at 16 h post-invasion and terminated immediately afterward (Fig. 2), which was obviously different from the other var genes [19], whose transcription could last until 30–36 h post-invasion [17, 30]. The two genes (PF3D7_0811600 and PF3D7_1361800) were found mainly expressed at 40 and 48 h p.i., respectively (data not shown). Further, earlier studies found that, in most cases, only one var gene was activated in an erythrocytic cycle [4, 24]. The data from this study found that the expression of the three var3 genes might not follow the rule of mutual exclusion, since the encoded proteins were expressed in a major population of the parasites in the same culture, which will be further discussed later.
Figure 2.
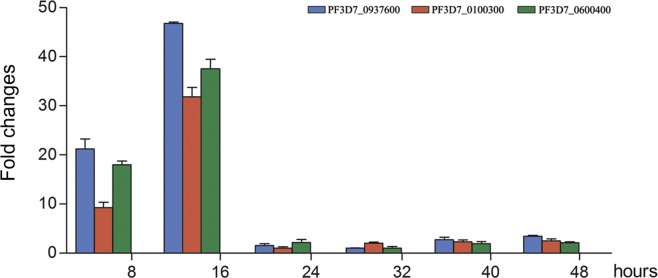
Transcriptions of the three var3 genes in the P. falciparum 3D7 clone. The transcriptions of the three var3 genes at 8, 16, 24, 32, 40, and 48 h p.i. are shown. Transcript levels relative to that of the internal control gene were calculated as 2−ΔCt (var genes).
The three var-like genes were differentially expressed in either the late trophozoite or schizont stages
The expression of the var3 genes were, in the first step, analyzed by Western blots using total proteins of infected erythrocytes as antigens, including the ring, late trophozoite, and schizont stage, respectively. The results showed that a single protein band was recognized by the variant-specific antibodies, except that with the anti-ATS antibody, which can literally recognize all PfEMP1 variants. Further, the var3 genes were found to be differentially expressed within three different time windows. The gene PF3D7_0600400 was expressed in both the late trophozoite stage and schizont stage, which was in a similar way to other var genes [14, 25]. However, the molecular size of the protein was much smaller than the theoretical value of the encoded protein. The reason is a mystery, since no sequence mutation was observed in either gDNA or cDNA. It was likely that the translation of the corresponding mRNA was terminated earlier controlled by an unknown mechanism, which needs to be further studied. PF3D7_0937600 was only expressed in the late trophozoite stage (24–32 h p.i.), but not in the schizont stage (36–44 h p.i.). In contrast, the protein encoded by PF3D7_0100300 could be detected in the schizont stage (36–44 h p.i.), but not in either the ring or late trophozoite stages (Fig. 3). Thus, the var3 genes were activated in a similar way to other var genes, but the corresponding mRNAs were likely less stable. In an earlier study, Wang et al. found that proteins encoded by the var3 genes were expressed on some of the unselected parasites from malaria patients [30]. Here, our data further showed that the three var3 genes were indeed differentially expressed by the parasites, which strongly indicated that the “mini-PfEMP1” also participated in the immune evasion of P. falciparum.
Figure 3.
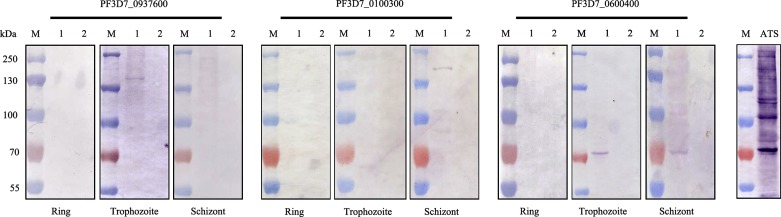
Western blot analysis of native proteins in three developmental stages. The expression of the native proteins encoded by the three var3 genes (PF3D7_0100300, PF3D7_0600400, and PF3D7_0937600) in the ring, trophozoite, and schizont stages were, respectively, detected by the variant-specific antibodies generated in rats. Lanes 1 and 2 represent results from infected red blood cells and uninfected red blood cells, respectively. A band of approximately 152 kDa was detected in the late trophozoite stage (24–30 h p.i.) with the antibody specific to PF3D7_0937600. A band of approximately 153 kDa was detected in the schizont stage (36–44 h p.i.) with the antibody specific to PF3D7_0100300. And a band of approximately 70 kDa was detected in both the trophozoite stage and schizont stage (24–44 h p.i.) for the PF3D7_0600400-specific antibody.
To further confirm and localize the proteins expressed by the three var3 genes in the infected erythrocytes, an immunofluorescence assay (IFA) was performed using the anti-PF3D7_0100300-His, anti-PF3D7_0600400-His, and anti-PF3D7_0937600-His IgGs as primary antibodies; meanwhile, an anti-ATS IgG was used as a control. The results clearly showed that the distribution of the proteins on the infected red blood cells was in a typical punctuated pattern of PfEMP1 [12]. The pre-immune IgG of the immunized rats did not show any surface reactivity with the pRBC. Further, as observed in the Western blot assays, PF3D7_0600400 was expressed in both the late trophozoite stage and schizont stage, while PF3D7_0937600 was only expressed in the late trophozoite stage and PF3D7_0100300 could only be detected in the schizont stage (Fig. 4). The punctuated patterns of the fluorescence of the proteins recognized with the variant-specific antibodies as well as the anti-ATS antibodies suggested that they followed the same translocation pathway as other PfEMP1s in the infected red blood cells. Thus, the three var3 genes were transcribed at the same time but expressed in different time windows in the infected erythrocytes, indicating they are controlled by a distinct mechanism of posttranscriptional regulation, which requires further investigation.
Figure 4.
Immunofluorescence assays with variant-specific antibodies. An immunofluorescence assay (IFA) was performed using anti-PF3D7_0100300-His, anti-PF3D7_0600400-His, and anti-PF3D7_0937600-His IgG (1:50) as primary antibodies and anti-ATS IgG as the control. Specific staining of infected erythrocytes (IEs) is observed as punctuated fluorescence patterns over the IE surface using a secondary antibody labeled with Alexa 488 (green). The pre-immune sera of the immunized rats did not result in any surface reactivity with the IE. DAPI (5 μg/mL) staining of DNA in the nuclei is blue. The anti-ATS IgG and anti-PF3D7_0600400-His IgG stained IEs of both the trophozoite and schizont stages, the anti-PF3D7_0937600-His IgG only reacted with IEs of the trophozoite stage, while the anti-PF3D7_0100300-His IgG only reacted with IEs of the schizont stage.
In conclusion, the expression of the three atypical var3 genes of the 3D7 P. falciparum clone was investigated by quantitative RT-PCR, Western blot, and immunofluorescence assay. It was found that the var3 genes were differentially expressed during the erythrocytic stage of the parasite with features which differed from the typical var genes.
Conflict of interest: All authors have declared no conflict of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81130033, 81171592).
Cite this article as: Zhang Y, Jiang N, Chang Z, Wang H, Lu H, Wahlgren M & Chen Q: The var3 genes of Plasmodium falciparum 3D7 strain are differentially expressed in infected erythrocytes. Parasite, 2014, 21, 19.
References
- 1.Anonymous. 2013. World Malaria Report 2013. World Health Organization [Google Scholar]
- 2.Barry AE, Trieu A, Fowkes FJ, Pablo J, Kalantari-Dehaghi M, Jasinskas A, Tan X, Kayala MA, Tavul L, Siba PM, Day KP, Baldi P, Felgner PL, Doolan DL. 2011. The stability and complexity of antibody responses to the major surface antigen of Plasmodium falciparum are associated with age in a malaria endemic area. Molecular & Cellular Proteomics, 10, M111.008326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cham GK, Turner L, Lusingu J, Vestergaard L, Mmbando BP, Kurtis JD, Jensen AT, Salanti A, Lavstsen T, Theander TG. 2009. Sequential, ordered acquisition of antibodies to Plasmodium falciparum erythrocyte membrane protein 1 domains. Journal of Immunology, 183, 3356–3363 [DOI] [PubMed] [Google Scholar]
- 4.Chen Q, Fernandez V, Sundström A, Schlichtherle M, Datta S, Hagblom P, Wahlgren M. 1998. Developmental selection of var gene expression in Plasmodium falciparum. Nature, 394, 392–395 [DOI] [PubMed] [Google Scholar]
- 5.Chen Q. 2007. The naturally acquired immunity in severe malaria and its implication for a PfEMP-1 based vaccine. Microbes and Infection, 9, 777–783 [DOI] [PubMed] [Google Scholar]
- 6.Dahlbäck M, Lavstsen T, Salanti A, Hviid L, Arnot DE, Theander TG, Nielsen MA. 2007. Changes in var gene mRNA levels during erythrocytic development in two phenotypically distinct Plasmodium falciparum parasites. Malaria Journal, 6, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. 2009. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA, 15, 116–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flick K, Chen Q. 2004. var genes, PfEMP1 and the human host. Molecular and Biochemical Parasitology, 134, 3–9 [DOI] [PubMed] [Google Scholar]
- 9.Fried M, Nosten F, Brockman A, Brabin BJ, Duffy PE. 1998. Maternal antibodies block malaria. Nature, 395, 851–852 [DOI] [PubMed] [Google Scholar]
- 10.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature, 419, 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinterberg K, Scherf A, Gysin J, Toyoshima T, Aikawa M, Mazie JC, da Silva LP, Mattei D. 1994. Plasmodium falciparum: the Pf332 antigen is secreted from the parasite by a brefeldin A-dependent pathway and is translocated to the erythrocyte membrane via the Maurer’s clefts. Experimental Parasitology, 79, 279–291 [DOI] [PubMed] [Google Scholar]
- 12.Joergensen L, Bengtsson DC, Bengtsson A, Ronander E, Berger SS, Turner L, Dalgaard MB, Cham GK, Victor ME, Lavstsen T, Theander TG, Arnot DE, Jensen AT. 2010. Surface co-expression of two different PfEMP1 antigens on single Plasmodium falciparum-infected erythrocytes facilitates binding to ICAM1 and PECAM1. PLoS Pathogens, 6, e1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Juillerat A, Lewit-Bentley A, Guillotte M, Gangnard S, Hessel A, Baron B, Vigan-Womas I, England P, Mercereau-Puijalon O, Bentley GA. 2011. Structure of a Plasmodium falciparum PfEMP1 rosetting domain reveals a role for the N-terminal segment in heparin-mediated rosette inhibition. Proceedings of the National Academy of Sciences of the United States of America, 108, 5243–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinyanjui SM, Howard T, Williams TN, Bull PC, Newbold CI, Marsh K. 2004. The use of cryopreserved mature trophozoites in assessing antibody recognition of variant surface antigens of Plasmodium falciparum-infected erythrocytes. Journal of Immunological Methods, 288, 9–18 [DOI] [PubMed] [Google Scholar]
- 15.Kirchgatter K, Portillo Hdel A. 2002. Association of severe noncerebral Plasmodium falciparum malaria in Brazil with expressed PfEMP1 DBL1 alpha sequences lacking cysteine residues. Molecular Medicine, 8, 16–23 [PMC free article] [PubMed] [Google Scholar]
- 16.Kraemer SM, Smith JD. 2006. A family affair: var genes, PfEMP1 binding, and malaria disease. Current Opinion in Microbiology, 9, 374–380 [DOI] [PubMed] [Google Scholar]
- 17.Kyes S, Horrocks P, Newbold C. 2001. Antigenic variation at the infected red cell surface in malaria. Annual Review of Microbiology, 55, 673–707 [DOI] [PubMed] [Google Scholar]
- 18.Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, Thera MA, Kone AK, Doumbo OK, Plowe CV, Rowe JA. 2006. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Molecular and Biochemical Parasitology, 150, 211–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lavstsen T, Salanti A, Jensen AT, Arnot DE, Theander TG. 2003. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and non-coding regions. Malaria Journal, 2, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods, 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 21.Moll K, Chene A, Ribacke U, Kaneko O, Nilsson S, Winter G, Haeggstrom M, Pan W, Berzins K, Wahlgren M, Chen Q. 2007. A novel DBL-domain of the P. falciparum 332 molecule possibly involved in erythrocyte adhesion. PLoS One, 2, e477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rask TS, Hansen DA, Theander TG, Gorm Pedersen A, Lavstsen T. 2010. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes – divide and conquer. PLoS Computational Biology, 6, e1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rorick MM, Rask TS, Baskerville EB, Day KP, Pascual M. 2013. Homology blocks of Plasmodium falciparum var genes and clinically distinct forms of severe malaria in a local population. BMC Microbiology, 13, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO Journal, 17, 5418–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scherf A, Pouvelle B, Buffet PA, Gysin J. 2001. Molecular mechanisms of Plasmodium falciparum placental adhesion. Cellular Microbiology, 3, 125–131 [DOI] [PubMed] [Google Scholar]
- 26.Smith DB, Johnson KS. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene, 67, 31–40 [DOI] [PubMed] [Google Scholar]
- 27.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell, 82, 89–100 [DOI] [PubMed] [Google Scholar]
- 28.Trager W, Jensen JB. 2005. Human malaria parasites in continuous culture. Journal of Parasitology, 91, 484–486 [DOI] [PubMed] [Google Scholar]
- 29.Trimnell AR, Kraemer SM, Mukherjee S, Phippard DJ, Janes JH, Flamoe E, Su XZ, Awadalla P, Smith JD. 2006. Global genetic diversity and evolution of var genes associated with placental and severe childhood malaria. Molecular and Biochemical Parasitology, 148, 169–180 [DOI] [PubMed] [Google Scholar]
- 30.Wang CW, Lavstsen T, Bengtsson DC, Magistrado PA, Berger SS, Marquard AM, Alifrangis M, Lusingu JP, Theander TG, Turner L. 2012. Evidence for in vitro and in vivo expression of the conserved var3 (type 3) plasmodium falciparum erythrocyte membrane protein 1. Malaria Journal, 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warimwe GM, Fegan G, Musyoki JN, Newton CR, Opiyo M, Githinji G, Andisi C, Menza F, Kitsao B, Marsh K, Bull PC. 2012. Prognostic indicators of life-threatening malaria are associated with distinct parasite variant antigen profiles. Science Translational Medicine, 4, 129ra45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Warimwe GM, Recker M, Kiragu EW, Buckee CO, Wambua J, Musyoki JN, Marsh K, Bull PC. 2013. Plasmodium falciparum var gene expression homogeneity as a marker of the host-parasite relationship under different levels of naturally acquired immunity to malaria. PLoS One, 8, e70467. [DOI] [PMC free article] [PubMed] [Google Scholar]