Abstract
Genome-wide association studies (GWAS) provide an important avenue for undertaking an agnostic evaluation of the association between common genetic variants and risk of disease. Recent advances in our understanding of human genetic variation and the technology to measure such variation have made GWAS feasible. Over the past few years a multitude of GWAS have identified and replicated many associated variants. These findings are enriching our knowledge about the genetic basis of disease and leading some to advocate using GWA study results for genetic testing. For many of the GWA study results, however, the underlying mechanisms remain unclear and the findings explain only a limited amount of heritability. These issues may be clarified by more detailed investigations, including analyses of less common variants, sequence-level data, and environmental exposures. Such studies should help clarify the potential value of genetic testing to the public’s health.
Keywords: copy number, linkage disequilibrium, population stratification, single nucleotide polymorphism, whole genome
INTRODUCTION
Genome-wide association studies (GWAS) compare common genetic variants in large numbers of affected cases to those in unaffected controls to determine whether an association with disease exists (34, 55). GWAS have been made possible by the identification of millions of single nucleotide polymorphisms (SNPs) across the human genome and the realization that a subset of these SNPs can capture (“tag”) common genetic variation via linkage disequilibrium (16). In parallel, advances in microarray-based technology have allowed investigators to genotype efficiently enormous numbers of SNPs (77).
Before the recent flood of GWA study projects, linkage and candidate gene studies were used to try to decipher the genetic basis of disease. Linkage analysis evaluates markers widely spaced across the genome to determine whether they are inherited along with disease in families with numerous affected individuals (2). Linkage analysis, however, can have low power to detect common genetic risk factors for disease and has low resolution owing to the limited number of meioses within families (55). Candidate gene studies can overcome these issues, focusing directly on the association between disease and variants in particular genes that have a priori biological support for being involved with disease. This focus comes at a cost: Candidate gene studies ignore much of the genome, and thus are likely to miss many causal regions or genes and instead find many false-positive associations (21, 36).
GWAS improve on these approaches, leveraging their strengths while overcoming weaknesses. In particular, GWAS have greater power than linkage studies to detect small to modest effects, even with an extremely strict alpha-level for statistical significance (55). Moreover, by casting a wide net of genetic markers across the entire genome, this approach does not require one to prespecify particular candidate genes for study and examines much of the common variation across the human genome. GWAS have convincingly detected hundreds of variants associated with a large number of diseases (19). Many of these findings are novel; associated SNPs in genes or chromosomal regions were not previously implicated in disease. These results are especially exciting in light of the previous difficulties replicating genetic findings for many diseases. For example, linkage and candidate gene studies of prostate cancer have had limited successes in replicating findings across studies, whereas GWAS have detected more than a dozen highly replicated genetic variants associated with this disease (80).
The enthusiasm surrounding GWA study findings is tempered, however, by the observation that genetic variants detected by most of these studies may not be causal for disease, may explain only a little of disease heritability, and may have limited public health impact (45). These issues have prompted some to question the value of current GWAS and to advocate shifting future research efforts to the study of less common genetic variation (12). In light of the mixed opinions ofGWAS, we consider here the following important aspects of these studies: design and analysis, findings and implications, limitations, and future prospects.
OVERALL STRATEGY AND METHODS
Laying the Foundation
The ability to undertake GWAS is a direct result of a number of important developments over the past decade. Sequencing of the human genome provided the initial foundation for GWAS (35, 74). Substantial efforts detected and validated more than ten million SNPs, which brought to light the common genetic variation across the human genome. It was then determined that much of this variation could be efficiently captured by a subset of “tag” SNPs via the phenomenon of linkage disequilibrium (LD) among neighboring SNPs (6, 11). The International Haplotype Map (HapMap) Consortium proceeded to measure the LD structure across multiple ancestral populations (10, 16, 17).
In conjunction with the increasing understanding of the human genome, technological advances in array-based genotyping of SNPs made feasible the simultaneous measurement of hundreds of thousands of SNPs (77). The number of variants assayed by these SNP arrays has rapidly increased, whereas the array prices have steadily decreased. At present, the arrays directly measure approximately one million SNPs while providing relatively high coverage of the common genetic variation across the human genome (26).
Multistage Study Designs
Sample sizes in the thousands are generally required for GWAS to have sufficient statistical power to detect the expected modest associations (e.g., odds ratios <1.5) while evaluating hundreds of thousands of SNPs (81). The large sample sizes and initial high cost of SNP arrays helped motivate the development and use of multistage GWA study designs (66). First, a subset of the study sample is genotyped in a discovery stage using the genome-wide SNP arrays. Then the most strongly associated SNPs are genotyped with a less-expensive genotyping platform in the remaining samples. This narrowing of the most promising SNPs can continue with additional replication stages, along with fine-mapping of associated regions.
The optimal division of samples across stages depends on a number of factors, but in general, the most efficient approach entails including approximately one-third to one-half of the samples in the initial stage and the remaining in the follow-up stages (57). How many of the most noteworthy SNPs should be subsequently tested depends on the sample sizes in the respective stages and how many false-negative results one is willing to accept. That said, the initial stages of GWAS may not clearly pinpoint SNPs that will be highly replicated by latter stages; therefore, as many SNPs as feasible should be carried over from one GWA study stage to the next (e.g., >1% of the first stage SNPs should be typed in the second stage).
One must also decide whether the early follow-up stages are considered part of a replication or a joint analysis. It seems most intuitive to use the follow-up data as a replication study, with the goal of confirming the initial findings (66). However, in light of the modest SNP associations and enormous multiple comparisons issue, to obtain sufficient power one is generally better off combining data from the first couple of stages in a joint analysis (57). One can view the joint analysis approach as a less expensive, single-stage GWA study: Fewer SNPs are typed in a second stage simply to reduce cost, but data from the first and second stages are combined, analyzed, and penalized for multiple comparisons as though a single-stage GWA study had been undertaken (28). Of course, even with a joint analysis of the first couple of stages, one must still replicate results with additional samples.
Note that decreasing SNP array costs have made multistage GWA study designs less essential. Genotyping 10,000–20,000 SNPs in a follow-up stage can cost just about as much as genotyping a genome-wide SNP array. Therefore, many of the most recent GWAS simply genotype all samples initially available with a SNP array. This practice also allows for simultaneous consideration of SNPs and other forms of genetic variation in one’s entire study sample (e.g., copy number variants). Nevertheless, new technologies for genotyping millions of SNPs or sequencing entire genomes may be sufficiently expensive, whereby multistage designs are again vital to genome-wide studies (discussed further below).
Subject Selection
Most GWAS select cases with a particular disease and compare them with unaffected controls. Of course, GWAS can also evaluate continuous traits, studying entire groups of subjects or selecting those at the extremes of the trait distribution to increase power for detecting associations (22). Whatever the phenotype, study subjects should be representative of their source population (76). In a case-control study, this requirement implies that controls be those individuals who, if diseased, would be cases, and controls are commonly selected to match the cases with respect to ethnicity, age, and sex.
Many GWAS, however, have been successful without overly rigorous control selection. In fact, owing to the high cost of subject recruitment and genotyping, there is a growing movement toward using existing genotype information among controls, who were recruited into previous studies and have been made available to researchers (37). The potential bias arising from using such convenience controls is tempered by the low measurement error in SNP genotyping, the lack of recall bias when studying inherited variants, the large sample sizes, the stringent criterion for statistical significance, and the rigorous replication of findings. In addition, if such controls result in population stratification bias—confounding of associations due to case-control differences in genetic ancestry (67)—this can be addressed analytically with genomic information (8, 50, 51).
Although most GWAS use unrelated controls, some use family members such as unaffected siblings or parents. Family-based designs directly control for population stratification and for some potential confounding by environmental exposures (i.e., those shared by family members). The increased sharing of genetic information among family members, however, can result in this design having substantially lower power for detecting main effects—although increased power for detecting gene-environment interactions—than studies of unrelated individuals would have (83).
Statistical Analysis
Once genotyping is complete, SNPs are subject to a number of quality-control checks— such as the proportion of samples successfully genotyped and testing for Hardy-Weinberg equilibrium—and those that fail are removed from further consideration. With SNP genotypes and external linkage disequilibrium information on the underlying structure among neighboring SNPs (e.g., from the HapMap project), one can impute some of the common untyped variants; this option allows for a more thorough and powerful evaluation of potential associations across the genome (23, 41, 42).
The relationship with disease is generally evaluated for each SNP using a trend test across the number of minor alleles. This allelic trend test provides relatively good properties, even if the true mode of inheritance is recessive or dominant. The analysis ofGWA study data can also test multimarker combinations of SNPs, haplotypes, or interactions for their association with disease (7, 47). Moreover, the statistical analysis can also leverage additional information about the SNPs, for example, whether they are part of a known pathway or are potentially functional (4).
To determine the overall statistical significance of GWA study results, one must address the issue of multiple comparisons arising from evaluating up to 1 million SNPs. The simplest approach is to use a Bonferroni correction, in which the conventional alpha level of p<0.05 is divided by the number of tests performed (e.g., 0.05/1,000,000 (5 × 10−8). This approach may be conservative because some assayed SNPs are correlated, owing to their linkage disequilibrium. But this conservatism is offset by the fact that the measured SNPs also represent unmeasured SNPs, so the effective number of independent tests in current GWAS is ~1 million (48). Some GWAS also calculate the false discovery rate (FDR) to assess the strength of associations (59, 65, 75). Note that while adhering to strict significance cut points is helpful to address issues of multiple comparisons, they are somewhat arbitrary and do not reflect the potential clinical or biological importance of an association (82).
GWA STUDY RESULTS
General Summary
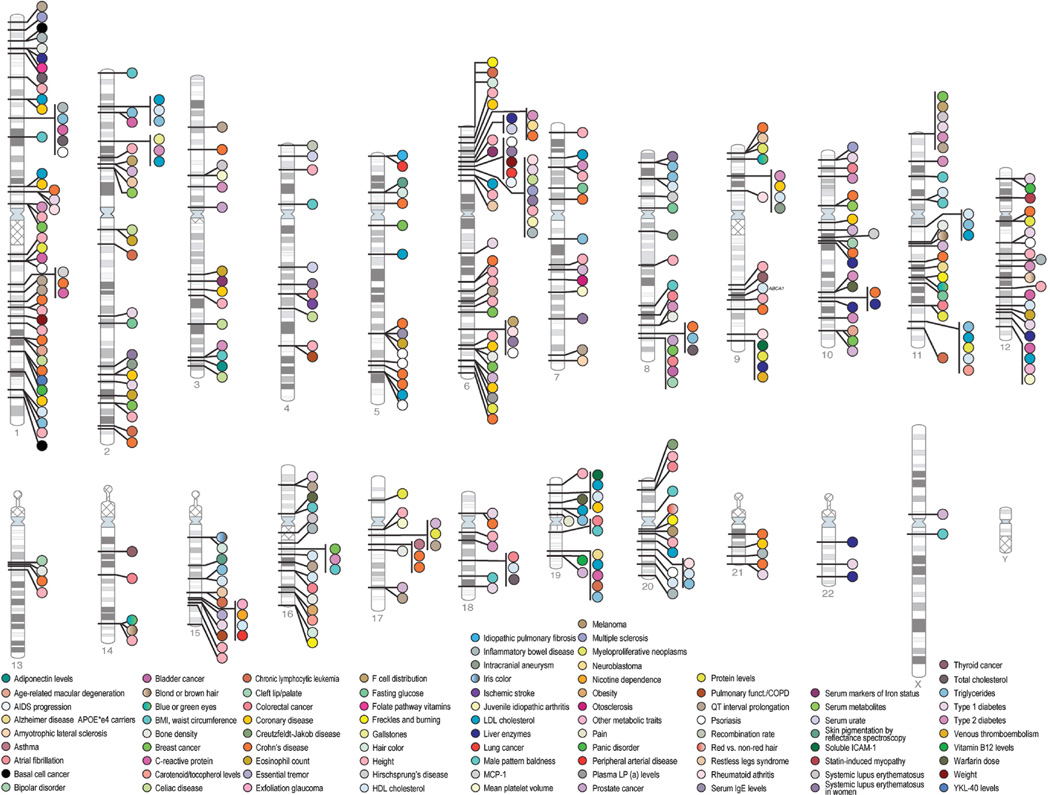
GWA study findings are collated and updated in the National Human Genome Research Institute’s “Catalog of Published Genome-Wide Association Studies” (http://www.genome.gov/gwastudies) (19). This catalog presents results from GWAS that evaluated at least 100,000 SNPs in the initial stage and gives details on associated SNPs with p-values <10−5. As of June 2009, the catalog includes more than 350 GWA study publications on more than 1600 distinct SNPs associated with more than 200 phenotypes. The chromosomal locations for many of these findings are highlighted in Figure 1.
Figure 1.
Chromosomal locations of genome-wide association (GWA) study results through March 2009. Results are given for 398 publications with p-values ≤ 5 × 10−8. Reproduced from the National Human Genome Research Institute’s GWAS Catalog: http://www.genome.gov/gwastudies. Credit: D. Leja and T. Manolio
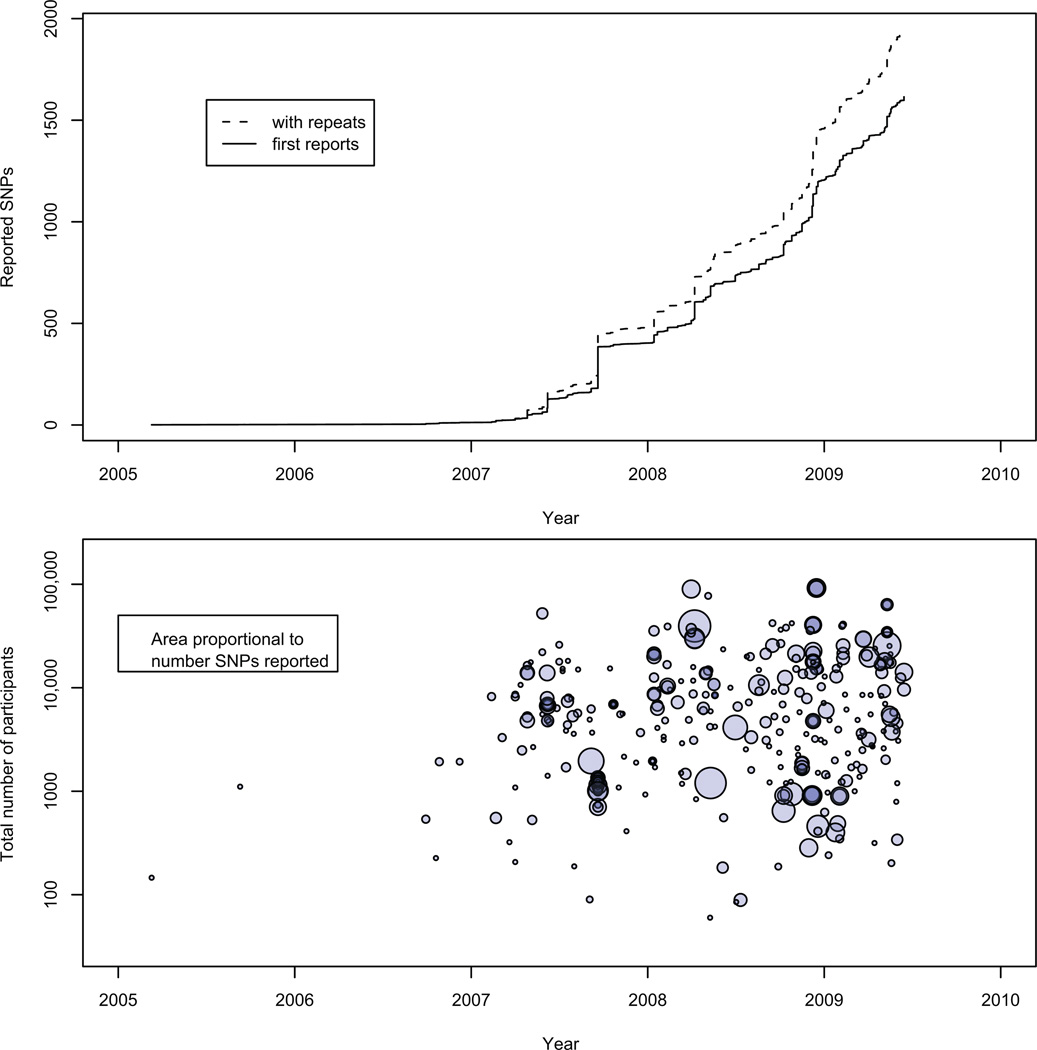
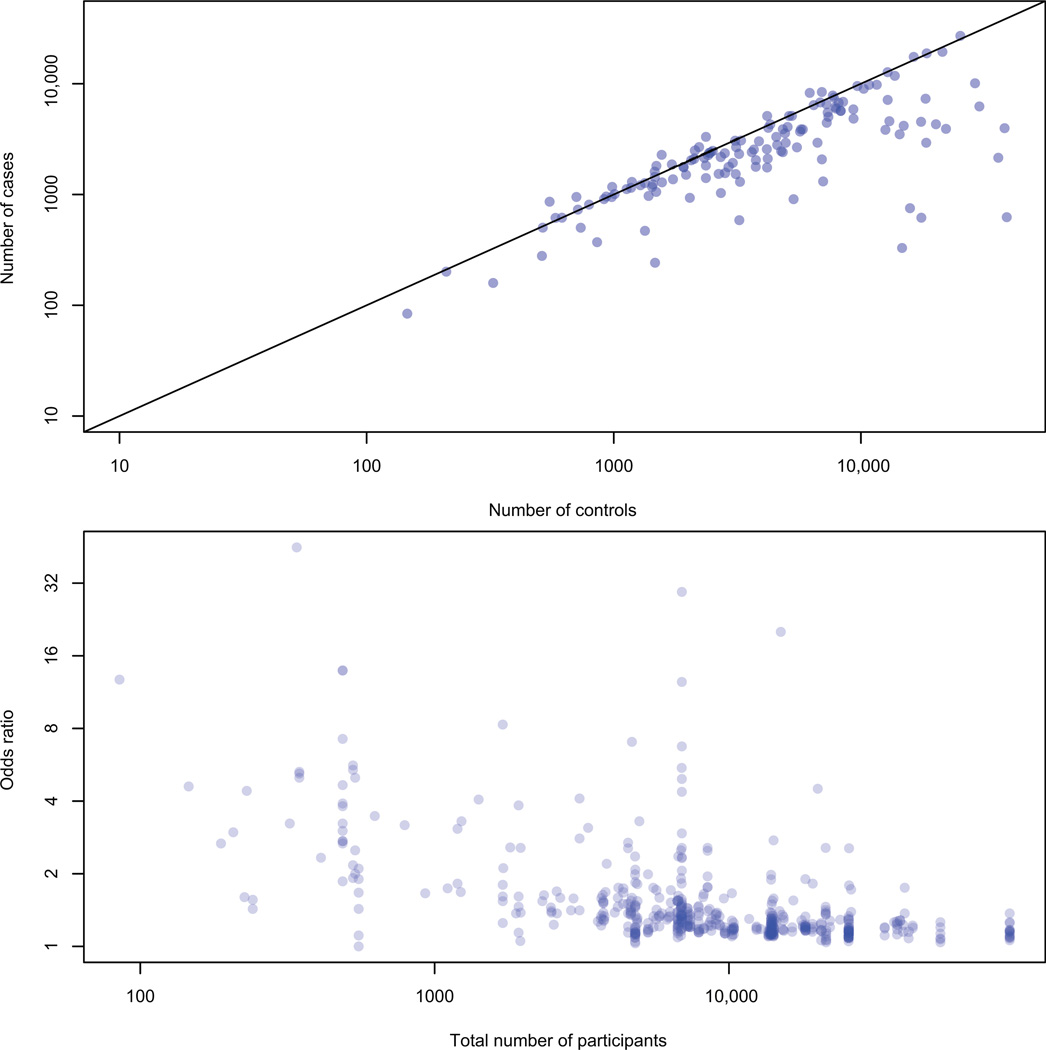
There are some interesting patterns in the first few years’ worth of GWA study results. As expected, common diseases are the most frequently studied byGWAS, including more than half a dozen publications on type 1 and 2 diabetes; prostate, breast, colorectal, and lung cancer; amyotrophic lateral sclerosis; cholesterol and triglyceride levels; Alzheimer’s disease; bipolar disorder and schizophrenia; Crohn’s disease; and rheumatoid arthritis (Figure 1) (19). Over the past few years, the cumulative number of associated SNPs detected by GWAS has exponentially increased; the number of statistically significant SNPs reported per GWA study and the sample size of the GWAS are also increasing with time (Figure 2). The GWAS have had samples sizes in the hundreds to tens of thousands of subjects; the initial stages had a median sample size of 1,752 individuals (interquartile range: 809 to 4763 people) and the follow-up stages had a median sample size of 3,671 (interquartile range: 1649 to 8968). Although some of the studies have similar numbers of cases and controls, many include a smaller number of cases than controls (Figure 3, top panel).
Figure 2.
Top panel: cumulative number of GWA study SNPs reported with p-values <10−5 over time. Bottom panel: GWA study findings by study sample size and number of SNPs per study over time. Each circle indicates a single publication, and the area of the circle reflects the number of associated SNPs in that study. From these plots we can see the rapid increase in GWA study SNPs and a slight trend toward larger studies and more noteworthy SNPs per study (19).
Figure 3.
Top panel: number of cases and controls in each published GWA study; many GWAS have fewer cases than controls. Bottom panel: total GWA study sample size (cases and controls) versus effect size for studies of binary traits; larger studies are better powered to detect smaller odds ratios (19).
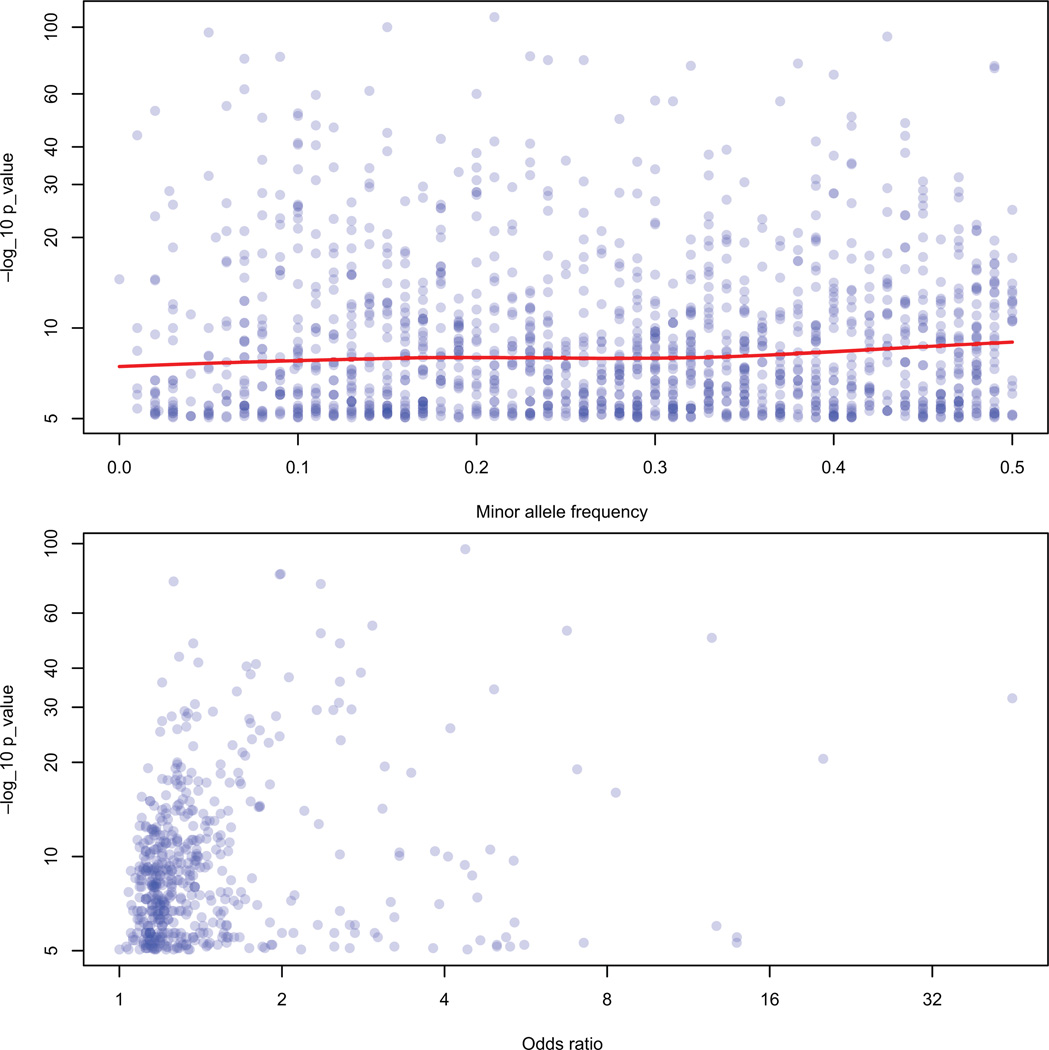
The effect sizes for the GWA study associations are generally quite modest: The median odds ratio = 1.28 (interquartile range = 1.17 to 1.55, for binary traits). There is an inverse relationship between sample size and effect sizes for the GWAS: Studies with larger sample sizes can detect smaller associations, which is expected because they have higher power than do smaller studies (Figure 3, bottom panel). Interestingly, the larger GWAS are unlikely to detect larger effect sizes; the ability of smaller GWAS to detect larger associations may reflect the winner’s curse or false-positive results (32). The minor allele frequencies of significant GWA study SNPs are relatively common (median = 0.28, interquartile range = 0.16 to 0.39)—again as expected on the basis of the design of GWAS and the measurement of common genetic variation by SNP arrays. The median p-value for associated SNPs is 1×10−7 (interquartile range = 3×10−6 to 9×10−12). The association p-values do not appear correlated with the corresponding SNP’s minor allele frequencies; the p-values do, however, appear correlated with the corresponding odds ratios (Figure 4).
Figure 4.
Top panel: GWA study p-values and associated SNP minor allele frequencies (MAF). The red line is a smoothed curve across these values and highlights that there is little impact of MAF on p-values for the strongest SNPs from GWAS. Bottom panel: GWA study p-values and corresponding odds ratios. There is a slight trend toward smaller odds ratios having smaller p-values (19).
About 70% of the GWAS-associated SNPs are in genes or genic regions, and many of the findings pertain to loci that have not been previously implicated in disease. To date, GWA study findings appear to be overrepresented in genes involved with cell adhesion, signal transduction, transport activity, and protein phosphorylation (25). Finally, it is worth noting that the SNP array platforms used in the GWAS to date are closely split between those offered by Illumina and Affymetrix (19).
Specific Examples
There are far too many GWAS to discuss each in much detail here. Therefore, we highlight results from three large and highly successful projects: The Wellcome Trust Case Control Consortium (WTCCC) (79), the de-CODE/Icelandic studies (15), and the Cancer Genetic Markers of Susceptibility (CGEMS) GWAS of prostate cancer (68, 85).
The WTCCC encompassed GWAS of seven major diseases: bipolar disorder, coronary artery disease, Crohn’s disease, hypertension, rheumatoid arthritis, type 1 diabetes, and type 2 diabetes (79). The initial stage included 14,000 affected individuals total from the United Kingdom, 2000 with each disease. For comparison, two sets of control groups, each containing 1500 individuals, were used: one from the 1958 British Birth Cohort and one from the U.K. National Blood Service (79). All study subjects were genotyped using the Affymetrix GeneChip 500K array. Analyses of the resulting data and additional follow-up work detected and replicated many promising SNP-disease associations, for example, a novel association between FTO and type 2 diabetes and associations for coronary artery disease on chromosome 9 (79). The importance of this project is highlighted by the fact that more than 700 other papers have cited the original publication as of June 2009.
The deCODE GWAS arise from studies that include more than 40,000 individuals from Iceland (15). This infrastructure allows for swiftly studying any phenotype for which de-CODE has access to a sufficiently large sample size. The deCODE GWAS have looked at a large number of phenotypes using nested case-control studies in Iceland with overlapping control groups and collaborative follow-up and replication studies on subjects from outside of Iceland. They have detected GWA study associations for many different phenotypes, including cancer (13, 14, 30, 58, 69); heart disease (18); obesity (71); glaucoma (70); and traits such as age at menarche (61), bone mineral density (60), and pigmentation, hair, and eye color (60). These successes illustrate the value of establishing large, well-characterized populations for evaluating many different diseases.
CGEMS is a multistage GWAS of prostate and breast cancer (http://cgems.cancer.gov). Focusing on the prostate cancer study, 1172 cases and 1157 controls of European-American ancestry were selected from the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial and genotyped using the Illumina 550K array (68, 85). Almost 27,000 of the most strongly associated SNPs were followed up in a second stage comprised of 4 other study populations with a total of 3941 cases and 3964 controls. Noteworthy findings from the CGEMS of prostate cancer included detecting associated SNPs in distinct loci on chromosome 8q24 and a risk SNP in the MSMB gene, which encodes beta-microseminoprotein, a primary constituent of semen (68, 85). Interestingly, the strongly associated SNP in MSMB (rs10993994) had only the 24,223rd smallest p-value in the initial stage (68), which illustrates that replicated SNPs may not initially have exceptionally small p-values and that it is important to follow up a large number of SNPs in the latter stages of GWAS.
The WTCCC and deCODE studies exhibited that GWAS can successfully use shared controls that are well matched on ethnicity for comparison with multiple phenotypes, even though they may not be fully representative of the cases’ source population. Nevertheless, recruiting controls in the same manner as cases—including obtaining detailed nongenetic information—allows investigators to evaluate gene-environment interactions appropriately; of course, one might simply decide to evaluate such associations with a case-only study design (64). The WTCCCGWAS also found that processing arrays at different laboratories can result in false-positive findings (5).
Researchers can request use of the WTCCC and CGEMS GWA study data. Sharing GWA study data is vital because it allows others to replicate findings, combine data, and examine phenotypic clustering with particular genetic variants, maximizing the use of this valuable information. To this end, National Institutes of Health grantees undertaking GWAS are required to develop a data-sharing plan and to deposit their data for use by other scientists (NIH Notice:NOT-OD-08–013). Many GWAS thus far have deposited data in dbGaP for use by the scientific community (40).
IMPLICATIONS OF GWA STUDY FINDINGS
New Insights
The highly replicated results from GWAS can help clarify the biological basis of disease, providing information about the mechanisms underlying the disease process. If multiple confirmed SNPs arise from a particular biological pathway, this implies that something unique to that pathway may help drive the etiology of disease. Similarly, if multiple diseases are associated with a particular locus, this suggests a common genetic basis for such diseases (56). For example, the SNPs in the chromosome 8q24 loci are associated with cancer of the prostate, breast, and colon, indicating that this locus is acutely involved with the carcinogenic process (9, 13, 72, 84, 86). In fact, more recent mechanistic work shows that the 8q24 prostate and colorectal cancer locus containing the risk SNP rs6983267 is in a transcriptional enhancer and interacts with the proto-oncogene MYC (49, 73).
From an epidemiologic perspective, GWA study SNPs may give relatively large estimates of the population-attributable fraction (e.g., 40% for multiple prostate cancer SNPs). These estimates reflect the fairly high-risk allele frequencies for these SNPs, which are commonly between 0.15 and 0.40 (Figure 4). Such population-attributable fraction estimates imply that the associated SNPs account for a sizeable proportion of disease, although these calculations make a number of assumptions and may overestimate or underestimate the true population-attributable fraction (43).
Genetic Testing
Another implication of GWA study results is the rapidly escalating availability of genetic testing. Currently available tests range from those prescribed by a physician for high-penetrance disease genes to those marketed direct-to-consumer for measuring variants that span the entire genome (29).With directto- consumer tests, individuals pay to have variation in their genomes assayed with the same SNP arrays used in GWAS. Results from these assays are returned to the consumer along with varying levels of additional information (e.g.,Web-based interpretation of some results, genetic counseling).
Some of the tests include information about the genetic basis of drug response, which is now being studied using GWAS (54). Pharmacogenomics has important near-term implications for which drug and dose an individual should receive. For example, GWAS confirmed that variants in the gene CYP2C9 impact metabolism of the anticoagulant drug warfarin (63); such findings suggest that individuals who carry the CYP2C9 variant that results in slow metabolism could be prescribed lower doses of warfarin, reducing potential side effects of severe bleeding and unnecessary health-care costs. In light of such results, personalized medicine is commonly touted as a major potential benefit of pharmacogenomics and GWAS, albeit with some reservations (46).
Although most individualGWA study SNPs are not effective for genetic testing owing to their modest associations with disease and low penetrance, one might consider genetic tests based on combinations of associated SNPs. For example, when looking at the distribution of five associated SNPs for prostate cancer, men in the top decile of risk alleles carried have an approximate two- to fourfold increase in risk in comparison to men in the lowest decile (31, 68, 87). Based on this increased risk, some investigators advocate a multiple-SNP screening test for prostate cancer (87), although there are some serious limitations with such tests (discussed below).
LIMITATIONS OF GWAS
Not Causal
Although many GWA study results have been highly replicated, quite a few of the variants are only associated with, not causal for, disease. Determining the causal factors underlying GWA study results can be extremely challenging, requiring fine mapping and mechanistic studies—which are underway for many findings (24). This is further complicated by the fact that ~30% of the associations detected to date are not even in gene regions (19). These issues, of course, limit our ability to understand the biological basis of GWA study results and to implement preventive or therapeutic measures.
Little Heritability
GWA study findings often account for only a limited amount of disease heritability (39), which in part reflects the small magnitude of effect for most SNPs detected by GWAS. Even if large effects are found (e.g., for combinations of SNPs), these SNPs may not have high penetrance and so do not confer a high risk of disease. This dark matter of unexplained heritability has raised concerns about the ultimate value of GWAS (12).
The original hypothesis motivating GWAS is that common diseases may be caused, in part, by common genetic variants (53). Thus, GWAS were designed to detect associations between disease and common SNPs (e.g., minor allele frequency >5%). The current SNP arrays measure variants primarily at or above this frequency and may even miss some common variation (26). Common diseases, however, are undoubtedly also due to rare variants. These variants can act alone or reflect allelic heterogeneity, whereby many different rare alleles within a particular locus each increase risk. Therefore, the inability of existing GWAS to evaluate rare variants may help explain why they account for little heritability.
Another possible explanation is that most GWAS have evaluated genetic variation due only to SNPs. Although SNPs comprise the most common form of genetic variation, copy number variants (CNVs) also give rise to substantial variability and account for some heritability of disease (20, 44, 52). GWAS are now evaluating CNVs, and CNV probes are being incorporated into the most current GWA study arrays. In addition, it has been difficult for investigators to detect gene-gene and gene-environment interactions by GWAS because this practice requires extremely large sample sizes and well-characterized environmental exposures (64). As sample sizes increase, GWAS will also be able to detect additional SNP associations that have even smaller effect sizes than those observed to date.
Not Very Predictive
Another important limitation of GWA study results—which is especially pertinent in light of the growing direct-to-consumer tests—is that they may not sufficiently distinguish between individuals with low and high risk of disease. For example, the five-SNP test noted above for prostate cancer provides only a slight increase in the area under the receiver operating characteristic curve (AUC) for classifying cases and controls (0.61 to 0.63) (87). In general, screening tests based on most of the GWA study SNPs detected to date will likely have low positive (and negative) predictive value for disease and have limited usefulness in a diagnostic setting (33, 78). Adding more GWA study SNPs with modest disease associations may not much improve the discriminatory ability of such tests. Moreover, few individuals will carry large numbers of GWA study risk alleles, so screening for these in the general population would not be cost-effective. Note also that justification for genetic testing also depends on the existence of effective interventions.
NEXT-GENERATION GWAS
Although the successes ofGWAS are tempered by their limitations, they do provide an important advance in our efforts toward deciphering the genetic basis of disease (1).GWAS highlight the value of agnostic approaches to the search for disease genes. Taking a broad genome-wide view is essential for achieving a more complete understanding of the genetic architecture of complex phenotypes. GWAS also emphasize the significance of undertaking complementary replication and validation studies across multiple populations (3).
Continued scientific and technological advances will allow investigators to study less common and different sources of genetic variation. Results from the 1000 Genomes project (http://www.1000genomes.org) can be used to assay less common SNPs with more sizeable genotyping platforms (e.g., 10 million SNPs). Sequencing technologies are rapidly decreasing in cost, and genome-wide sequence studies will eventually become feasible. Before sequencing all study subjects, future work may use a sequence/genotype hybrid design in which the initial phase sequences a subset of subjects, and based on these results, large-scale genotyping will be undertaken on the remaining study subjects.
As data on less common variants become available, and interest in detecting interactions and pathway effects on disease grows, there will be an increasing need for more complex statistical analysis tools. Methods that maximize the strengths of both agnostic genome-wide and knowledge-based biological approaches may help clarify potential associations such as explicitly incorporating into analysis additional existing information about the properties of genetic variants (4, 27). New statistical methods for evaluating rare variants will also be crucial as such data are generated on a wider scale (38).
The next generation of genome-wide studies will further improve our understanding of the disease process, risks, and response to therapy. The impact of these studies on a person’s and the public’s health will vary substantially. In some cases, the information will have little actionable value, and any corresponding genetic tests could ultimately increase health care costs by prompting individuals to obtain unnecessary medical care. In other situations, the knowledge gained will be extremely valuable and provide great benefit; hopefully this will encompass a large majority of genome-wide findings.
ACKNOWLEDGMENTS
I thank Drs. Iona Cheng and Inga Hallgrimsdottir for helpful comments on this manuscript, and Joel Mefford for creating Figures 2–4. This work was supported by grants R01 CA88164 and U01 CA127298 from the National Institutes of Health.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Altshuler D, Daly MJ, Lander ES. Genetic Mapping in Human Disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- 3.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 4.Chen GK, Witte JS. Enriching the analysis of genomewide association studies with hierarchical modeling. Am. J. Hum. Genet. 2007;81:397–404. doi: 10.1086/519794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat. Genet. 2005;37:1243–1246. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- 6.Daly MJ, Rioux JD, Schaffner SF, Hudson TJ, Lander ES. High-resolution haplotype structure in the human genome. Nat. Genet. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 7.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat. Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 8.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 9.Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447:1087–1093. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein DB. Common genetic variation and human traits. N. Engl. J. Med. 2009;360:1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 13.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat. Genet. 2007;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 14.Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat. Genet. 2007;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 15.Gulcher J, Stefansson K. Population genomics: laying the groundwork for genetic disease modeling and targeting. Clin. Chem. Lab. Med. 1998;36:523–527. doi: 10.1515/CCLM.1998.089. [DOI] [PubMed] [Google Scholar]
- 16.HapMap. The international hapmap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 17.HapMap. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Sci. N.Y. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 19.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinds DA, Kloek AP, Jen M, Chen X, Frazer KA. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nat. Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 21.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet. Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Huang BE, Lin DY. Efficient association mapping of quantitative trait loci with selective genotyping. Am. J. Hum. Genet. 2007;80:567–576. doi: 10.1086/512727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang L, Li Y, Singleton AB, Hardy JA, Abecasis G, et al. Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ioannidis JP, Thomas G, Daly MJ. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 2009;10(5):318–329. doi: 10.1038/nrg2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson AD, O’Donnell CJ. An open access database of genome-wide association results. BMC Med. Genet. 2009;10:6. doi: 10.1186/1471-2350-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jorgenson E, Witte JS. Coverage and power in genomewide association studies. Am. J. Hum. Genet. 2006;78:884–888. doi: 10.1086/503751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgenson E, Witte JS. A gene-centric approach to genome-wide association studies. Nat. Rev. Genet. 2006;7:885–891. doi: 10.1038/nrg1962. [DOI] [PubMed] [Google Scholar]
- 28.Jorgenson E, Witte JS. Genome-wide association studies of cancer. Fut. Oncol. 2007;3:419–427. doi: 10.2217/14796694.3.4.419. [DOI] [PubMed] [Google Scholar]
- 29.Kaye J. The regulation of direct-to-consumer genetic tests. Hum. Mol. Genet. 2008;17:R180–R183. doi: 10.1093/hmg/ddn253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiemeney LA, Thorlacius S, Sulem P, Geller F, Aben KK, et al. Sequence variant on 8q24 confers susceptibility to urinary bladder cancer. Nat. Genet. 2008;40:1307–1312. doi: 10.1038/ng.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kote-Jarai Z, Easton DF, Stanford JL, Ostrander EA, Schleutker J, et al. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol. Biomarkers Prev. 2008;17:2052–2061. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kraft P. Curses—winner’s and otherwise—in genetic epidemiology. Epidemiology. 2008;19:649–651. doi: 10.1097/EDE.0b013e318181b865. discussion 57–58. [DOI] [PubMed] [Google Scholar]
- 33.Kraft P, Wacholder S, Cornelis MC, Hu FB, Hayes RB, et al. Beyond odds ratios—communicating disease risk based on genetic profiles. Nat. Rev. Genet. 2009;10:264–269. doi: 10.1038/nrg2516. [DOI] [PubMed] [Google Scholar]
- 34.Lander ES. The new genomics: global views of biology. Science. 1996;274:536–539. doi: 10.1126/science.274.5287.536. [DOI] [PubMed] [Google Scholar]
- 35.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 36.Lohmueller KE, Pearce CL, Pike M, Lander ES, Hirschhorn JN. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33:177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- 37.Luca D, Ringquist S, Klei L, Lee AB, Gieger C, et al. On the use of general control samples for genome-wide association studies: Genetic matching highlights causal variants. Am. J. Hum. Genet. 2008;82:453–463. doi: 10.1016/j.ajhg.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maher B. Personal genomes: the case of the missing heritability. Nature. 2008;456:18–21. doi: 10.1038/456018a. [DOI] [PubMed] [Google Scholar]
- 40.Manolio TA. Collaborative genome-wide association studies of diverse diseases: programs of the NHGRI’s office of population genomics. Pharmacogenomics. 2009;10:235–241. doi: 10.2217/14622416.10.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchini J, Howie B. Comparing algorithms for genotype imputation. Am. J. Hum. Genet. 2008;83:535–539. doi: 10.1016/j.ajhg.2008.09.007. author reply 539–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 43.Mason CA, Tu S. Partitioning the population attributable fraction for a sequential chain of effects. Epidemiol. Perspect. Innov. 2008;5:5. doi: 10.1186/1742-5573-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, et al. Common deletion polymorphisms in the human genome. Nat. Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 45.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 46.Nebert DW, Zhang G, Vesell ES. From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab. Rev. 2008;40:187–224. doi: 10.1080/03602530801952864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pe’er I, de Bakker PI, Maller J, Yelensky R, Altshuler D, Daly MJ. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nat. Genet. 2006;38:663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 48.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet. Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 49.Pomerantz MM, Ahmadiyeh N, Jia L, Herman P, Verzi MP, et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. 2009;41(8):882–884. doi: 10.1038/ng.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price AL, Patterson NJ, Plenge RM,Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 51.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am. J. Hum. Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 54.Rieder MJ, Livingston RJ, Stanaway IB, Nickerson DA. The environmental genome project: reference polymorphisms for drug metabolism genes and genome-wide association studies. Drug Metab. Rev. 2008;40:241–261. doi: 10.1080/03602530801952880. [DOI] [PubMed] [Google Scholar]
- 55.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 56.Rzhetsky A, Wajngurt D, Park N, Zheng T. Probing genetic overlap among complex human phenotypes. Proc. Natl. Acad. Sci. USA. 2007;104:11694–11699. doi: 10.1073/pnas.0704820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 58.Stacey SN, Manolescu A, Sulem P, Thorlacius S, Gudjonsson SA, et al. Common variants on chromosome 5p12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2008;40:703–706. doi: 10.1038/ng.131. [DOI] [PubMed] [Google Scholar]
- 59.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Styrkarsdottir U, Halldorsson BV, Gretarsdottir S, Gudbjartsson DF, Walters1 GB, et al. Multiple genetic loci for bone mineral density and fractures. N. Engl. J. Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 61.Sulem P, Gudbjartsson DF, Rafnar T, Holm H, Olafsdottir EJ, et al. Genome-wide association study identifies sequence variants on 6q21 associated with age at menarche. Nat. Genet. 2009;41:734–738. doi: 10.1038/ng.383. [DOI] [PubMed] [Google Scholar]
- 62.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007;39:1443–1452. doi: 10.1038/ng.2007.13. [DOI] [PubMed] [Google Scholar]
- 63.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thomas D. Methods for investigating gene-environment interactions in candidate pathway and genome-wide association studies. Annu. Rev. Public Health. 2010;31:21–36. doi: 10.1146/annurev.publhealth.012809.103619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas DC, Clayton DG. Betting odds and genetic associations. J. Natl. Cancer Inst. 2004;96:421–423. doi: 10.1093/jnci/djh094. [DOI] [PubMed] [Google Scholar]
- 66.Thomas DC, Haile RW, Duggan D. Recent developments in genomewide association scans: a workshop summary and review. Am. J. Hum. Genet. 2005;77:337–345. doi: 10.1086/432962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas DC, Witte JS. Point: population stratification: a problem for case-control studies of candidate-gene associations? Cancer Epidemiol. Biomarkers Prev. 2002;11:505–512. [PubMed] [Google Scholar]
- 68.Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, et al. Multiple loci identified in a genomewide association study of prostate cancer. Nat. Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 69.Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature. 2008;452:638–642. doi: 10.1038/nature06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thorleifsson G, Magnusson KP, Sulem P, Walters GB, Gudbjartsson DF, et al. Common sequence variants in the LOXL1 gene confer susceptibility to exfoliation glaucoma. Science. 2007;317:1397–1400. doi: 10.1126/science.1146554. [DOI] [PubMed] [Google Scholar]
- 71.Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 72.Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007;39:984–988. doi: 10.1038/ng2085. [DOI] [PubMed] [Google Scholar]
- 73.Tuupanen S, Turunen M, Lehtonen R, Hallikas O, Vanharanta S, et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 2009;41(8):885–890. doi: 10.1038/ng.406. [DOI] [PubMed] [Google Scholar]
- 74.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 75.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wacholder S, McLaughlin JK, Silverman DT, Mandel JS. Selection of controls in case-control studies. I. Principles. Am. J. Epidemiol. 1992;135:1019–1028. doi: 10.1093/oxfordjournals.aje.a116396. [DOI] [PubMed] [Google Scholar]
- 77.Wang DG, Fan JB, Siao CJ, Berno A, Young P, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–1082. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 78.Ware JH. The limitations of risk factors as prognostic tools. N. Engl. J. Med. 2006;355:2615–2617. doi: 10.1056/NEJMp068249. [DOI] [PubMed] [Google Scholar]
- 79.Wellcome Trust Case Control Consort. (WTCCC) Genome-wide association study of 14000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Witte JS. Prostate cancer genomics: towards a new understanding. Nat. Rev. Genet. 2009;10:77–82. doi: 10.1038/nrg2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Witte JS, Elston RC, Cardon LR. On the relative sample size required for multiple comparisons. Stat. Med. 2000;19:369–372. doi: 10.1002/(sici)1097-0258(20000215)19:3<369::aid-sim335>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 82.Witte JS, Elston RC, Schork NJ. Genetic dissection of complex traits. Nat. Genet. 1996;12:355–356. doi: 10.1038/ng0496-355. author reply 357–358. [DOI] [PubMed] [Google Scholar]
- 83.Witte JS, Gauderman WJ, Thomas DC. Asymptotic bias and efficiency in case-control studies of candidate genes and gene-environment interactions: basic family designs. Am. J. Epidemiol. 1999;149:693–705. doi: 10.1093/oxfordjournals.aje.a009877. [DOI] [PubMed] [Google Scholar]
- 84.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 85.Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007;39:870–874. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 86.Zanke BW, Greenwood CM, Rangrej J, Kustra R, Tenesa A, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2007;39:989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 87.Zheng SL, Sun J, Wiklund F, Smith S, Stattin P, et al. Cumulative association of five genetic variants with prostate cancer. N. Engl. J. Med. 2008;358:910–919. doi: 10.1056/NEJMoa075819. [DOI] [PubMed] [Google Scholar]