Abstract
The following mini review summarizes recent research from the Author’s laboratory as presented to the Environmental Mutagen Society in October 2012. It provides an overview of the DNA glycosylases that recognize oxidized DNA bases using the Fpg/Nei family of DNA glycosylases as models for how structure can inform function. For example, even though human NEIL1 and the plant and fungal orthologs lack the zinc finger shown to be required for binding, DNA crystal structures revealed a “zincless finger” with the same properties. Also the “lesion recognition loop” is not involved in lesion recognition rather stabilization of 8-oxoG in the active site pocket. Unlike the other Fpg/Nei family members, Neil3 lacks two of the three void-filling residues that stabilize the duplex and interact with the opposite strand which may account for its preference for lesions in single stranded DNA. We also showed, using single molecule approaches, that DNA glycosylases search for their substrates in a sea of undamaged DNA by using a wedge residue that is inserted into the DNA helix to probe for the presence of damage.
Keywords: DNA base damage, DNA glycosylase, oxidative DNA damage, Fpg/Nei family of DNA glycosylases, Single molecule studies, Glycosylase search, Glycosylase substrate specificity, Glycosylase crystal structures, Eukaryotic Neil glycosylases, Neil3
Oxidation of DNA Bases
Reactive oxygen species (ROS) cause a broad spectrum of damage to DNA bases and as well produce strand breaks and sites of base loss (for reviews see(Duclos et al. 2012b; Halliwell and Gutteridge 2002; Jezek and Hlavata 2005)). The most common source of ROS is the mitochondria, where about 1% of the oxygen intake results in the production of the superoxide anion radical O2•−. This radical can either be hydrated to form the highly reactive hydroxyl radical, •OH, or be converted by superoxide dismutase to hydrogen peroxide. Hydrogen peroxide can also produce hydroxyl radicals in a Fenton reaction which occurs in the presence of transition metals. Superoxide anion radical can react with •NO to produce peroxynitrite and in the presence of other reactants can also lead to the formation of hydroxyl radicals. Hydroxyl radicals themselves are highly reactive and generally interact with the molecule closest to them after their formation. In contrast to hydroxyl radicals, hydrogen peroxide and •NO can travel long distances throughout the cell. Low levels of hydrogen peroxide and other ROS are used in a variety of cell signaling processes (for reviews see (Azad et al. 2010; Funato and Miki 2010; Hamanaka and Chandel 2010; Kar et al. 2010; Morgan and Liu 2011; Pendyala and Natarajan 2010)), therefore, a steady state level must be maintained by enzymes such as glutathione reductase and catalase.
Cytochrome P450-dependent reactions produce ROS by peroxisomes, macrophages, neutrophils, endothelial cells and lipoperoxidation. Higher levels of ROS are also formed by a number of drugs and xenobiotics, ultraviolet light and ionizing radiation and by any agents that induce inflammation. Ionizing radiation is the most well-studied source of hydroxyl radical production which occurs during the radiolysis of water (von Sonntag 1987).
The structures of lesions produced by ROS were determined in the middle of the last century by radiation chemists who analyzed the damaged bases resulting from irradiation of DNA in vitro (for reviews see (Breen and Murphy 1995; Dizdaroglu 1992; Dizdaroglu et al. 2002). The major pyrimidine products come from the C5-OH− adduct radical with the thymine radical leading to thymine glycol (Tg) and the cytosine radical leading to cytosine glycol. Cytosine glycol is unstable and deaminates to uracil glycol which can then dehydrate to 5-hydroxyuracil (5-OHU) or cytosine glycol can directly dehydrate to 5-hydroxycytosine (5-OHC) (Dizdaroglu et al. 1986). The major purine products are derived from the C5-OH− adduct radicals of guanine and adenine (for reviews see (Breen and Murphy 1995; Dizdaroglu 1992; Dizdaroglu 1998; Steenken 1989)) and, depending on whether the conditions are oxidizing or reducing, lead to 8-oxoguanine (8-oxoG) and 8-oxoadenine or 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and 4,6-diamino-5-formamidopyrimidine (FapyA). Guanine has the lowest redox potential of the four bases and readily oxidizes to 8-oxoG which is probably the most highly studied base lesion although FapyG is likely to be at least as important (Liu et al. 2010). 8-oxoG also turns out to have a lower redox potential than any of the four normal bases and can be further oxidized to a number of products, the most studied are spiroiminodihydantoin (Sp) and guanidinohydantoin (Gh) (for reviews see (Burrows 2009; Neeley and Essigmann 2006)). 2-hydroxyadenine can also be formed by hydroxyl radical attack at the C2 position of adenine (Lesiak and Wheeler 1990)
Oxidized DNA base lesions are recognized in cells by enzymes called DNA glycosylases which are the first enzymes in the base excision repair pathway, a repair pathway that is highly conserved from bacteria to humans (Duclos et al. 2012b). Although some base lesions are benign, most oxidized DNA base lesions if left unrepaired can be lethal and/or mutagenic (Duclos et al. 2012b) and thus initiate carcinogenesis (for a review see (Wallace et al. 2012)). Over the years our laboratory has been asking how oxidized DNA bases are repaired with an emphasis on glycosylase structures and substrate preferences (for a review see (Prakash et al. 2012)) including how glycosylases see lesions in nucleosomes (for a review see (Odell et al. 2013)). More recently we have been examining how glycosylases find their substrates in undamaged DNA (Dunn et al. 2011). We have also spent time attempting to elucidate what happens when a polymerase encounters an unrepaired lesion. This has led in several directions including structural studies of polymerase interactions with unrepaired lesions (for reviews see (Hogg et al. 2005; Zahn et al. 2011)) as well as defining the consequences of individual lesions in terms of potential lethality and/or mutagenesis (for a review see (Wallace 2002)). In this review I will focus on how structure provides insights into function using the Fpg/Nei family of DNA glycosylases as an example and as well how the DNA glycosylases search for their substrates in DNA.
The Base Excision DNA Repair Pathway and the DNA Glycosylases that Recognize Oxidized DNA Bases
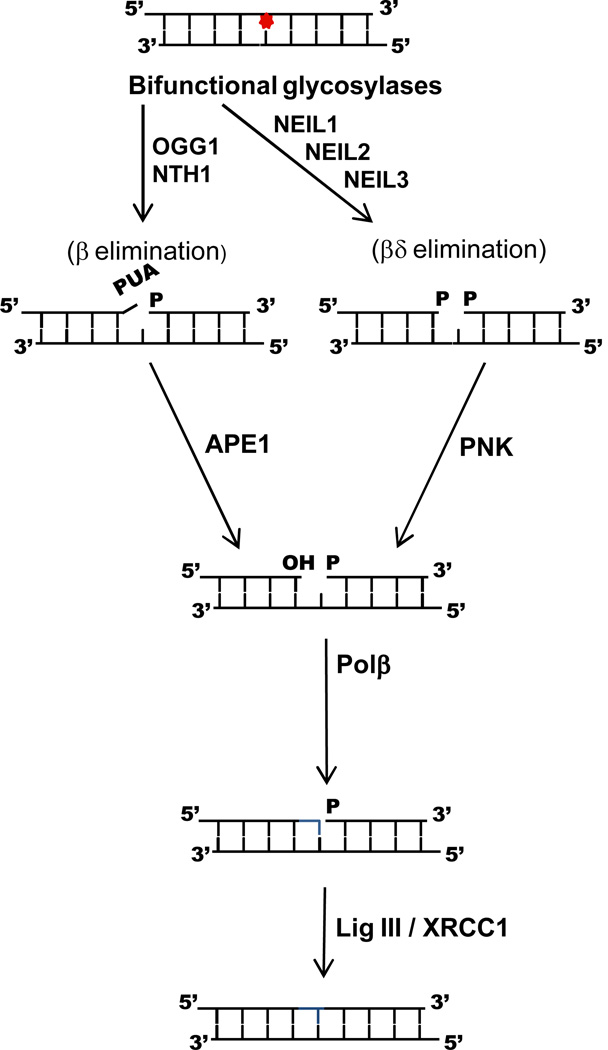
The vast majority of endogenous DNA damages, including oxidative DNA damages, are repaired by the base excision repair (BER); a schematic of short patch BER initiated by the glycosylases that remove oxidized bases is shown in Figure 1 (for reviews see (Barnes and Lindahl 2004; Duclos et al. 2012b; Fromme and Verdine 2004; Izumi et al. 2003; Krokan et al. 2000; Mitra et al. 2002)). There are five distinct enzymatic steps in this pathway, the first of which is catalyzed by a DNA glycosylase. DNA glycosylases cleave the N-glycosyl bond, releasing the base and leaving an abasic site in the DNA. In the second step of the reaction, an AP endonuclease, APE1, creates a nick in the DNA 5’ of the AP site resulting in 3’-hydroxyl and 5’-deoxyribosephosphate termini. In this case, removal of the 5’ dRP block is carried out by the intrinsic lyase activity of the next enzyme in the pathway, DNA polymerase β. However, all of the glycosylases that remove oxidized bases also cleave the DNA backbone on the 3’ side of the abasic site via a lyase reaction. The 3’ blocking sugar that results from β-elimination can be removed by the phosphodiesterase activity of APE1; if the glycosylase utilizes a β,δ-elimination mode, the phosphate group is removed by polynucleotide kinase PNK. To facilitate hand-off to the next enzyme in the pathway and reduce the accumulation of toxic intermediates, the eukaryotic enzymes tend to bind to their products. In the case of the bifunctional glycosylases, this can result in cleavage of the AP site by APE instead of by the intrinsic lyase activity of the glycosylase. In all scenarios, Pol β fills in the gap with the complementary nucleotide which creates the appropriate substrate for DNA ligase III to seal the nick. The DNA glycosylases that recognize oxidized DNA bases and their substrates are summarized in Table 1.
Figure 1.
Schematic of the short patch base excision repair process initiated by the bifunctional mammalian DNA glycosylases that recognize oxidative DNA lesions.
Table 1.
The DNA Glycosylases the Recognize Oxidized DNA Bases
| DNA Glycosylases | Lesions Recognized | Proposed Cellular Role |
|---|---|---|
| Endonuclease III, Nth | Oxidized pyrimidines, formamidopyrimidines | Housekeeping glycosylases for oxidized pyrimidines |
| hNTH1 | ||
| Endonuclease VIII, Nei | Oxidized pyrimidines, formamidopyrimidines | Appears to be housekeeping glycosylase in E. coli |
| NEIL1 | Recognizes lesions in duplex DNA and single-stranded DNA and appears to act as a “cowcatcher” to remove lesions in front of the replication fork (Dou et al. 2008; Hegde et al. 2013) | |
| NEIL2 | Prefers lesions in single-stranded DNA and data suggest that it repairs lesions during transcription (Chang and Lu 2002) | |
| NEIL3 | Prefers lesions in single-stranded DNA, telomere sequences, and G4 quadruplex structures and is found in proliferating cells (Liu et al. 2010; Liu et al. 2013; Zhou et al. 2013) | |
| Formamidopyrimidine DNA glycosylase, Fpg (MutM) | 8-oxoG formamidopyrimidines | Housekeeping glycosylases for oxidized purines |
| hOGG1 | ||
| MutY | A opposite 8-oxoG A opposite FapyG |
Acts post-replication and removes misincorporated A from the newly synthesized strand and in eukaryotes is associated with PCNA (Chang and Lu 2002; Parker and Eshleman 2003) |
| MUTYH |
References
Chang DY, Lu AL. 2002. Functional interaction of MutY homolog with proliferating cell nuclear antigen in fission yeast, Schizosaccharomyces pombe. J Biol Chem 277(14):11853-8.
Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. 2008. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem 283(6):3130-40.
Hegde ML, Hegde PM, Bellot LJ, Mandal SM, Hazra TK, Li GM, Boldogh I, Tomkinson AE, Mitra S. 2013. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc Natl Acad Sci U S A 110(33):E3090-9.
Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. 2010. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A 107(11):4925-30.
Liu M, Doublie S, Wallace SS. 2013. Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat Res 743-744:4-11.
Parker AR, Eshleman JR. 2003. Human MutY: gene structure, protein functions and interactions, and role in carcinogenesis. Cell Mol Life Sci 60(10):2064-83.
Zhou J, Liu M, Fleming AM, Bujrrows CJ, Wallace SS. 2013. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem:in press.
Our laboratory became interested in base excision repair many decades ago, when a postdoctoral fellow in the lab, Gary Strniste, isolated an enzyme from Escherichia coli capable of cleaving x-ray-induced DNA base damages (Strniste and Wallace 1975). We called this enzyme the x-ray endonuclease which was subsequently named endonuclease III or Nth. N stands for endonuclease and th stands for three. Nth is the founding grandmother of the helix-hairpin-helix (HhH) superfamily of DNA glycosylases that is highly conserved from bacteria to humans (Aspinwall et al. 1997; Hilbert et al. 1997; Sarker et al. 1998). Ogg1, a eukaryotic DNA glycosylase which removes oxidized purines (Aburatani et al. 1997; Arai et al. 1997; Radicella et al. 1997; Roldan-Arjona et al. 1997; Rosenquist et al. 1997), and MutY, which removes adenine misinserted opposite 8-oxoG or FapyG (McGoldrick et al. 1995; Pope and David 2005; Slupska et al. 1996; Slupska et al. 1999) are also members of the HhH structural family.
Formamidopyrimidine DNA glycosylase or Fpg was originally isolated from E. coli in Tomas Lindahl’s laboratory as a DNA glycosylase which removes methyl FapyG from alkylated DNA (Chetsanga and Lindahl 1979). Finally, almost two decades ago, Bob Melamede in the laboratory identified a second E. coli activity that had the same substrate specificity as endo III or Nth and named it endo VIII or Nei (Jiang et al. 1997b; Melamede et al. 1994). When we cloned the E. coli gene for endo VIII (Nei) by reverse genetics, we were surprised to find that the protein sequence of Nei had significant homology to Fpg DNA glycosylase (Jiang et al. 1997a). We went on to show that E. coli double mutants lacking both Nth and Nei, therefore lacking the ability to remove oxidized pyrimidines, exhibited about a 30-fold increase in spontaneous mutation frequency (Jiang et al. 1997a). Interestingly, even though both of these proteins recognize thymine glycol and other oxidized thymine products, all of the mutations we observed were C to T transitions (Jiang et al. 1997a), indicating that the premutagenic oxidative pyrimidine lesions in E. coli arose from cytosines not thymines. We had shown earlier that thymine glycol was not a mutagenic lesion (Hayes et al. 1988) which is in keeping with the fact that the pairing surface of the oxidized thymine residues remains intact. More recently George Teebor’s group showed that Nth Neil1 nullizygous mice exhibit substantially increased pulmonary and hepatocellular tumors (Chan et al. 2009).
A little over 10 years ago in silico analysis allowed our laboratory and others to identify three novel members of the Fpg/Nei family from humans, the so-called Neil or Nei-like proteins (Bandaru et al. 2002; Hazra et al. 2002a; Hazra et al. 2002b; Morland et al. 2002; Wallace et al. 2003). At the same time, a backup enzyme was identified in Nth nullizygous mice (Takao et al. 2002). Two of these three enzymes, NEIL1 and NEIL2, were cloned, expressed, purified and characterized by a number laboratories, including our own (Bandaru et al. 2002; Dou et al. 2003; Hailer et al. 2005; Hazra et al. 2002a; Hazra et al. 2002b; Jaruga et al. 2004; Katafuchi et al. 2004; Krishnamurthy et al. 2008; Morland et al. 2002; Wallace et al. 2003). Like their bacterial orthologs, the substrate specificities of NEIL1 and NTH1 substantially overlap. Both recognize Tg, 5-OHC, dihydrothymine, dihydrouracil, 5-OHU but not 8-oxoG. NEIL1 forms specific interactions with a number of the replication proteins, appears to be cell cycle regulated and has been postulated to act as a cowcatcher ahead of the replication fork, eliminating potentially mutagenic lesions (Dou et al. 2008; Hegde et al. 2013; Hegde et al. 2008; Theriot et al. 2010; Wiederhold et al. 2004). NEIL2 prefers lesions in single-stranded DNA over those in duplex DNA and interacts with a number of transcription factors including RNA polymerase II. NEIL2 has been suggested to act in transcription-coupled repair of oxidative DNA damage (Banerjee et al. 2011). The mouse ortholog of NEIL3 has only recently been purified and characterized (Liu et al. 2010; Takao et al. 2009), (see below). Neil3 is expressed in neural stem cells, hematopoietic tissues and in cancer cells (for a review see (Liu et al. 2013a)). Neil3 also appears to be cell cycle regulated and localized in the nucleus. Although Neil1 (Hu et al. 2005) and Neil2 (Mandal et al. 2012) have been found in mitochondria thus far Neil3 has not (Liu et al. 2013a; Torisu et al. 2005).
The Fpg/Nei Family: How Structure Informs Function
In contrast to the HhH superfamily members, Nth and MutY, which have sequences, structures and functions that do not significantly differ across all three phyla, a number of the subfamilies of the Fpg family have changed significantly in sequence from their common ancestor, thus making it difficult to understand the evolution of these enzymes (Pumo et al. 2010). For example, essentially all bacteria have an Fpg protein while only a small subset have Nei proteins. However, in the Actinobacteria there are six Fpg/Nei gene clades, two within the Nei clade and four within the Fpg clade. In eukaryotes, the plant/fungal clade is clearly part of the Fpg family (Kathe et al. 2009) while in metazoans, Neil2 and Neil3 seem to form their own clade separate from Neil1. Horizontal gene transfer appears to be the likely event in the initial evolution of Nei proteins from a common ancestor which contained at least one Fpg/Nei homolog with features similar to Fpg. Vertical evolution may have been responsible for the transfer of an early Fpg/Nei gene to early eukaryotes followed by the diversification of these proteins in higher eukaryotes.
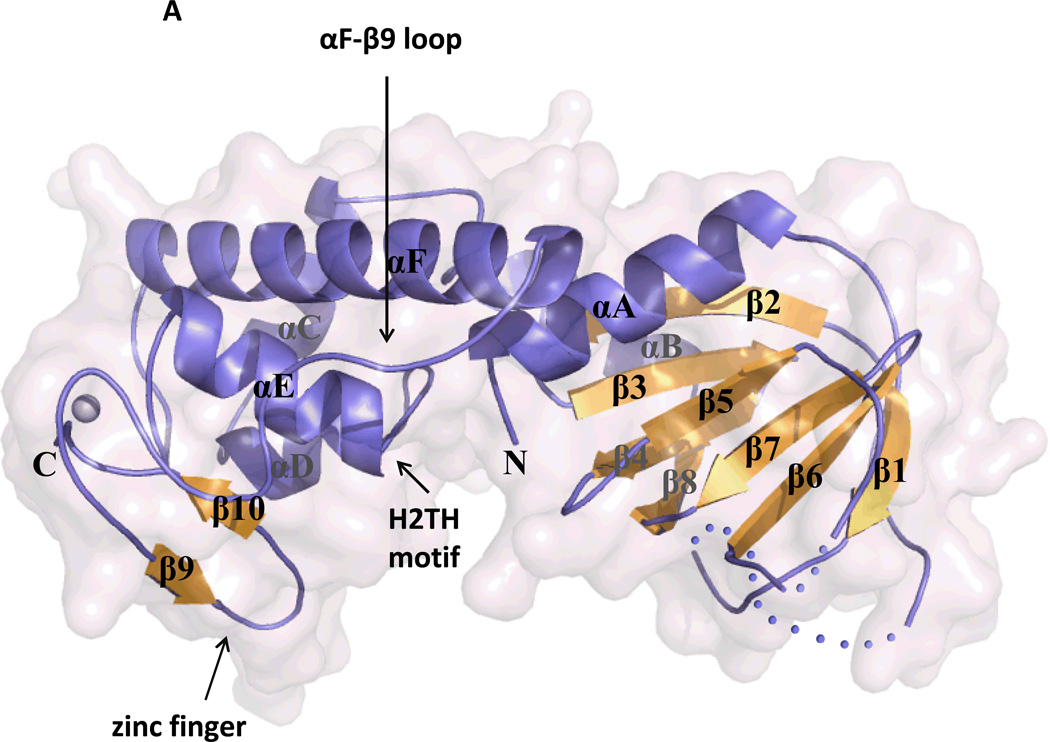
There are a number of crystal structures of the Fpg/Nei glycosylases and overall, the structures of the Fpg and Nei proteins are similar (Banerjee et al. 2006; Coste et al. 2004; Coste et al. 2008; Doublié et al. 2004; Fromme and Verdine 2002; Fromme and Verdine 2003; Gilboa et al. 2002; Golan et al. 2005; Imamura et al. 2009; Le Bihan et al. 2011; Pereira de Jesus et al. 2005; Qi et al. 2009; Qi et al. 2010; Serre et al. 2002; Sugahara et al. 2000; Sung et al. 2013; Zharkov et al. 2002) (and for reviews see (Fromme et al. 2004; Prakash et al. 2012; Verdine and Norman 2003)). They contain a two-domain architecture which is connected by a flexible hinge region. The C-terminal domain contains predominately α helices, two of which form the conserved helix-two-turns-helix motif of the Fpg family as well as two anti-parallel β strands that fold into a zinc finger motif. Both of these are required for DNA binding (Kropachev et al. 2006; O'Connor et al. 1993; Tchou et al. 1993). An additional feature in the C-terminal domain is the so called “lesion recognition loop” (Banerjee et al. 2006; Fromme and Verdine 2002; Fromme and Verdine 2003; Qi et al. 2009; Sugahara et al. 2000). This loop was shown to wrap around and form hydrogen bonds with both 8-oxoG (Fromme and Verdine 2003) and FapyG (Coste et al. 2004) in Fpg structures containing the lesion in the active site pocket. The N-terminal domain also contains three void-filling residues. One of these, to be discussed later, is involved in the lesion search process, while all three aid in everting the lesion into the active site pocket of the glycosylase and remain in the DNA duplex to stabilize the DNA (Doublié et al. 2004; Gilboa et al. 2002; Imamura et al. 2009; Zaika et al. 2004; Zharkov et al. 2002). The N-terminus of the Fpg/Nei family of proteins also contains the catalytic residues including an N-terminal proline that attacks the C1’ position of the target nucleotide (Kropachev et al. 2006; Tchou and Grollman 1995).
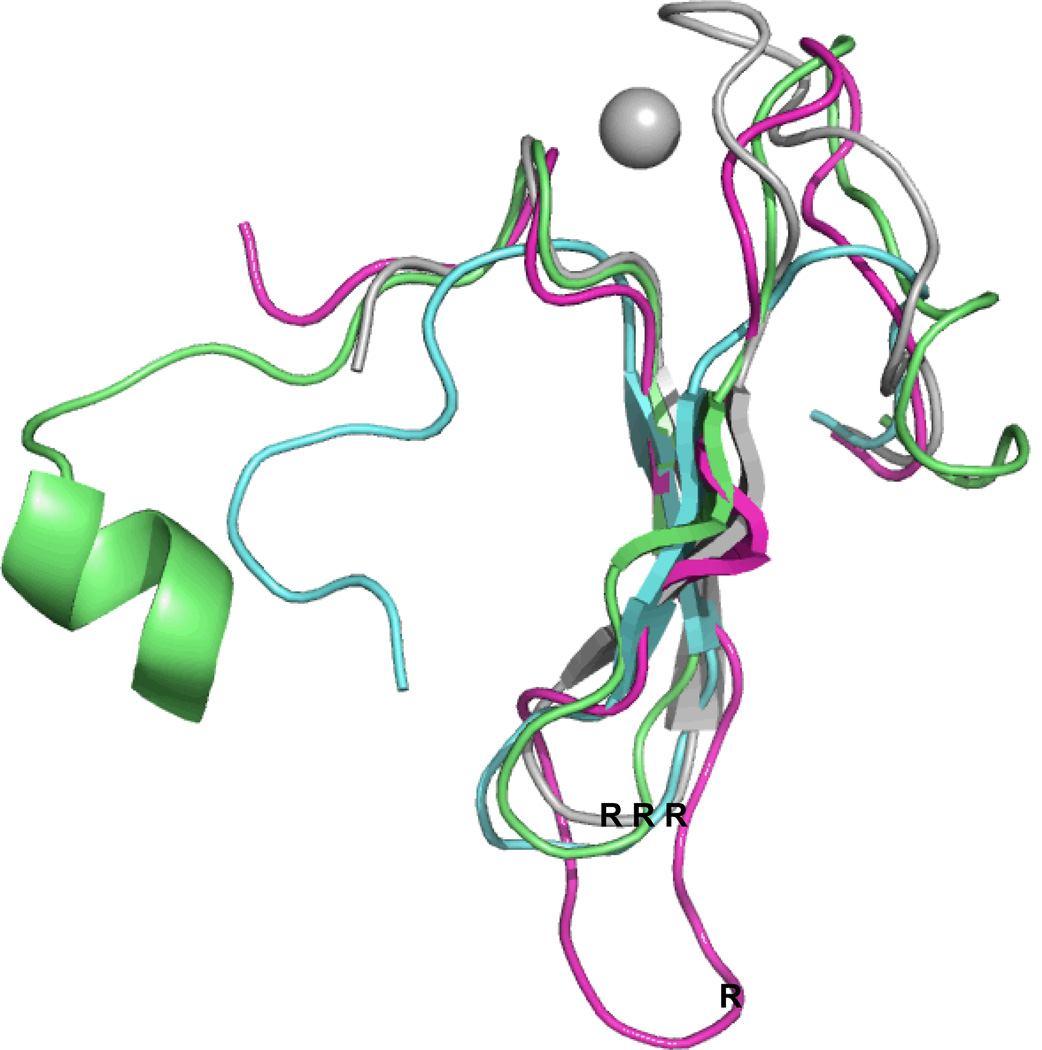
Once we had cloned and sequenced a number of eukaryotic members of the Fpg/Nei Family, it became clear that NEIL1 as well as the plant, fungal and one of the mimivirus members of this family were lacking the zinc finger which had been shown to be essential for DNA binding by the bacterial proteins (Kropachev et al. 2006; O'Connor et al. 1993; Tchou et al. 1993) (and for a review see (Wallace et al. 2003)). Although we had purified an enzymatically active human NEIL1 lacking a zinc finger, it took a crystal structure to tell us why it was active (Doublié et al. 2004). It turns out that the NEIL1 zinc finger region was superimposable upon that of the E. coli protein but instead of the anti-parallel β strands being coordinated by zinc, the human NEIL1 motif contained two anti-parallel β strands that mimicked the zinc finger fold only there were no longer loops present that needed to be coordinated by zinc. Moreover, this “zincless finger” motif contained the highly conserved arginine, which was present in the bacterial proteins containing a zinc finger and was required for glycosylase activity (Doublié et al. 2004; Zharkov et al. 2002). We observed similar zincless finger motifs in the Arabidopsis thaliana Fpg (Duclos et al. 2012a) and the mimivirus Nei1 proteins (Imamura et al. 2009). A superposition of the three zincless fingers from these crystal structures on the Lactus lactococcus zinc finger is shown in Figure 2.
Figure 2.
Superposition of the zincless fingers of human NEIL1, blue, PDB ID code: 1TDH (Doublié et al. 2004), Mimivirus Nei1 (MvNei1), dark pink, PDB ID code: 3A46 (Imamura et al. 2009), A. thaliana (AthFpg), green, PDB ID code: 3TWL (Duclos et al. 2012a), with the zinc finger of L. lactus Fpg, grey, PDB ID code: 1TD2, (Serre et al. 2002).
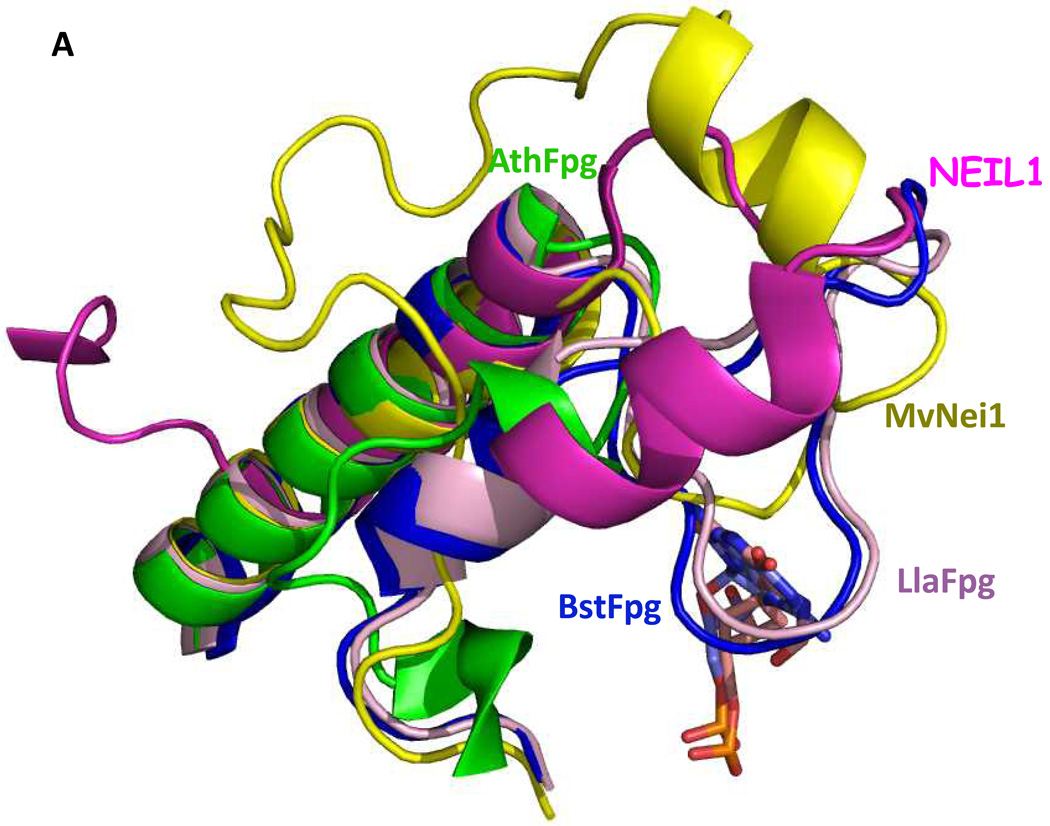
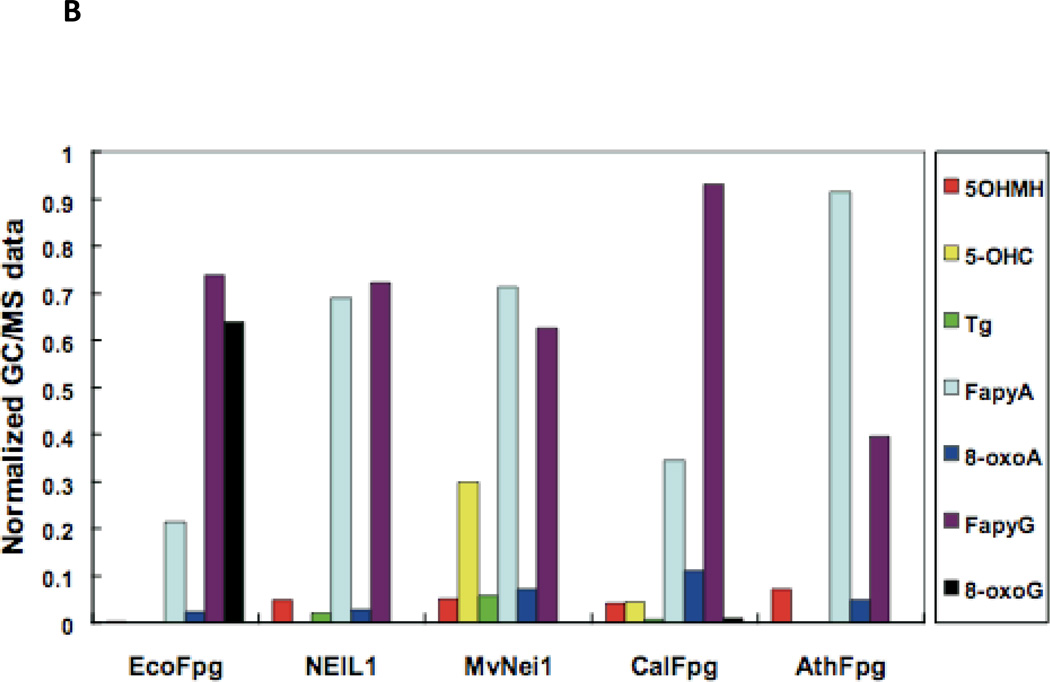
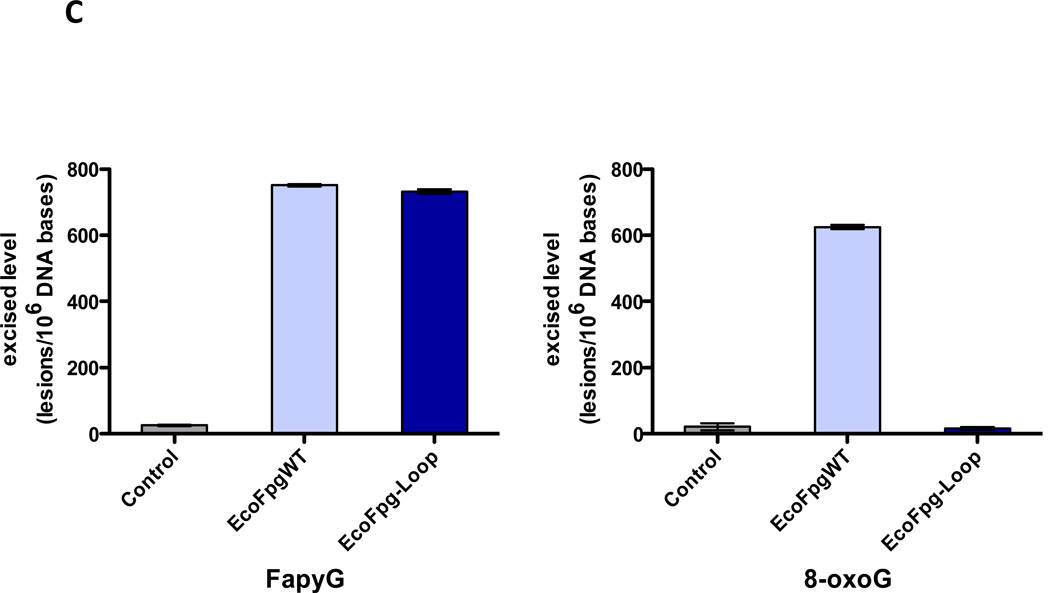
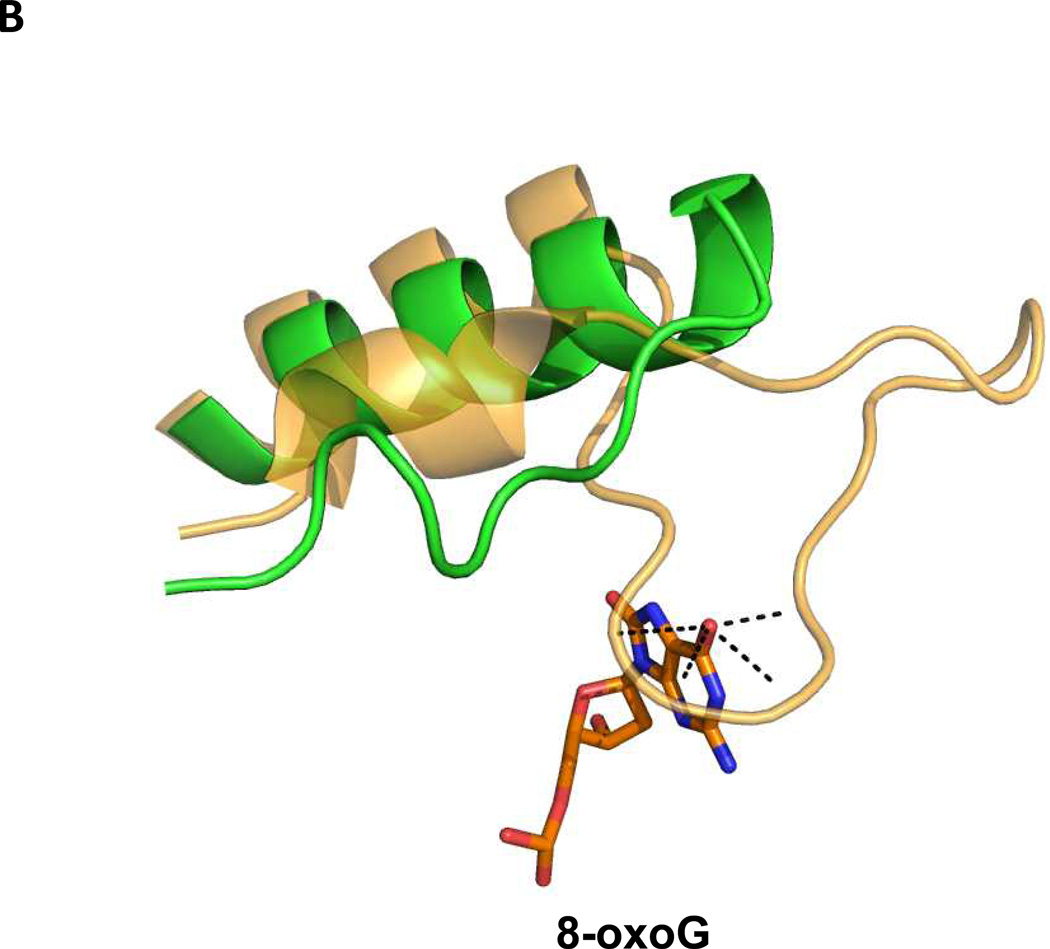
In the α-helical C-terminal domain of the Fpg protein there is a loop region called the α-F-β9/10 loop which had been presumed to be involved in lesion recognition, the so-called “lesion recognition loop” (Banerjee et al. 2006; Fromme and Verdine 2002; Fromme and Verdine 2003; Qi et al. 2009; Sugahara et al. 2000). This loop reads the pronation state of N7, thus distinguishing G from 8-oxoG and makes contacts with 8-oxoG and FapyG in the substrate binding pocket, primarily by main chain interactions. In the bacterial Fpg proteins these loops appear to be functionally similar and are of comparable length, about 27 amino acid residues. This loop is missing in E. coli Nei (Golan et al. 2005) as well as from the eukaryotic proteins, A. thaliana Fpg (Duclos et al. 2012a) and NEIL1 (Doublié et al. 2004), and is significantly shortened in the mimivirus Nei1 (Imamura et al. 2009). For a superposition of the structures of the loop regions of NEIL1, AthFpg, and MvNei1 with the L. lactococcus and Bacillus stereothermopolus α-F-β9/10 loops see Figure 3A. NEIL1, AthFpg and MvNei1 do not recognize 8-oxoguanine. This is true not only for oligonucleotide substrates containing 8-oxoG but for 8-oxoG in γ-irradiated calf thymus DNA (Kathe et al. 2009; Liu et al. 2010) (Figure 3B). However, FapyG was still cleaved. This observation was surprising since the crystal structure of L. lactus Fpg bound to FapyG showed that the α-F-β9/10 loop hydrogen bonded with the FapyG in the substrate binding pocket (Coste et al. 2004). However, when we deleted this “lesion recognition loop” from E. coli Fpg, we found that Fpg still recognized its substrates including FapyG, but it did not recognize 8-oxoguanine (Duclos et al. 2012a)(Figure 3C). Taken together the data confirm that this misnamed “lesion recognition loop” is only required for stabilizing 8-oxoguanine in the active site pocket, but not FapyG, FapyA or 8-oxoadenine (Duclos et al. 2012a).
Figure 3.
“The Lesion Recognition Loop”. (A) Close up view of the αF-β9/10 loop of the bacterial Fpg/Nei proteins compared to their eukaryotic counterparts. Human NEIL1, dark pink, PDB ID code: 1TDH (Doublié et al 2004), A. thaliana (AthFpg) green, PDB ID code: 3TWL (Duclos et al 2012a) (MvNei1) (gold, PDB ID code: 3A46, (Imamura et al 2009) B. stereothermophilus Fpg (BstFpg)-DNA complex blue, PDB ID code: IR2YY (Fromme and Verdine 2002), L. lactus Fpg (LlaFpg) –DNA complex, Light pink, PDB ID code: 1TDZ (Coste et al 2004).
(B) GC/MS analysis of damaged bases released from γ-irradiated calf thymus DNA by E. coli Fpg (EcoFpg), human NEIL1, Mimivirus Nei1 (MvNei1), Candida albicans Fpg (CalFpg) and A. thaliana (AthFpg). Amounts of damaged bases were normalized to illustrate the preferences of the enzymes (Kathe et al. 2009) (5-OHMH is 5-hydroxy-5-methylhydantoin). (C) GC/MS analysis of the release of FapyG and 8-oxoG from γ-irradiated calf thymus DNA by E. coli wild type Fpg (EcoFpgWT) and E. coli Fpg missing the “lesion recognition loop” (EcoFpg-Loop) (Duclos et al. 2012a).
Neil3: Substrates and Structure
Neil 3, which is more than twice the size of the bacterial Fpg/Nei proteins, also proved to be the most intractable to purify and characterize (for a review see (Liu et al. 2013a)). Although Seeberg and co-workers found that lysates from insect cells overexpressing NEIL3 were able to excise methyl FapyG when compared to unfected cell lysates (Morland et al. 2002), subsequent studies failed to detect any glycosylase activity in either purified or in vitro translated Neil3 proteins (Krokeide et al. 2009; Takao et al. 2002; Takao et al. 2009; Torisu et al. 2005) although a weak lyase activity was demonstrated on a single-stranded substrate (Takao et al. 2009). Our laboratory developed the protocol (Liu et al. 2010; Liu et al. 2012) for purifying full-length mouse Neil3, a truncated version containing only the glycosylase domain, as well as the glycosylase domain of human NEIL3. All three versions of Neil3 exhibit a strong preference for lesions in single-stranded DNA and recognize both oxidized pyrimidines and oxidized purines (Liu et al. 2010). Neil3 also prefers substrates that contain single-stranded regions such as looped structures and structures representing replication forks (Liu et al. 2010). In duplex DNA, the further oxidation products of guanine, Sp and Gh, are by far the best substrates as are FapyG and FapyA in γ-irradiated DNA (Liu et al. 2010). Although bifunctional, Neil3 has a very weak lyase activity (Krokeide et al. 2013; Liu et al. 2010) and as with the monofunctional DNA glycosylases, the abasic site remaining after Neil3 glycosylase activity is likely cleaved in the cell by APE1. Neil3 can also function as a glycosylase in vivo as demonstrated by its ability to substantially reduce the spontaneous mutation frequency in E. coli triple mutants lacking Fpg Nei and MutY glycosylases (Liu et al. 2010). In addition, NEIL3 substantially reduced the levels of FapyG in genomic DNA, suggesting that Neil3 plays an important role in removing this lesion in vivo (Liu et al. 2010). Others have shown that expression of the NEIL3 glycosylase domain in an E. coli fpg nei double mutant partially restores the wild type hydrogen peroxide sensitivity of these cells (Takao et al. 2009).
We recently used DNA containing the human telomeric sequence in duplex, single-stranded and G4 quadruplex DNA structures as substrates for the various human glycosylases (Zhou et al. 2013). Neil3 preferentially excises Tg from quadrupex DNA and from double–stranded and single-stranded DNA containing Tg in the telomeric sequence context. Neil3 and NEIL1 also recognize and remove Sp and Gh from quadruplex structures but sequence context does not appear to play a role in the removal of Sp and Gh from single stranded and duplex DNA. NTH1, NEIL1 and NEIL2 do not recognize Tg in quadruplex DNA nor does OGG1 recognize 8-oxoG in quadruplex DNA.
We also were able to solve the unliganded structure of the glycosylase portion of Neil3 (Liu et al. 2013b) which showed NEIL3 to have the same overall fold as other Fpg/Nei family members including the H2TH motif and the zinc finger motif (Figure 4A). Rather than proline, however, Neil3 has a unique N-terminal valine that we had previously shown to be required for catalysis (Liu et al. 2010). In keeping with its lack of recognition of 8-oxoG, Neil3 has a severely truncated lesion recognition loop (Figure 4B).
Figure 4.
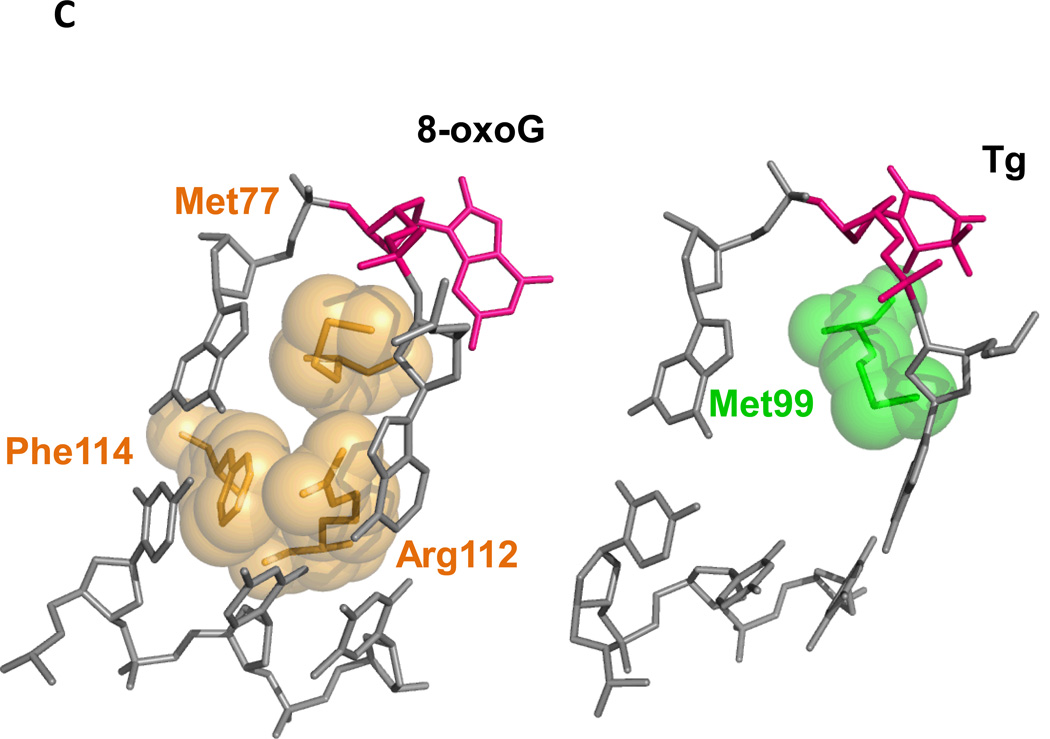
(A) Ribbon diagram of the truncated version of mouse Neil3. PDB ID Code: 3WOF (Liu et al. 2013b) (B) Neil3 lacks the so-called “lesion recognition loop.” Neil3 in green (Liu et al. 2013b), B. stereothermophilus Fpg PDB ID Code: 3WOF, (BstFpg) in gold (Liu et al. 2013b) , PDB ID code: 1R2Y (Fromme and Verdine 2002). (C) Void-filling residues of BstFpg, PDB ID code: 1R2Y (Fromme and Verdine 2003) compared to mouse Neil3 PDB ID Code: 3WOF (Liu et al. 2013b) with Tg DNA from MvNei1 PDB ID code: 3VK8 (Imamura et al. 2012).
The structural features of Neil3 do, however, provide some insight as to why it prefers lesions in DNA containing single-stranded regions (Liu et al. 2013b). First, Neil3 lacks two of the three canonical void-filling residues that stabilize the duplex and interact with opposite strand after eversion of the base lesion. Figure 4C shows a comparison between the three void-filling residues of BstFpg (Fromme and Verdine 2003) to the single methionine of mouse Neil3 (Liu et al. 2013b). Also, unlike any other Fpg/Nei family member, Neil3 has negatively charged residues repulsive to the opposite strand. Despite what we know about Neil3, the question arises as to why vertebrates are equipped with Neil3 while other organisms can live without it. Furthermore, since the discovery of Neil3, its biological role remains an enigma even though the data indicate that Neil3 is required in proliferating cells (Hildrestrand et al. 2009; Kauffmann et al. 2008; Morland et al. 2002; Rolseth et al. 2008; Torisu et al. 2005).
How Do the DNA Glycosylases Find their Substrates and Distinguish Them from Normal Bases?
Although there have been a number of crystal structures solved of Fpg protein with 8-oxoG (Fromme and Verdine 2002; Fromme and Verdine 2003) or FapyG (Serre et al. 2002) in the substrate binding pocket, until recently there were no structures of Nth or Nei proteins complexed to DNA containing lesions. Nth and Nei have large substrate binding pockets and very broad substrate specificities (Duclos et al. 2012b). The question is, how do they find their substrates and distinguish them from normal bases? To try and answer this question the structure of mimivirus Nei1, a NEIL1 ortholog, was solved with DNA containing thymine glycol or 5-hydroxyuracil in the substrate binding pocket (Imamura et al. 2009). What was found was that the binding pocket was highly hydrophobic and very few interactions with thymine glycol or 5-hydroxyuracil were observed. Moreover, when these potentially interacting residues were mutated there was no loss in glycosylase activity indicating no relationship to specificity. Taken together these crystallographic data strongly suggest that lesion recognition likely happens before the base is everted into the substrate binding pocket.
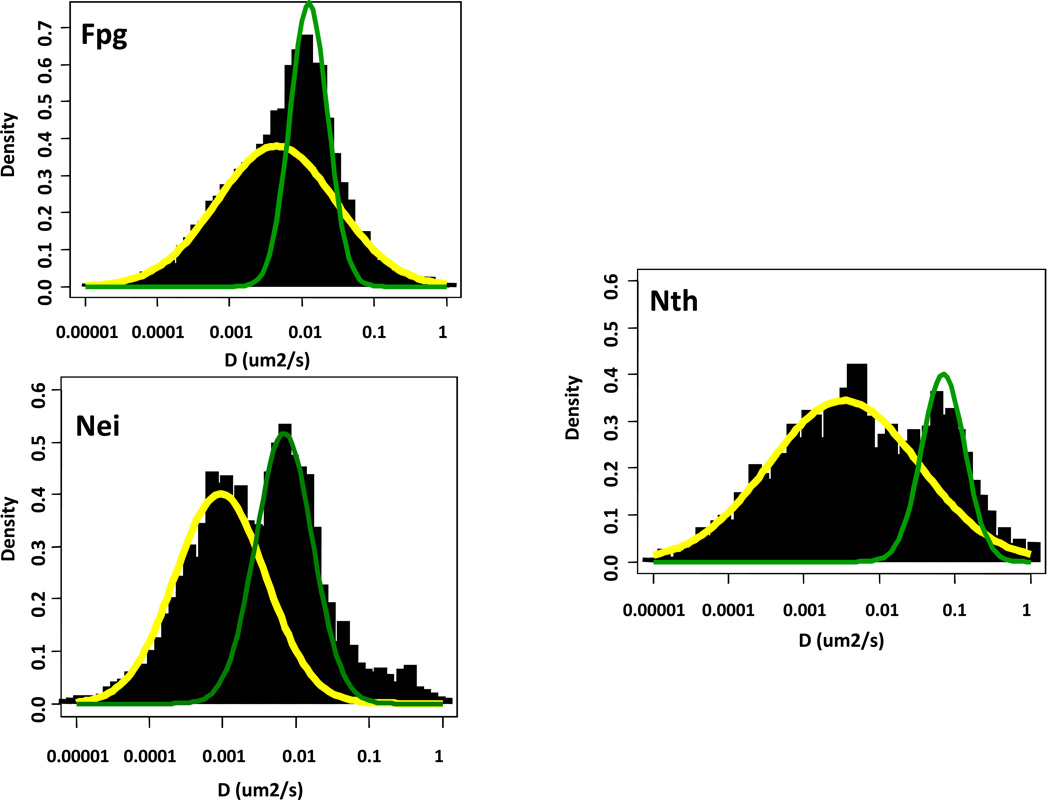
To address the search question we used a single molecule TIRF approach with a YOYO-1-labeled λDNA tightrope which was flow-elongated between polylysine-coated silica beads (Dunn et al. 2011; Kad et al. 2010). Once the DNA was suspended, quantum dot-labeled DNA glycosylases were added to the tightrope and their diffusive behavior examined in the absence of flow. DNA glycosylases do not require ATP and use thermal energy to diffuse along the DNA molecule searching for their targets. We found the wild type bacterial glycosylases Fpg, Nei and Nth, to diffuse along the DNA with a broad distribution of rates that ranged over two orders of magnitude (Dunn et al. 2011). The majority, however, were below 0.05 µm2 per second which is consistent with the glycosylase rotating around the DNA molecule (Bagchi et al. 2008; Schurr 1979). When the individual trajectories were evaluated on a time-weighted basis (Nelson et al. 2013) the diffusive behavior of all three enzymes showed both slow and fast peaks of motion (Figure 5).
Figure 5.
Time-weighted distribution of diffusion constants of Fpg, Nei and Nth on undamaged DNA demonstrating that the diffusive behavior of both families show both slow and fast peaks of motion (Nelson et al. 2013).
As described above, members of the Fpg/Nei family of DNA glycosylases have three so-called void-filling residues that are inserted into the DNA helix, help flip out the damaged base, interact with the opposite strand and stabilize the DNA helix (Fromme and Verdine 2002; Fromme and Verdine 2003; Zharkov et al. 2002). The HhH superfamily, represented by Nth, uses a similar mechanism. Interestingly, the crystal structure of Bacillus stearothermophilus Fpg covalently cross-linked to undamaged DNA showed one of these residues, phenylalanine, to be wedged into the helix occupying a position analogous to its position in the Fpg complex with DNA containing a damage (Banerjee et al. 2006; Qi et al. 2012; Qi et al. 2010). When we mutated the analogous residue in E.coli Fpg we lost a substantial population of the slow moving glycosylases and the mean diffusion constant increased significantly (Dunn et al. 2011). We found the same results for Nei and Nth glycosylases (Nelson et al. 2013). Since the wedge mutants do not undergo the slower diffusive motion along the DNA, the data suggest that insertion of the wedge is responsible for this population of wild type glycosylases and that the residue may in fact be responsible for finding the lesion. The data also say that both families of the glycosylases appear to be behaving in a similar manner. This conclusion is supported not only by our single molecule data (Dunn et al. 2011; Nelson et al. 2013) and the crystal structures of Fpg cross-linked to undamaged DNA described above, but also by analysis of E. coli Fpg using stopped flow presteady state kinetics that divides the Fpg catalytic reaction into five steps with the second step after the initial binding of Fpg to DNA to be insertion of the phenylalanine wedge residue (Koval et al. 2010; Kuznetsov et al. 2007; Kuznetsov et al. 2012; Kuznetsov et al. 2009). Thus, all the data support the hypothesis that these glycosylases recognize the lesion to be excised prior to its insertion into the substrate binding pocket. Moreover, if one corrects for the estimated activation barrier, the slow scanning population of Fpg, Nei and Nth diffuse at about 1.3x106 bp2/sec with a binding lifetime of about 1.3 seconds. Thus the glycosylases would be able to scan about 1600 bp. Since there are about 400 molecules of each glycosylase per E. coli cell (Demple and Harrison 1994), it would take about 16 seconds for each population of glycosylases to interrogate the genome for the lesions they recognize (Dunn et al. 2011).
We then asked what happens when the wild type DNA glycosylases interact with DNA containing damages (Nelson et al. 2013) To create substrates for Fpg we treated DNA with methylene blue plus light to produce 8-oxoG and FapyG lesions (Boiteux et al. 1992; Czeczot et al. 1991; Muller et al. 1990). We also treated DNA with osmium tetroxide to produce thymine glycol substrates (Ide et al. 1985; Katcher and Wallace 1983; Laspia and Wallace 1988) for Nei and Nth. We compared undamaged DNA to two populations of damaged DNA, a low dose population containing between 100-120 damages per λ DNA molecule and a high dose population, containing between 270-340 damages per λ DNA molecule. What we found was that for all three enzymes, the mean diffusion constant significantly decreased as the number of damages increased, and at the same time, there was an increase in the number of paused DNA glycosylases. Finally, the binding lifetimes of the glycosylases increased with the number of DNA damages. These data are in keeping with the idea that when a glycosylase encounters a damage it stops to remove it.
Taken together our data support the hypothesis that glycosylases undergo several different types of motion. First there is a slow scanning phase where the glycosylase is inserting a wedge residue to determine if a lesion is present in the DNA molecule. There is also a faster phase that is consistent with the glycosylase rotating around the DNA molecule without or rarely inserting the wedge residue. Some molecules even appear to go faster and scan in a one-dimensional diffusive manner. Finally glycosylases can be observed to pause or stop when they encounter a DNA lesion.
PERSPECTIVES
During the past several decades we have learned a great deal about the DNA glycosylases that remove oxidized DNA bases but we have much left to learn. At the basic level, structures of human NTH and the NEIL proteins with lesions in the substrate binding pocket will help us more fully understand their substrate specificities. This is especially true for NEIL3 complexed with lesion-containing single stranded DNA which is its preferred substrate. We also need to elucidate the roles that the oxidative DNA glycosylases play in mammalian cells. Although progress has been made with NEIL1 and NEIL2, the cellular role of NEIL3 remains an enigma. Also do the mammalian DNA glycosylases search for damage in DNA in a manner similar to their bacterial counterparts? Our ultimate goal is to comprehend what roles mutant DNA glycosylases play in human disease.
ACKNOWLEDGMENTS
I am extremely grateful to my generous collaborators: The crystallographic studies are a collaboration with Dr. Sylvie Doublié, Professor, Department of Microbiology and Molecular Genetics, University of Vermont, the single molecule studies are a collaboration with Dr. David Warshaw, Professor and Chair, Department of Molecular Physiology and Biophysics, University of Vermont, and the GC/MS analysis was performed by Dr. Miral Dizdaroglu, Chemical Science and Technology Laboratory, National Institute of Standards and Technology, Gaithersburg, Maryland. I would also like to thank all of my students, postdoctoral associates and technicians without whom this work would not have been accomplished. I would like to especially acknowledge students, Minmin Liu, Andrew Dunn and Jia Zhou, and postdoctoral associates, Stephanie Duclos, Scott Kathe and Shane Nelson, whose work was highlighted in this manuscript. Financial support for this work was from National Institutes of Health grants R37 CA 33657, P01 CA098993 and R01 CA52040 awarded by the National Cancer Institute.
Biography

Footnotes
The Environmental Mutagenesis and Genomics Society conferred this award to Dr. Susan S. Wallace for her fundamental studies on DNA damage repair caused by environmental agents and for exemplary leadership in science. Dr. Wallace has contributed greatly to the discovery and understanding of how base excision repair enzymes protect the DNA from the deleterious consequences of oxidative lesions. Her work helped define the spectrum of DNA damage generated by reactive oxygen species and how persistent DNA modifications can lead to mutations. Throughout her career, Dr. Wallace has employed cutting-edge technologies to elucidate the mechanisms by which base excision repair enzymes function. Dr. Wallace is also recognized for her dedication to training and mentoring of young scientists and the many leadership roles in scientific societies, committees and journals.
REFERENCES
- Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res. 1997;57(11):2151–2156. [PubMed] [Google Scholar]
- Arai K, Morishita K, Shinmura K, Kohno T, Kim SR, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Cloning of a human homolog of the yeast OGG1 gene that is involved in the repair of oxidative DNA damage. Oncogene. 1997;14(23):2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- Aspinwall R, Rothwell DG, Roldan-Arjona T, Anselmino C, Ward CJ, Cheadle JP, Sampson JR, Lindahl T, Harris PC, Hickson ID. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc Natl Acad Sci U S A. 1997;94(1):109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad N, Iyer A, Vallyathan V, Wang L, Castranova V, Stehlik C, Rojanasakul Y. Role of oxidative/nitrosative stress-mediated Bcl-2 regulation in apoptosis and malignant transformation. Ann N Y Acad Sci. 2010;1203:1–6. doi: 10.1111/j.1749-6632.2010.05608.x. [DOI] [PubMed] [Google Scholar]
- Bagchi B, Blainey PC, Xie XS. Diffusion constant of a nonspecifically bound protein undergoing curvilinear motion along DNA. J Phys Chem B. 2008;112(19):6282–6284. doi: 10.1021/jp077568f. [DOI] [PubMed] [Google Scholar]
- Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1(7):517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Santos WL, Verdine GL. Structure of a DNA glycosylase searching for lesions. Science. 2006;311(5764):1153–1157. doi: 10.1126/science.1120288. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Mandal SM, Das A, Hegde ML, Das S, Bhakat KK, Boldogh I, Sarkar PS, Mitra S, Hazra TK. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J Biol Chem. 2011;286(8):6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- Boiteux S, Gajewski E, Laval J, Dizdaroglu M. Substrate specificity of the Escherichia coli Fpg protein (formamidopyrimidine-DNA glycosylase): excision of purine lesions in DNA produced by ionizing radiation or photosensitization. Biochemistry. 1992;31(1):106–110. doi: 10.1021/bi00116a016. [DOI] [PubMed] [Google Scholar]
- Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic Biol Med. 1995;18(6):1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- Burrows CJ. Surviving an Oxygen Atmosphere: DNA Damage and Repair. ACS Symp Ser Am Chem Soc. 2009;2009:147–156. doi: 10.1021/bk-2009-1025.ch008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MK, Ocampo-Hafalla MT, Vartanian V, Jaruga P, Kirkali G, Koenig KL, Brown S, Lloyd RS, Dizdaroglu M, Teebor GW. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst) 2009;8(7):786–794. doi: 10.1016/j.dnarep.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetsanga CJ, Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6(11):3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J Biol Chem. 2004;279(42):44074–44083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- Coste F, Ober M, Le Bihan YV, Izquierdo MA, Hervouet N, Mueller H, Carell T, Castaing B. Bacterial base excision repair enzyme Fpg recognizes bulky N7-substituted-FapydG lesion via unproductive binding mode. Chem Biol. 2008;15(7):706–717. doi: 10.1016/j.chembiol.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Czeczot H, Tudek B, Lambert B, Laval J, Boiteux S. Escherichia coli Fpg protein and UvrABC endonuclease repair DNA damage induced by methylene blue plus visible light in vivo and in vitro. J Bacteriol. 1991;173(11):3419–3424. doi: 10.1128/jb.173.11.3419-3424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B, Harrison L. Repair of oxidative damage to DNA: enzymology and biology. Annu Rev Biochem. 1994;63:915–948. doi: 10.1146/annurev.bi.63.070194.004411. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M. Oxidative damage to DNA in mammalian chromatin. Mutat Res. 1992;275(3–6):331–342. doi: 10.1016/0921-8734(92)90036-o. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M. Mechanisms of free radical damage to DNA. In: Aruoma O, Halliwell B, editors. DNA and Free Radicals: Techniques, Mechanisms and Applications. St. Lucia: OICA International; 1998. pp. 3–26. [Google Scholar]
- Dizdaroglu M, Holwitt E, Hagan MP, Blakely WF. Formation of cytosine glycol and 5,6-dihydroxycytosine in deoxyribonucleic acid on treatment with osmium tetroxide. Biochem J. 1986;235(2):531–536. doi: 10.1042/bj2350531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizdaroglu M, Jaruga P, Birincioglu M, Rodriguez H. Free radical-induced damage to DNA: mechanisms and measurement. Free Radic Biol Med. 2002;32(11):1102–1115. doi: 10.1016/s0891-5849(02)00826-2. [DOI] [PubMed] [Google Scholar]
- Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem. 2003;278(50):49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- Dou H, Theriot CA, Das A, Hegde ML, Matsumoto Y, Boldogh I, Hazra TK, Bhakat KK, Mitra S. Interaction of the human DNA glycosylase NEIL1 with proliferating cell nuclear antigen. The potential for replication-associated repair of oxidized bases in mammalian genomes. J Biol Chem. 2008;283(6):3130–3140. doi: 10.1074/jbc.M709186200. [DOI] [PubMed] [Google Scholar]
- Doublié S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc Natl Acad Sci U S A. 2004;101(28):10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos S, Aller P, Jaruga P, Dizdaroglu M, Wallace SS, Doublie S. Structural and biochemical studies of a plant formamidopyrimidine-DNA glycosylase reveal why eukaryotic Fpg glycosylases do not excise 8-oxoguanine. DNA Repair (Amst) 2012a;11(9):714–725. doi: 10.1016/j.dnarep.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duclos S, Doublié S, Wallace SS. Consequences and repair of oxidative DNA damage. In: Greim H, Albertini RJ, editors. The Cellular Response to the Genotoxic Insult: The Question of Threshold for Genotoxic Carcinogens. Cambridge, United Kingdom: The Royal Society of Chemistry; 2012b. pp. 109–153. [Google Scholar]
- Dunn AR, Kad NM, Nelson SR, Warshaw DM, Wallace SS. Single Qdot-labeled glycosylase molecules use a wedge amino acid to probe for lesions while scanning along DNA. Nucleic Acids Res. 2011;39(17):7487–7498. doi: 10.1093/nar/gkr459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Banerjee A, Verdine GL. DNA glycosylase recognition and catalysis. Curr Opin Struct Biol. 2004;14(1):43–49. doi: 10.1016/j.sbi.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat Struct Biol. 2002;9(7):544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J Biol Chem. 2003;278(51):51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Verdine GL. Base excision repair. Adv Protein Chem. 2004;69:1–41. doi: 10.1016/S0065-3233(04)69001-2. [DOI] [PubMed] [Google Scholar]
- Funato Y, Miki H. Redox regulation of Wnt signalling via nucleoredoxin. Free Radic Res. 2010;44(4):379–388. doi: 10.3109/10715761003610745. [DOI] [PubMed] [Google Scholar]
- Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J Biol Chem. 2002;277(22):19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- Golan G, Zharkov DO, Feinberg H, Fernandes AS, Zaika EI, Kycia JH, Grollman AP, Shoham G. Structure of the uncomplexed DNA repair enzyme endonuclease VIII indicates significant interdomain flexibility. Nucleic Acids Res. 2005;33(15):5006–5016. doi: 10.1093/nar/gki796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 2005;4(1):41–50. doi: 10.1016/j.dnarep.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge JMC. Free Radicals in Biology and Medicine. Oxford: Oxford Universit Press; 2002. [Google Scholar]
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35(9):505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes RC, Petrullo LA, Huang HM, Wallace SS, LeClerc JE. Oxidative damage in DNA. Lack of mutagenicity by thymine glycol lesions. J Mol Biol. 1988;201(2):239–246. doi: 10.1016/0022-2836(88)90135-0. [DOI] [PubMed] [Google Scholar]
- Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci U S A. 2002a;99(6):3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem. 2002b;277(34):30417–30420. doi: 10.1074/jbc.C200355200. [DOI] [PubMed] [Google Scholar]
- Hegde ML, Hegde PM, Bellot LJ, Mandal SM, Hazra TK, Li GM, Boldogh I, Tomkinson AE, Mitra S. Prereplicative repair of oxidized bases in the human genome is mediated by NEIL1 DNA glycosylase together with replication proteins. Proc Natl Acad Sci U S A. 2013;110(33):E3090–E3099. doi: 10.1073/pnas.1304231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J Biol Chem. 2008;283(40):27028–27037. doi: 10.1074/jbc.M802712200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert TP, Chaung W, Boorstein RJ, Cunningham RP, Teebor GW. Cloning and expression of the cDNA encoding the human homologue of the DNA repair enzyme, Escherichia coli endonuclease III. J Biol Chem. 1997;272(10):6733–6740. doi: 10.1074/jbc.272.10.6733. [DOI] [PubMed] [Google Scholar]
- Hildrestrand GA, Neurauter CG, Diep DB, Castellanos CG, Krauss S, Bjoras M, Luna L. Expression patterns of Neil3 during embryonic brain development and neoplasia. BMC Neurosci. 2009;10:45. doi: 10.1186/1471-2202-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Wallace SS, Doublie S. Bumps in the road: how replicative DNA polymerases see DNA damage. Curr Opin Struct Biol. 2005;15(1):86–93. doi: 10.1016/j.sbi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280(49):40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- Ide H, Kow YW, Wallace SS. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 1985;13(22):8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Averill A, Wallace SS, Doublie S. Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA. J Biol Chem. 2012;287(6):4288–4298. doi: 10.1074/jbc.M111.315309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura K, Wallace SS, Doublie S. Structural characterization of a viral NEIL1 ortholog unliganded and bound to abasic site-containing DNA. J Biol Chem. 2009;284(38):26174–26183. doi: 10.1074/jbc.M109.021907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193(1–2):43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43(50):15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- Jezek P, Hlavata L. Mitochondria in homeostasis of reactive oxygen species in cell, tissues, and organism. Int J Biochem Cell Biol. 2005;37(12):2478–2503. doi: 10.1016/j.biocel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- Jiang D, Hatahet Z, Blaisdell JO, Melamede RJ, Wallace SS. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J Bacteriol. 1997a;179(11):3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Hatahet Z, Melamede RJ, Kow YW, Wallace SS. Characterization of Escherichia coli endonuclease VIII. J Biol Chem. 1997b;272(51):32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- Kad NM, Wang H, Kennedy GG, Warshaw DM, Van Houten B. Collaborative dynamic DNA scanning by nucleotide excision repair proteins investigated by single- molecule imaging of quantum-dot-labeled proteins. Mol Cell. 2010;37(5):702–713. doi: 10.1016/j.molcel.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S, Subbaram S, Carrico PM, Melendez JA. Redox-control of matrix metalloproteinase-1: a critical link between free radicals, matrix remodeling and degenerative disease. Respir Physiol Neurobiol. 2010;174(3):299–306. doi: 10.1016/j.resp.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J Biol Chem. 2004;279(14):14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- Katcher HL, Wallace SS. Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry. 1983;22(17):4071–4081. doi: 10.1021/bi00286a013. [DOI] [PubMed] [Google Scholar]
- Kathe SD, Barrantes-Reynolds R, Jaruga P, Newton MR, Burrows CJ, Bandaru V, Dizdaroglu M, Bond JP, Wallace SS. Plant and fungal Fpg homologs are formamidopyrimidine DNA glycosylases but not 8-oxoguanine DNA glycosylases. DNA Repair (Amst) 2009;8(5):643–653. doi: 10.1016/j.dnarep.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Rosselli F, Lazar V, Winnepenninckx V, Mansuet-Lupo A, Dessen P, van den Oord JJ, Spatz A, Sarasin A. High expression of DNA repair pathways is associated with metastasis in melanoma patients. Oncogene. 2008;27(5):565–573. doi: 10.1038/sj.onc.1210700. [DOI] [PubMed] [Google Scholar]
- Koval VV, Kuznetsov NA, Ishchenko AA, Saparbaev MK, Fedorova OS. Real-time studies of conformational dynamics of the repair enzyme Ecoli formamidopyrimidine-DNA glycosylase and its DNA complexes during catalytic cycle. Mutat Res. 2010;685(1–2):3–10. doi: 10.1016/j.mrfmmm.2009.08.018. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry. 2008;47(27):7137–7146. doi: 10.1021/bi800160s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan HE, Nilsen H, Skorpen F, Otterlei M, Slupphaug G. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476(1–2):73–77. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- Krokeide SZ, Bolstad N, Laerdahl JK, Bjoras M, Luna L. Expression and purification of NEIL3, a human DNA glycosylase homolog. Protein Expr Purif. 2009;65(2):160–164. doi: 10.1016/j.pep.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Krokeide SZ, Laerdahl JK, Salah M, Luna L, Cederkvist FH, Fleming AM, Burrows CJ, Dalhus B, Bjoras M. Human NEIL3 is mainly a monofunctional DNA glycosylase removing spiroimindiohydantoin and guanidinohydantoin. DNA Repair (Amst) 2013 doi: 10.1016/j.dnarep.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropachev KY, Zharkov DO, Grollman AP. Catalytic mechanism of Escherichia coli endonuclease VIII: roles of the intercalation loop and the zinc finger. Biochemistry. 2006;45(39):12039–12049. doi: 10.1021/bi060663e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov NA, Koval VV, Zharkov DO, Vorobjev YN, Nevinsky GA, Douglas KT, Fedorova OS. Pre-steady-state kinetic study of substrate specificity of Escherichia coli formamidopyrimidine--DNA glycosylase. Biochemistry. 2007;46(2):424–435. doi: 10.1021/bi060787r. [DOI] [PubMed] [Google Scholar]
- Kuznetsov NA, Vorobjev YN, Krasnoperov LN, Fedorova OS. Thermodynamics of the multi-stage DNA lesion recognition and repair by formamidopyrimidine-DNA glycosylase using pyrrolocytosine fluorescence--stopped-flow pre-steady-state kinetics. Nucleic Acids Res. 2012;40(15):7384–7392. doi: 10.1093/nar/gks423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov NA, Zharkov DO, Koval VV, Buckle M, Fedorova OS. Reversible chemical step and rate-limiting enzyme regeneration in the reaction catalyzed by formamidopyrimidine-DNA glycosylase. Biochemistry. 2009;48(48):11335–11343. doi: 10.1021/bi901100b. [DOI] [PubMed] [Google Scholar]
- Laspia MF, Wallace SS. Excision repair of thymine glycols, urea residues, and apurinic sites in Escherichia coli. J Bacteriol. 1988;170(8):3359–3366. doi: 10.1128/jb.170.8.3359-3366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bihan YV, Angeles Izquierdo M, Coste F, Aller P, Culard F, Gehrke TH, Essalhi K, Carell T, Castaing B. 5-Hydroxy-5-methylhydantoin DNA lesion, a molecular trap for DNA glycosylases. Nucleic Acids Res. 2011;39(14):6277–6290. doi: 10.1093/nar/gkr215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesiak KB, Wheeler KT. Formation of alpha-deoxyadenosine in polydeoxynucleotides exposed to ionizing radiation under anoxic conditions. Radiat Res. 1990;121(3):328–337. [PubMed] [Google Scholar]
- Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci U S A. 2010;107(11):4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Holmes A, Averill AM, Cannan W, Wallace SS. Expression and purification of active mouse and human NEIL3 proteins. Protein Expr Purif. 2012;84(1):130–139. doi: 10.1016/j.pep.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Doublie S, Wallace SS. Neil3, the final frontier for the DNA glycosylases that recognize oxidative damage. Mutat Res. 2013a;743–744:4–11. doi: 10.1016/j.mrfmmm.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Imamura K, Averill AM, Wallace SS, Doublié S. Structural characterization of a mouse ortholog of human NEIL3 with a marked preference for single-stranded DNA. Structure. 2013b;21:1–10. doi: 10.1016/j.str.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal SM, Hegde ML, Chatterjee A, Hegde PM, Szczesny B, Banerjee D, Boldogh I, Gao R, Falkenberg M, Gustafsson CM, et al. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3'-phosphatase in maintenance of mitochondrial genome. J Biol Chem. 2012;287(4):2819–2829. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick JP, Yeh YC, Solomon M, Essigmann JM, Lu AL. Characterization of a mammalian homolog of the Escherichia coli MutY mismatch repair protein. Mol Cell Biol. 1995;15(2):989–996. doi: 10.1128/mcb.15.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melamede RJ, Hatahet Z, Kow YW, Ide H, Wallace SS. Isolation and characterization of endonuclease VIII from Escherichia coli. Biochemistry. 1994;33(5):1255–1264. doi: 10.1021/bi00171a028. [DOI] [PubMed] [Google Scholar]
- Mitra S, Izumi T, Boldogh I, Bhakat KK, Hill JW, Hazra TK. Choreography of oxidative damage repair in mammalian genomes. Free Radic Biol Med. 2002;33(1):15–28. doi: 10.1016/s0891-5849(02)00819-5. [DOI] [PubMed] [Google Scholar]
- Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21(1):103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30(22):4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E, Boiteux S, Cunningham RP, Epe B. Enzymatic recognition of DNA modifications induced by singlet oxygen and photosensitizers. Nucleic Acids Res. 1990;18(20):5969–5973. doi: 10.1093/nar/18.20.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem Res Toxicol. 2006;19(4):491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- Nelson SR, Dunn AR, Wallace SS, Warshaw DM. DNA Glycosylases Search for Oxidized DNA Bases: Single molecule approaches to understanding search mechanisms and lesion encounters. 2013 Submitted. [Google Scholar]
- O'Connor TR, Graves RJ, de Murcia G, Castaing B, Laval J. Fpg protein of Escherichia coli is a zinc finger protein whose cysteine residues have a structural and/or functional role. J Biol Chem. 1993;268(12):9063–9070. [PubMed] [Google Scholar]
- Odell ID, Wallace SS, Pederson DS. Rules of engagement for base excision repair in chromatin. J Cell Physiol. 2013;228(2):258–266. doi: 10.1002/jcp.24134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendyala S, Natarajan V. Redox regulation of Nox proteins. Respir Physiol Neurobiol. 2010;174(3):265–271. doi: 10.1016/j.resp.2010.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005;33(18):5936–5944. doi: 10.1093/nar/gki879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope MA, David SS. DNA damage recognition and repair by the murine MutY homologue. DNA Repair (Amst) 2005;4(1):91–102. doi: 10.1016/j.dnarep.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Prakash A, Doublie S, Wallace SS. The Fpg/Nei family of DNA glycosylases: substrates, structures, and search for damage. Prog Mol Biol Transl Sci. 2012;110:71–91. doi: 10.1016/B978-0-12-387665-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumo DE, Barrantes-Reynolds R, Kathe SD, Wallace SS, Bond JP. Evolution of the Fpg/Nei family of DNA glycosylases. In: Kovalchuk I, Kovalchuk O, editors. Genome Instability and Transgenerational Effects. New York: Nova Science Publishers, Inc.; 2010. pp. 71–88. [Google Scholar]
- Qi Y, Nam K, Spong MC, Banerjee A, Sung RJ, Zhang M, Karplus M, Verdine GL. Strandwise translocation of a DNA glycosylase on undamaged DNA. Proc Natl Acad Sci U S A. 2012;109(4):1086–1091. doi: 10.1073/pnas.1111237108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Spong MC, Nam K, Banerjee A, Jiralerspong S, Karplus M, Verdine GL. Encounter and extrusion of an intrahelical lesion by a DNA repair enzyme. Nature. 2009;462(7274):762–766. doi: 10.1038/nature08561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Spong MC, Nam K, Karplus M, Verdine GL. Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM. J Biol Chem. 2010;285(2):1468–1478. doi: 10.1074/jbc.M109.069799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae . Proc Natl Acad Sci U S A. 1997;94(15):8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94(15):8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolseth V, Runden-Pran E, Luna L, McMurray C, Bjoras M, Ottersen OP. Widespread distribution of DNA glycosylases removing oxidative DNA lesions in human and rodent brains. DNA Repair (Amst) 2008;7(9):1578–1588. doi: 10.1016/j.dnarep.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist TA, Zharkov DO, Grollman AP. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc Natl Acad Sci U S A. 1997;94(14):7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker AH, Ikeda S, Nakano H, Terato H, Ide H, Imai K, Akiyama K, Tsutsui K, Bo Z, Kubo K, et al. Cloning and characterization of a mouse homologue (mNthl1) of Escherichia coli endonuclease III. J Mol Biol. 1998;282(4):761–774. doi: 10.1006/jmbi.1998.2042. [DOI] [PubMed] [Google Scholar]
- Schurr JM. The one-dimensional diffusion coefficient of proteins absorbed on DNA. Hydrodynamic considerations. Biophys Chem. 1979;9(4):413–414. [PubMed] [Google Scholar]
- Serre L, Pereira de Jesus K, Boiteux S, Zelwer C, Castaing B. Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 2002;21(12):2854–2865. doi: 10.1093/emboj/cdf304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupska MM, Baikalov C, Luther WM, Chiang JH, Wei YF, Miller JH. Cloning and sequencing a human homolog (hMYH) of the Escherichia coli mutY gene whose function is required for the repair of oxidative DNA damage. J Bacteriol. 1996;178(13):3885–3892. doi: 10.1128/jb.178.13.3885-3892.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slupska MM, Luther WM, Chiang JH, Yang H, Miller JH. Functional expression of hMYH, a human homolog of the Escherichia coli MutY protein. J Bacteriol. 1999;181(19):6210–6213. doi: 10.1128/jb.181.19.6210-6213.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenken S. Purine bases, nucleosides, and nucleotides: aqueous solution redox chemistry and transformation reactions of their radical cations and e- and OH adducts. Chem Rev. 1989;89:503–520. [Google Scholar]
- Strniste GF, Wallace SS. Endonucleolytic incision of x-irradiated deoxyribonucleic acid by extracts of Escherichia coli. Proc Natl Acad Sci U S A. 1975;72(6):1997–2001. doi: 10.1073/pnas.72.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S. Crystal structure of a repair enzyme of oxidatively damaged DNA MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J. 2000;19(15):3857–3869. doi: 10.1093/emboj/19.15.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung RJ, Zhang M, Qi Y, Verdine GL. Structural and biochemical analysis of DNA helix invasion by the bacterial 8-oxoguanine DNA glycosylase MutM. J Biol Chem. 2013;288(14):10012–10023. doi: 10.1074/jbc.M112.415612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem. 2002;277(44):42205–42213. doi: 10.1074/jbc.M206884200. [DOI] [PubMed] [Google Scholar]
- Takao M, Oohata Y, Kitadokoro K, Kobayashi K, Iwai S, Yasui A, Yonei S, Zhang QM. Human Nei-like protein NEIL3 has AP lyase activity specific for single-stranded DNA and confers oxidative stress resistance in Escherichia coli mutant. Genes Cells. 2009;14(2):261–270. doi: 10.1111/j.1365-2443.2008.01271.x. [DOI] [PubMed] [Google Scholar]
- Tchou J, Grollman AP. The catalytic mechanism of Fpg protein. Evidence for a Schiff base intermediate and amino terminus localization of the catalytic site. J Biol Chem. 1995;270(19):11671–11677. doi: 10.1074/jbc.270.19.11671. [DOI] [PubMed] [Google Scholar]
- Tchou J, Michaels ML, Miller JH, Grollman AP. Function of the zinc finger in Escherichia coli Fpg protein. J Biol Chem. 1993;268(35):26738–26744. [PubMed] [Google Scholar]
- Theriot CA, Hegde ML, Hazra TK, Mitra S. RPA physically interacts with the human DNA glycosylase NEIL1 to regulate excision of oxidative DNA base damage in primer-template structures. DNA Repair (Amst) 2010;9(6):643–652. doi: 10.1016/j.dnarep.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torisu K, Tsuchimoto D, Ohnishi Y, Nakabeppu Y. Hematopoietic tissue-specific expression of mouse Neil3 for endonuclease VIII-like protein. J Biochem. 2005;138(6):763–772. doi: 10.1093/jb/mvi168. [DOI] [PubMed] [Google Scholar]
- Verdine GL, Norman DP. Covalent trapping of protein-DNA complexes. Annu Rev Biochem. 2003;72:337–366. doi: 10.1146/annurev.biochem.72.121801.161447. [DOI] [PubMed] [Google Scholar]
- von Sonntag C. The Chemical Basis of Radiation Biology. London: Taylor and Francis; 1987. [Google Scholar]
- Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33(1):1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- Wallace SS, Bandaru V, Kathe SD, Bond JP. The enigma of endonuclease VIII. DNA Repair (Amst) 2003;2(5):441–453. doi: 10.1016/s1568-7864(02)00182-9. [DOI] [PubMed] [Google Scholar]
- Wallace SS, Murphy DL, Sweasy JB. Base excision repair and cancer. Cancer Lett. 2012;327(1–2):73–89. doi: 10.1016/j.canlet.2011.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, Wilson SH, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol Cell. 2004;15(2):209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Zahn KE, Wallace SS, Doublie S. DNA polymerases provide a canon of strategies for translesion synthesis past oxidatively generated lesions. Curr Opin Struct Biol. 2011;21(3):358–369. doi: 10.1016/j.sbi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaika EI, Perlow RA, Matz E, Broyde S, Gilboa R, Grollman AP, Zharkov DO. Substrate discrimination by formamidopyrimidine-DNA glycosylase: a mutational analysis. J Biol Chem. 2004;279(6):4849–4861. doi: 10.1074/jbc.M310262200. [DOI] [PubMed] [Google Scholar]
- Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21(4):789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liu M, Fleming AM, Bujrrows CJ, Wallace SS. Neil3 and NEIL1 DNA glycosylases remove oxidative damages from quadruplex DNA and exhibit preferences for lesions in the telomeric sequence context. J Biol Chem. 2013 doi: 10.1074/jbc.M113.479055. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]