Abstract

Systems toxicology is a broad-based approach to describe many of the toxicological features that occur within a living system under stress or subjected to exogenous or endogenous exposures. The ultimate goal is to capture an overview of all exposures and the ensuing biological responses of the body. The term exposome has been employed to refer to the totality of all exposures, and systems toxicology investigates how the exposome influences health effects and consequences of exposures over a lifetime. The tools to advance systems toxicology include high-throughput transcriptomics, proteomics, metabolomics, and adductomics, which is still in its infancy. A well-established methodology for the comprehensive measurement of DNA damage resulting from every day exposures is not fully developed. During the past several decades, the 32P-postlabeling technique has been employed to screen the damage to DNA induced by multiple classes of genotoxicants; however, more robust, specific, and quantitative methods have been sought to identify and quantify DNA adducts. Although triple quadrupole and ion trap mass spectrometry, particularly when using multistage scanning (LC–MSn), have shown promise in the field of DNA adductomics, it is anticipated that high-resolution and accurate-mass LC–MSn instrumentation will play a major role in assessing global DNA damage. Targeted adductomics should also benefit greatly from improved triple quadrupole technology. Once the analytical MS methods are fully mature, DNA adductomics along with other -omics tools will contribute greatly to the field of systems toxicology.
1. Systems Toxicology of DNA Damage
Traditional toxicology is rapidly evolving into a systems-based approach whereby many toxicological interactions that occur within a living system under stress or subjected to a variety of endogenous and/or exogenous exposures are captured. Systems toxicology approaches have been developed as a means to understand better and predict potential toxicities of specific compounds (e.g., early-stage drug development), to perform more accurate and comprehensive assessment of environmental exposures, or to assess the gene–environment interaction in disease causation.1−5
It is now well-recognized that understanding the etiology of complex diseases such as cancer requires understanding the human body’s response to exposures to many hazardous chemicals. The identification of toxic chemicals that enter the body from exogenous sources such as air pollutants, radiation, water contaminants, food, and drugs must be considered together with endogenous chemicals derived from cellular metabolism or endogenous processes, including inflammation, oxidative stress, infection, and chemicals derived from complex interactions with the gut flora.4−6
The recognized impact of chemicals in the environment and within the body on human health has led to the development of the exposome concept. The exposome refers to the totality of exposures received by a person during life, including exposures from life-style factors, from the prenatal period to death. This concept was developed to underline the need for better tools and techniques to provide a more accurate environmental exposure assessment in human health studies.7 The notion of the exposome leads to a new definition of the measurement of environmental exposures that includes the body’s internal chemical environment and the external environmental influences as well as their associated biological responses.8,9 The exposome is highly dynamic, evolving over time, and thus can only be measured as a cumulative exposure at a certain point in time.
The ability to investigate the exposome is possible due to the development of several new powerful technologies such as high-throughput transcriptomics, proteomics, and metabolomics.4,10,11 These techniques can be used to investigate an individual’s response to exposure to complex mixtures taking into account interindividual variations in bioavailability, absorption, metabolism, and excretion of compounds. The presence of endogenous reactive electrophiles, formed as a consequence of cellular processes such as inflammation, can also impact health and may trigger responses in the body such as formation of modified proteins or DNA. The investigation of these effects has led to the development of adductomic approaches for the investigation of protein adducts12 and DNA adducts.13
The exposure to genotoxic chemicals and their reactive metabolites induces chemical modifications of DNA, known as DNA adducts, which have been studied extensively and play a key role in chemically induced carcinogenesis.14−16 DNA adducts, if not repaired, can lead to mutations during cell division and may ultimately disrupt the regular functioning of the systems regulating normal cell growth. Quantitation of the levels of specific DNA adducts can provide valuable information about the biologically effective dose of the procarcinogen. The measurement of DNA adducts resulting from the exposure to a specific carcinogen can provide crucial information about the genotoxic potential of the chemical and its mechanism of carcinogenesis.17,18 The DNA adducts of specific chemicals have been employed as biomarkers in molecular epidemiology and cancer prevention studies.19−25
Studies on Aflatoxin B1 (AFB1) and aristolochic acid (AA) are two prime examples where the molecular epidemiologic data on chemical exposures, chemical-specific DNA adducts in target tissues, and mutation spectra in tumor-related genes have provided a mechanistic understanding of the causal role for a chemical in the development of cancer.26−29 AFB1 is a fungal toxicant and a potent animal and human hepatocellular carcinogen that can be found as a contaminant in various crops.26,27 AA, a carcinogen present in the plant species of the genus Aristolochia, is a causal agent of upper urothelial tract cancer in subjects of the Balkans and Taiwan.28,30−32 The positive associations observed between dietary AFB1 exposure and the incidence of hepatocellular carcinoma in Asia and Africa were greatly strengthened by the application of validated DNA adduct biomarkers; specifically, when a characteristic G:C → T:A transversion at the third base in codon 249 of the TP53 tumor-suppressor gene was attributed to the N7 guanine adduct of AFB1.26,33,34 There is also compelling molecular epidemiology data on AA exposure and cancer risk: the N6 deoxyadenosine adduct of AA, which is responsible for otherwise rare A:T → T:A transversions, is frequently observed in the TP53 gene in urothelial carcinomas of patients.28,31,32
The data of these molecular epidemiologic studies combined with the mechanistic information on chemical carcinogenesis have firmly established causative linkages between exposure to AFB1 and AA and cancer risk. These investigations,26−28,100 are the gold standard in epidemiological studies that seek to identify chemical exposures and increased cancer risk. AFB1 and AA are highly potent human carcinogens, where elevated levels of exposure and unique mutational signatures were used to establish the causative role of these chemicals in human cancer. Many genotoxicants do not display such unique or characteristic mutational fingerpints in tumor-related genes, and demonstrating causality of specific chemicals in cancer will be difficult.
The ability to biomonitor many DNA adducts may permit the identification of exposures to certain hazardous chemicals in the environment and permit strategies to mitigate the exposures and the risk of cancer. These considerations of exposures to multiple genotoxicants in the environment and diet together with the need to develop methods to screen for unknown DNA adducts formed after exposure to specific toxicants have prompted researchers to develop a new DNA adductomic approach to screen for many DNA lesions simultaneously,35,36 ultimately aiming at developing a sensitive, versatile, and robust DNA adductomic approach, which would be a very powerful tool for exposome characterization and cancer-based systems toxicology studies.
2. Methods for Screening DNA Adducts
Several methods for DNA adduct analysis exist, including 32P-postlabeling, LC–MS, GC–MS, CE–MS, fluorescence, immunoassay, and electrochemical detection.37,38 Many DNA adduct investigations from the 1980s until the present were predominately done using the 32P-postlabeling methodology, which is well-suited to a broad-based DNA adduct analysis because of its inherent ability to monitor many adducted nucleotides simultaneously in a given sample. (39) More recently, MS analysis has emerged as the preferred technique because it provides a selective and quantitative measure of specific DNA modifications.40−42 The recent improvements in MS scanning acquisition capabilities and instrument sensitivity have set the stage for the use of MS in high-throughput applications in DNA adductomic approaches. Both 32P-postlabeling and MS-based DNA adductomic methodologies are discussed in the following sections.
2.1. 32P-Postlabeling
The 32P-postlabeling assay was developed by Kurt and Erica Randerath, Ramesh Gupta, and Vijay Reddy at Baylor College of Medicine in Houston more than 30 years ago.43−45 This technique employs T4 polynucleotide kinase to incorporate enzymatically [γ-32P]ATP of very high specific activity (>5000 Ci/mmol) into carcinogen-adducted 3′ nucleotides to form the radiolabeled 3′,5′-bisnucleotide adducts (Figure 1). The technique has been applied to screen for DNA adducts formed by more than 100 chemicals that include aristolochic acids, aromatic amines, azo compounds, estrogens, heterocyclic aromatic amines, lipid peroxidation products, methylating agents, mycotoxins, nitro aromatics, and polycyclic aromatic hydrocarbons and other genotoxicants.19,31,39,46−51
Figure 1.
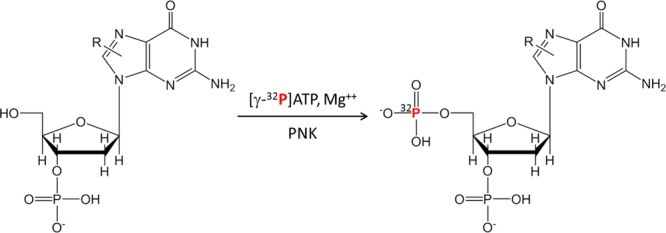
DNA is enzymatically digested to 3′ mononucleotides, and the postlabeling is then achieved by transfer of 32P-orthophosphate from [γ-32P]ATP to the 5′-OH position of the deoxyribonucleotide adduct. This reaction is mediated by polynucleotide kinase (PNK).
The 32P-postlabeling assay was a great advancement in the biomonitoring of DNA adducts because it was the first method that could be employed in human studies where there are potentially many hazardous chemicals present in the environment. Several versions of the 32P-postlabeling method were established, employing limiting concentrations of [γ-32P]ATP to label DNA adducts selectively over nonmodified nucleotides, different conditions of enzymatic digestion of DNA, or pre-enrichment of DNA adducts from nonmodified 3′ nucleotides by extraction with n-butanol prior to postlabeling. These assays resulted in the enhancement of the sensitivity of DNA adduct detection. Despite the fact that these modifications to the original assay were made more than 20 years ago, they still remain the primary methods for postlabeling today.39,45,52 Certain DNA adducts can be detected at levels approaching 1 adduct per 1010 nucleotides. The 32P-labeled adducts are separated from nonmodified radiolabeled 3′,5′ nucleotides (pNps) by two-dimensional TLC, and the adducts are either visualized as a pattern of spots by autoradiography or seen as chromatographic peaks by HPLC with radioactive detection.18,43,45,53
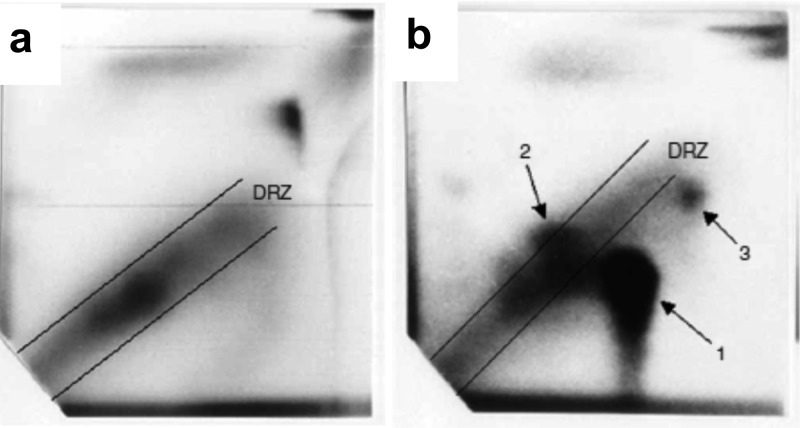
The 32P-postlabeling method has been successfully employed to screen for DNA adducts in lung, liver, esophagus, pancreas, colon, prostate, mammary gland, white blood cells,50,54−56 and exfoliated epithelial cells in urine of smokers57 or epithelial cells in the milk of lactating mothers.58 Other studies have screened for DNA adducts derived from tobacco genotoxicants or lipid peroxidation products in the oral cavity and sputum from lung cancer patients.59−61 The analysis of DNA of smokers by 32P-postlabeling has revealed the presence of smoking-related adducts in many tissues.55,62 The complex array of lesions is often seen as a diffuse diagonal radioactive zone (DRZ) on TLC plates and probably represents a mixture of DNA adducts derived from many genotoxicants in tobacco smoke, as shown in Figure 2a.55,62,63 The chemical structures of the smoking-related adducts observed in the DRZ are unknown.64 Some of the lesions were thought to be derived from PAHs or aromatic amines; however, Arif et al. recently reported that many of the cigarette smoke-associated lung DNA adducts appearing on their chromatograms as DRZs were not related to either PAHs or aromatic amines.59 In other studies, putative adducts of aromatic amines, heterocyclic aromatic amines, polycyclic aromatic hydrocarbons, and aristolochic acid (Figure 2b) have been tentatively identified by comigration of the 32P-labeled adduct with synthetic DNA adduct standards using HPLC with UV detection.46,58,63,65,66
Figure 2.
(a) Specimen obtained from a smoker in which a diagonal radioactive zone (DRZ) is present (the typical pattern of smoking-related DNA adducts). This analysis is from an autoradiogram of DNA adducts in lymphocytes from a smoker. (b) Autoradiogram of DNA adducts in renal tissue from a patient with aristolochic acid nephropathy using the 32P-postlabeling method. In spot 1, the adduct was identified by cochromatography with a synthesized standard as 7-(deoxyadenosine-N6-yl)-aristolactam I, in spot 2 as 7 (deoxyguanosine-N2-yl)-aristolactam I, and in spot 3 as 7-(deoxyadenosine-N6-yl)-aristolactam II. Adapted with permission from ref (63), copyright 2007, Macmillan Publishers Ltd.
The DNA adduct biomarker data obtained from numerous studies employing the 32P-postlabeling assay reveal that human DNA is modified with many different environmental, dietary genotoxicants, and endogenous electrophiles and that the level of DNA damage can be potentially influenced by lifestyle and host factors.55,64 The 32P-postlabeling assay remains a mainstay for biomonitoring exposure and screening for DNA damage in humans because it is a highly sensitive technique and the cost of establishing an analytical laboratory is relatively low. However, the 32P-method has important limitations. The technique is labor-intensive and requires significant amounts of radioactive phosphorus, a strong β-emitter and potential health hazard; there are a lack of suitable internal standards to account for adduct recovery and labeling efficiency, which can vary by more than 100-fold;67 and there is no structural information about the lesion, which leaves the identity of the adduct ambiguous; in addition, different kinds of adducts tend to require different assay conditions. Finally, the identification and quantification of DNA adducts by the 32P-postlabeling method are particularly challenging in humans, where many overlapping lesions may be present.55,64
2.2. Mass Spectrometry Approaches for DNA Adductomics
Liquid chromatography–mass spectrometry (LC–MS) is currently the primary method for characterization and quantitation of covalent modification of DNA,13,37,68 commonly performed using tandem mass spectrometry and positive mode electrospray ionization. Gas chromatography–mass spectrometry (GC–MS) is less commonly used because many adducts are thermally unstable. Nevertheless, chemical derivatization of the polar nucleosides or more commonly the aglcyone adducts has made some types of DNA adducts amenable to GC–MS-based analysis.69 GC–MS has been used primarily for specific classes of DNA adducts, such as those resulting from the reaction of reactive oxygen species (•OH) with DNA. Care must be taken to avoid artifactual formation of oxidized bases during the derivatization at high temperature of the hydrolysate, thus potentially reducing the versatility of the application of this technique.70
Mass spectrometry-based analysis of DNA modifications traditionally has been a bottom-up approach where analyses focus on small numbers of anticipated DNA adducts based on a priori assumptions regarding the formation of specific adducts and in vitro analysis. Typically, studies focus on the identification and quantitation of a limited number of DNA modifications arising from a specific exposure and/or chemical interaction. However, to measure DNA damage comprehensively, which is required by the systems toxicology paradigm, a new top-down DNA adductomics approach is required that can simultaneously screen for multiple DNA adducts derived from known and unknown exposures and from biological responses to exposures such as induced formation of endogenous electrophiles. Initial investigations of various biological samples have been performed,35,36,68,71−80 and effective methodologies are poised to come to the forefront with rapidly improving instrumentation and additional development work aimed at optimizing chromatography, sample preparation, and data collection and analysis.
LC–MSn-based DNA adductomics investigations take advantage of the common structural feature of deoxyribonucleosides: a deoxyribose moiety bound to the nucleobase through a glycosidic bond.81 The product ion spectra of structurally modified DNA nucleosides in the positive ionization mode (A–B–dR, where B = nucleobase, A = modification, and dR = 2′-deoxyribose sugar) acquired under low-energy collision-activated dissociation (CAD) are dominated by the cleavage of the glycosidic bond and a neutral loss of dR (116 amu), leading to protonated nucleobases ([B–A] + H)+, as shown in Figure 3.
Figure 3.
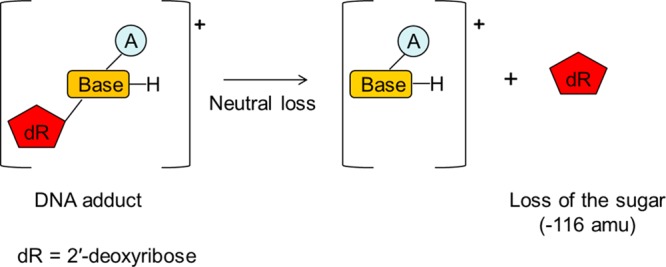
2′-Deoxyribose is the sugar moiety present in all DNA adducts. Low-energy CAD spectra of structurally modified DNA nucleosides (A–B–dR, where B = nucleobase, A = modification, and dR = 2′-deoxyribose sugar) are dominated by the cleavage of the glycosidic bond and a neutral loss of dR (116 amu), leading to protonated nucleobase ions ([B–A] + H)+. Monitoring the neutral loss can be used for DNA adduct screening via LC–MSn methodologies.
DNA adducts that are exceptions to this paradigm are those (1) formed on the phosphate portion of DNA, (2) excreted as free nucleobases in urine because of spontaneous hydrolysis, depurination, and/or active repair,82−84 and (3) structurally similar to the DB[a,I]P dihydroepoxide adduct of dA, which upon fragmentation was found to produce mostly ions corresponding to the DB[a,I]P moiety.78 In spite of these exceptions, the neutral loss of the deoxyribose group is widely universal and can be used for screening DNA adducts via LC–MSn methodologies.
3. Sample Source and Preparation
Common biological sources of DNA include tissues, blood, oral cells, and urine. For quantitative analysis of DNA adducts, 1–200 μg of DNA is required depending on the abundance of the lesion and the sensitivity of the analytical method. The amount of DNA required for successful adduct detection via DNA adductomic investigations will depend greatly on the level of adduct present, the analytical approach, the ionization efficiency of the adduct, and the MS instrumentation.13 More DNA is required for adductomic analysis than for targeted DNA adduct quantitation using the same instrumentation and analytical parameters. The breadth of DNA adductomics is dependent on the efficiency of digestion of chemically modified DNA, and adducts of different structures may require different cocktails of enzymes for quantitative digestion of DNA to the mononucleoside adducts (typical combinations of enzymes are nucleases or DNases used with phosphodiesterases and alkaline phosphatase).85
After DNA hydrolysis, DNA adducts must be enriched and purified from the nonmodified deoxynucleosides that are present in 106 to 109 excess. Therefore, the DNA hydrolysates are often processed by solid-phase extraction cleanup steps to enrich DNA adducts and to remove the bulk of unmodified nucleosides, proteins, inorganic salts, and other sample components that can interfere with MS analysis.37,41 The cleanup of DNA adductomic samples requires great care to avoid inadvertent loss of unknown or unanticipated adducts, especially hydrophilic adducts. If a specific class of adducts are the target of the analysis, then more selective enrichment procedures may be possible. Finally, the ionization efficiencies of different classes of DNA adducts vary and therefore pH and solvent compositions must be optimized for LC–MS measurements.
4. Scanning Modes
Constant neutral loss (CNL) scanning using triple quadrupole MS instrumentation has been a common approach for DNA adductomic analysis.73,77,78 In this scan mode, shown in Figure 4, quadrupole 1 (Q1) and Q3 scan in sync offset from each other by the user-defined mass difference (m/z = 116 amu). Voltage is applied to the collision cell (q2) filled with inert gas (typically argon) to induce fragmentation. Only ions that fragment with the specific neutral loss will pass through Q1, q2, and Q3 and be recorded as signal.
Figure 4.
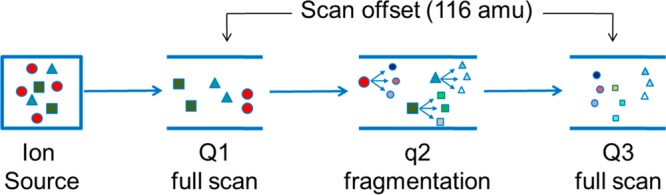
Constant neutral loss scanning method using triple quadrupole instrumentation. Quadrupoles Q1 and Q3 scan in sync offset from each other by the user-defined mass difference (m/z = 116 amu). Voltage is applied to the collision cell (q2) filled with argon to induce fragmentation. Only ions that fragment with the specific neutral loss will pass through Q1, q2, and Q3 and be recorded as signal.
A variation is a pseudo-CNL approach35,36,72,74,75,79 whereby instead of actually scanning the quadrupoles, the system is set to monitor a number of contiguous selected reaction monitoring (SRM) transitions all involving a loss of 116 amu. Multiple injections are made (typically 7–15), each covering a different mass range, so that a large range is ultimately covered for a given sample.
The pseudo-CNL approach may provide superior sensitivity over the traditional CNL approach because multiple transitions, all employing the loss of 116 in SRM mode and covering a small mass range, are monitored, as opposed to CNL, where scanning is performed over a large mass range. Scanning of the triple quadrupole mass spectrometer results in a significant loss in sensitivity, relative to SRM operation, because of the poor duty cycle of individual ion mass detection.
The pseudo-CNL approach, requiring multiple injections, is more time-consuming and susceptible to instrument variability than traditional CNL. The newer generation of triple quadrupole instruments are capable of rapid switching between SRM transitions and may permit a single analysis covering the entire mass range (e.g., 250–500 amu in 0.5 s) of interest. Conversely, the traditional CNL analysis can be performed over a smaller mass scan range with multiple injections to increase sensitivity. The relative merits of the traditional CNL and pseudo-CNL approaches require further evaluation with newer, more advanced triple quadrupole MS instrumentation. Relative ease of method set up and data analysis may provide an advantage of one approach over the other.
Another powerful MS-based DNA adductomics methodology uses data-dependent (DD) scanning, where repeated full-scan analysis is followed by fragmentation of the most abundant ions throughout the entire chromatographic run. DD scanning is performed in real time by the instrument software and can be optimized for improved DNA adduct coverage by avoiding repeated fragmentation of the most abundant ions. The acquisition method can be programmed such that masses of ions selected for fragmentation can be placed into a dynamic exclusion list that makes them no longer eligible for fragmentation for a predetermined length of time. In addition, a reject mass list can be programmed such that a list of interfering background ions can be excluded from analysis (fragmentation). Examples of the use of this approach include DD–MS2 analysis using quadrupole-time-of-flight instrumentation80 and DD–MS3 analysis using ion trap instrumentation, as illustrated in Figure 5.71,76
Figure 5.
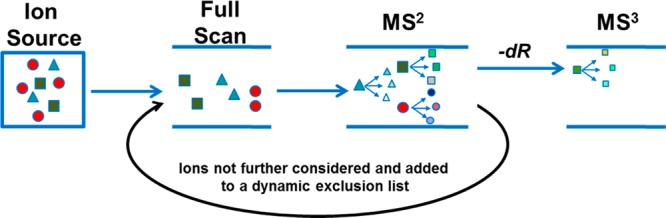
Data-dependent (DD) scanning method. Schematic representation of a DD–MS3 analysis. Repeated full-scan analysis is followed by fragmentation of the most abundant ions determined by the mass spectrometer software in real time throughout the entire chromatographic run based on the initial programming of the data-dependent method. The ions showing neutral loss of 116 amu (green) undergo additional fragmentation (MS3). The ion masses not showing neutral loss of 116 amu (red and blue) are put into an exclusion list so that they are no longer eligible for fragmentation for a predetermined length of time. To avoid repeated fragmentation of the most abundant ions, the method can be programmed such that masses of ions selected for fragmentation are also added to the exclusion list for a predetermined length of time. Adapted from ref (91). Copyright 2014 American Chemical Society.
5. DNA Adductomics Analysis Using Mass Spectrometry
The published studies of DNA adductomic analysis utilizing MS2 neutral loss of deoxyribose are summarized in Table 1. The efforts of the various research groups active in this field are briefly summarized in this section.
Table 1. Summary of Published LC–MSn-Based DNA Adductomics Studies.
| approach | instrument | sample type | adduct type | details | ref |
|---|---|---|---|---|---|
| CNL | triple quad | in vitro | PhIPa | early report of MS adductomics | (77) |
| triple quad | in vitro animal tissues | IQb | electrospray at ∼500 nL/min | (73) | |
| triple quad | in vitro | polyaromatic hydrocarbons | column switching 500–650 Da | (78) | |
| pseudo-CNL | triple quad | human lung | screening of unknowns | method development adductome map data analysis | (35) |
| triple quad | human lung and esophagus | screening of unknowns | seven adducts unambiguously detected | (74) | |
| triple quad | various human tissues | screening of unknowns | adducts detected at ∼1 adduct per 107 bases | (72) | |
| triple quad | food (Quorn, button mushrooms, brewer’s yeast) | screening of unknowns | 7 SRM transitions per injection | (79) | |
| triple quad | in vitro | micronucleus test-positive compounds | application to micronucleus test | (75) | |
| triple quad | human gastric mucosa | screening of unknowns | first report of lipid peroxidation-related adducts in human | (36) | |
| DD–MS2 | Q-TOF | in vitro cells | melphalan | column switching, 5 μl/min | (80) |
| DD–CNL–MS3 | ion trap | human hepatocytes (treated) | 4-ABPc | human samples examined MS3 data acquisition | (71) |
| rat liver | MeIQxd | ||||
| buccal cells (in vivo) | tobacco constituents | ||||
| ion trap | in vitro | illudin S | used similar method to ref (71) | (76) |
2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP).
2-Amino-3-methylimidazo[4,5-f]quinolone (IQ).
4-Aminobiphenyl (4-ABP).
2-Amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx).
Vouros and co-workers and Turesky and co-workers were the among the first researchers using this approach to screen for DNA adducts.73Already in 1994, Wolf and Vouros applied a CNL approach using a capillary liquid chromatography-continuous flow fast atom bombardment mass spectrometry method for the screening of adducts formed in vitro by the reaction of N-acetoxy-N-acetyl-2-aminofluorene with calf thymus DNA.81 Additional initial efforts by Turesky’s group77 investigated adducts formed in vitro from 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP), a carcinogen formed in cooked meats. CNL methodology with triple quadrupole instrumentation and capillary chromatography was used to identify three PhIP-related adduction products that were shown to be dG–C8 adducts of PhIP (dG–C8–PhIP) and two ring-opened oxidized derivatives of dG–C8–PhIP. This effort was followed by in vitro and in vivo investigations of DNA adducts derived from 2-amino-3-methylimidazo[4,5-f]quinoline (IQ), another structurally related food-derived mutagen.73 A similar CNL scanning methodology was employed with a triple quadrupole MS, and nanoflow conditions were used to improve sensitivity.86 The investigators directly compared the screening of DNA adducts of kidney tissues of monkeys chronically fed with IQ by SRM and CNL with 32P-postlabeling data; the same pattern of adducts was identified by both MS and 32P-postlabeling techniques.
Esmans and co-workers were the first researchers to demonstrate the use of data-dependent scanning for DNA adduct screening. They used quadrupole-TOF instrumentation to analyze DNA treated with melphalan in vitro, testing single nucleosides, calf thymus DNA, and DNA from Jurkat cells.80 A column-switching device was used for online sample cleanup and operated under capillary flow conditions (5 μL/min). In spite of acquiring data by high-resolution TOF MS, the results were reported with nominal masses, and the accurate mass capabilities of this instrumentation were not utilized.
Matsuda and co-workers have published a series of investigations20,35,36,72,74,75,80 after establishing a pseudo-CNL methodology.35 They employed a triple quadrupole MS and acquired 32 SRM transitions per injection with 12 injections per sample to cover a mass range of m/z 228.8 to 602.8 amu. The DNA from lung tissue of a nonsmoker and a smoker was analyzed to generate what the authors refer to as an adductome map (Figure 6). The adductome map is a plot of ion signal intensity (indicated as circles whose diameters are proportional to their relative intensities) with m/z as the y axis and retention time as the x axis. The comparison between maps obtained for the nonsmoker and smoker samples permits the visualization of putative adducts of different intensities in the two samples. This scanning method was then used to measure the intertissue DNA damage variation in human lung and esophagus,74 lipid peroxidation-induced DNA adducts in human autopsy tissues,72 DNA adducts induced by DNA-damaging capability of in vitro micronucleus test-positive compounds,75 and lipid peroxidation-induced DNA adducts in human gastric mucosa.36 More than 100 putative DNA lesions were detected by this method. Although the CNL and pseudo-CNL approaches do allow for the screening of multiple putative lesions in DNA, the observed signal could be a result of other compounds in the DNA digest matrix that lose 116 amu upon collision-induced dissociation.
Figure 6.
Two adductome maps of putative DNA adducts detected in central (A) and peripheral (B) human lung tissue DNA from the same individual. The neutral loss of 2′-deoxyribose from positively ionized 2′-deoxynucleoside putative adducts was analyzed by LC/ESI-MS/MS in MRM mode transmitting the [M + H]+ → [M + H – 116]+ transition over a total of 374 transitions in the mass range from m/z 228.8 to 602.8. The graph plots the m/z of the observed putative adducts versus the retention time (tR). The adducts whose size is proportional to the signal intensity are represented with a circle. An active zone in which most of the putative adducts are observed is indicated by the box on the map. Attention is drawn to the presence of four putative adducts that were detected with relatively similar area response values in the active zone of the two lung DNA samples. These four unidentified putative adducts are designated in the adductome maps by letters a–d. The four putative adducts, (a) m/z 307.8, tR 11.46–11.48; (b) m/z 285.8, tR 11.48; (c) m/z 265.8, tR 13.36–13.40; and (d) m/z 283.8, tR 11.44–11.48, all possessed relatively similar area response values in lung tissue. Adapted from ref (74), copyright 2007, used with permission from Elsevier.
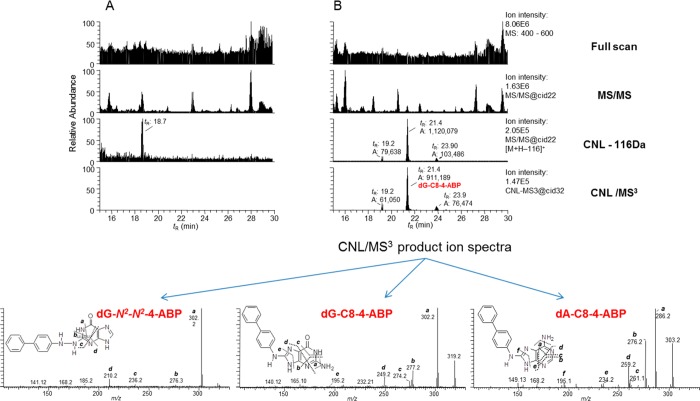
Turesky and co-workers used the DD–CNL–MS3 approach to screen for DNA adducts in human hepatocytes treated with 4-aminobiphenyl (4-ABP), in livers of rats exposed to 2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (MeIQx), and buccal cell DNA from tobacco smokers.71 The DD–CNL–MS3 approach involves the triggering of MS3 fragmentation following the neutral loss of 116 amu in the MS2 spectra of the putative adduct. They used a linear quadrupole ion trap MS and capillary column with a CaptiveSpray ion source (Bruker, Billerica, MA) to improve sensitivity. They reported that the DD–CNL–MS3 technique can be used to screen for DNA adducts derived from different classes of carcinogens at levels of adduct modification approaching 1 adduct per 108 unmodified DNA bases when 10 μg of DNA was employed. The data-dependent CNL–MS3 scan mode is a powerful postacquisition data-mining technique for discovery and structural elucidation of DNA adducts.71 The acquisition of the product ion spectrum at the MS3 scan stage provides spectra with rich structural information about the structure of the aglycone adduct, and multistage MSn scanning is a major advancement over the CNL scanning techniques, as shown in Figure 7.
Figure 7.
Example of a method used to screen for DNA adducts in human hepatocytes using data-dependent constant neutral loss–triple stage mass spectrometry performed on a linear quadrupole ion trap mass spectrometer. (A) Chromatograms obtained upon analysis of an untreated sample. (B) Chromatograms obtained upon analysis of a 4-ABP-treated sample; the [M + H – 116]+ ion, within the top 10 most abundant fragment ions in the MS/MS spectrum, triggers the acquisition of the MS3 spectrum, providing structural information on the detected DNA modifications. The full scan and MS/MS chromatograms are complex and show many features; however, the chromatogram displaying the ions that underwent a loss of 116 amu and the CNL-MS3 chromatogram are greatly simplified. Adapted from ref (71). Copyright 2009 American Chemical Society.
Berdal and co-workers used a pseudo-CNL method to investigate damaged deoxynucleosides present in a commercially available fungus-based food, Quorn, and in button mushrooms and dried powdered brewer’s yeast.79 They used a triple quadrupole instrument covering the mass range from m/z 240 to 406 amu with multiple injections (24) per sample and with seven SRM transitions per injection. They reported 90 putative DNA adducts.
Singh, Farmer, and co-workers developed a targeted adductomics approach for investigating in vitro the effects of exposure to polycyclic aromatic hydrocarbons (PAHs) present in industrial or urban air pollution, tobacco smoke, and cooked food.78 They used CNL (mass range of m/z 500–650 amu) with a triple quadrupole MS and online column switching along with software-based peak picking and integration. For method development, PAH-modified DNA samples were obtained by reaction of the antidihydrodiol epoxide metabolites of benzo[a]pyrene, benzo[b]fluoranthene, dibenzo[a,I]pyrene (DB[a,I]P), and dibenz[a,h]anthracene with calf thymus DNA.
Sturla and co-workers investigated the reaction of illudin S in the presence of prostaglandin reductase 1/NAD(P)H with calf thymus DNA using DD–CNL–MS3 with ion trap instrumentation.76 The methodology used was adapted from the work of Turesky and co-workers.71 No novel unknown DNA adducts were observed, however the analysis allowed for the observation that illudin S-derived DNA adducts are characterized by the same mass fragments as acylfulvene-derived DNA adducts.
6. Alternative Approaches
Inagaki suggested an alternative DNA adductomic approach whereby the aglycone guanine adducts can be identified by characteristic fragments of m/z 152 and 135 corresponding to protonated guanine and protonated [Gua + H–NH3]+, respectively.87 They suggested that a similar approach could be used for adenine adducts with characteristic fragments of m/z 136 and 119 (protonated adenine and protonated [Ade + H–NH3]+). They investigated the potential of this strategy by screening for acrylamide adducts formed from the reaction of glycidamide with dG and calf thymus DNA, and they report observing new DNA adducts where two molecules of acrylamide were attached to guanine and adenine. This method was done using precursor ion scanning and triple quadrupole instrumentation. One benefit of this approach is that it employs simple thermal or acid hydrolysis of DNA to release free nucleobases rather than enzymatic hydrolysis to nucleosides. However, many adducts formed at various sites of the guanine base do not undergo CID to produce protonated guanine [Gua + H–NH3]+ at m/z 152 or [Gua + H–NH3]+ at m/z 135. In addition, no discussion has been given to the detection of adducts of cytosine and thymine.
Giese and co-workers have developed a method for nontargeted analysis of modified nucleotides in DNA (and RNA) involving derivatization of the nucleotides with isotopologue benzoylhistamines followed by MALDI-TOF or MALDI-TOF/TOF analysis.88 Adduct identification is based on a combination of three features: phosphate-specificity of the tagging, detection of adducts as pair of signals, and measurement of a fragment that reveals whether a deoxyribose or ribose is present. The derivatization allows for adduct identification upon observation of a characteristic fragmentation pattern of the modified deoxyribose upon either postsource decay in the case of MALDI-TOF analysis or collision-induced dissociation with MALDI-TOF/TOF analysis. The derivatization also improves the sensitivity of adduct detection and allows for semiquantitation because of adduct signal responses that are comparable on a molar basis to within a factor of 3. The TOF analysis also provides the advantage of accurate mass analysis. The authors detected two known adducts, 5-hydroxymethylcytosine and 5-formylcytosine (fC), and one unknown adduct, 6-oxothymine, in a DNA sample from the human placenta of a smoker. This methodology was also used to profile adducts formed in vitro upon reaction of p-benzoquinone and DNA.89
7. Future Directions/Conclusions
DNA adductomics is a relatively new field, and with recent improvements in sensitivity, liquid chromatography–mass spectrometry (LC–MS) is primed to replace 32P-postlabeling as the preferred approach for DNA adduct screening in humans because of its selectivity and specificity and the structural information it provides. Rapidly improving high-resolution/accurate-mass LC–MS hybrid orbital trap and TOF instrumentation provides an extremely powerful platform for DNA adductomic analysis, providing both accurate-mass measurements for molecular formula determination and fragmentation information for adduct structural identification utilizing data-dependent methodologies. Castro-Perez et al. developed a CNL method utilizing Q-TOF technology for screening glutathione conjugates of various drugs in human and rat liver microsomes, illustrating the potential for the use of this approach for DNA adductomic analysis. (90) Recently, a high-resolution/accurate-mass DD–CNL–MS3 methodology for DNA adductomics has been developed using ion trap–orbital trap technology coupled with nano HPLC (300 nL/min).91
A new generation of triple quadrupole instrumentation has become dramatically more sensitive and capable of rapid switching between SRM transitions and faster scanning. Improvements in these capabilities make both the CNL and pseudo-CNL methodologies dramatically more powerful for the detection of low-level adducts, although the limitation of nominal mass determination still exists as does the lack of fragmentation beyond MS2. It is our experience that MS3 characterization of DNA adducts by linear quadrupole ion trap MS instrumentation provides high-quality spectra and structural features about the aglycone adducts. Also, it requires 10- to 100-fold less analyte than that required for triple quadrupole MS/MS scanning under elevated CAD conditions. Ion trap MS3 product ion spectra data can be acquired on DNA adducts in humans at levels of DNA modification as low as 3 adducts per 109 DNA bases.92,93
In addition, technological improvements, driven by the needs of proteomic analysis, have made nanospray operation increasingly routine. HPLC flow rates can relatively easily be reduced from the 100 to 500 μL/min employed in typical adductomics studies to date to capillary or nano HPLC rates, providing a significant increase in sensitivity because of the inverse relationship between electrospray sensitivity and flow rate.
Improvements in sample preparation and cleanup are also needed, especially when detection of hydrophilic adducts is considered. Data-dependent approaches will greatly benefit from less chemical noise because the ability to measure low-level adducts requires the instrument to dig through the background signal to sample the low-level adduct signals for MS2 analysis. Similarly, the CNL triple quadrupole methodology will be capable of lower levels of limits of detection with reduced background. Also, understanding the relative advantages and limitations of the various enzyme DNA hydrolysis cocktail protocols is critically important because of the potential difficulty in observing certain types of adducts resulting from incomplete enzyme hydrolysis.
The improvements in MS instrument performance are expected to result in the detection of many DNA adducts in humans. A bottleneck in DNA adductomics is expected to be the characterization of unknown adducts similar to that observed in metabolomics for characterization of unknown metabolites, even with accurate-mass measurements, product ion spectra, and available online metabolomics databases. A database of DNA adducts would be immensely helpful in the advancement of the field of DNA adductomics, but currently one does not exist in spite of the fact that hundreds of DNA adducts have been characterized by MS methods. The creation of a DNA adductomics database of accurate masses would require a thorough literature search of the molecular formulas of characterized DNA adducts and would be especially valuable for high-resolution/accurate-mass approaches. A compilation of fragmentation spectra by both ion trap and quadrupole-type fragmentation at the MS2 and MS3 levels at various collision energies would also be immensely helpful.
A potentially powerful new direction in DNA adduct screening would be targeted DNA adductomic analysis. One possible approach would be to use high-resolution/accurate-mass instrumentation whereby a large number of DNA adduct parent ion masses are put into a mass inclusion list to be selected under high mass accuracy criteria for MSn fragmentation. Another approach would be to take advantage of the rapid SRM switching capabilities (∼500/s) of the new generation of triple quadrupole instruments using a large list of potential DNA adducts to target for SRM detection. Both of these approaches will provide greater coverage of potential adducts in complex DNA digest mixtures by simultaneously targeting large numbers of suspected adducts or known DNA adducts of a particular class of genotoxicants.
Glossary
Abbreviations
- MS
mass spectrometry
- LC–MSn
liquid chromatography mass spectrometry with multistage scanning
- AB1
aflatoxin B1
- AA
aristolochic acid
- GC–MS
gas chromatography mass spectrometry
- CE–MS
capillary electrophoresis mass spectrometry
- pNps
nonmodified 3′,5′ nucleotides
- DRZ
diagonal radioactive zone
- TLC
thin-layer chromatography
- dR
2′-deoxyribose sugar
- DB[a,I]P
dibenzo[a,I]pyrene
- CNL
constant neutral loss
- SRM
selected reaction monitoring
- PhIP
2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine
- IQ
2-amino-3-methylimidazo[4,5-f]quinolone
- 4-ABP
4-aminobiphenyl
- MeIQx
2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline
- DD
data dependent
- TOF
time of flight
- PAH
polycyclic aromatic hydrocarbons
- MALDI
matrix-assisted laser desoption/ionization
This work was supported by NIH grants R01CA122320 (R.J.T.), R01ES0195 (R.J.T.), P01CA1600321 (R.J.T.), and P30CA077598-11 (Masonic Cancer Center Support grant, S.B. and P.W.V.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Waters M. D.; Fostel J. M. (2004) Toxicogenomics and systems toxicology: Aims and prospects. Nat. Rev. Genet. 5, 936–948. [DOI] [PubMed] [Google Scholar]
- Heijne W. H.; Kienhuis A. S.; van Ommen B.; Stierum R. H.; Groten J. P. (2005) Systems toxicology: Applications of toxicogenomics, transcriptomics, proteomics and metabolomics in toxicology. Expert Rev. Proteomics 2, 767–780. [DOI] [PubMed] [Google Scholar]
- Craig A.; Sidaway J.; Holmes E.; Orton T.; Jackson D.; Rowlinson R.; Nickson J.; Tonge R.; Wilson I.; Nicholson J. (2006) Systems toxicology: integrated genomic, proteomic and metabonomic analysis of methapyrilene induced hepatotoxicity in the rat. J. Proteome Res. 5, 1586–1601. [DOI] [PubMed] [Google Scholar]
- Patti G. J.; Yanes O.; Siuzdak G. (2012) Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 13, 263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. P.; Park Y.; Ziegler T. R. (2012) Nutritional metabolomics: Progress in addressing complexity in diet and health. Annu. Rev. Nutr. 32, 183–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J. K.; Holmes E.; Kinross J.; Burcelin R.; Gibson G.; Jia W.; Pettersson S. (2012) Host-gut microbiota metabolic interactions. Science 336, 1262–1267. [DOI] [PubMed] [Google Scholar]
- Wild C. P. (2005) Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol., Biomarkers Prev. 14, 1847–1850. [DOI] [PubMed] [Google Scholar]
- Wild C. P. (2012) The exposome: From concept to utility. Int. J. Epidemiol. 41, 24–32. [DOI] [PubMed] [Google Scholar]
- Wild C. P.; Scalbert A.; Herceg Z. (2013) Measuring the exposome: A powerful basis for evaluating environmental exposures and cancer risk. Environ. Mol. Mutagen. 54, 480–499. [DOI] [PubMed] [Google Scholar]
- Vogel C.; Marcotte E. M. (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bantscheff M.; Lemeer S.; Savitski M. M.; Kuster B. (2012) Quantitative mass spectrometry in proteomics: Critical review update from 2007 to the present. Anal. Bioanal. Chem. 404, 939–965. [DOI] [PubMed] [Google Scholar]
- Rappaport S. M.; Li H.; Grigoryan H.; Funk W. E.; Williams E. R. (2012) Adductomics: Characterizing exposures to reactive electrophiles. Toxicol. Lett. 213, 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretyakova N.; Villalta P. W.; Kotapati S. (2013) Mass spectrometry of structurally modified DNA. Chem. Rev. 113, 2395–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E. C. (1978) Some current perspectives on chemical carcinogenesis in humans and experimental animals: Presidential Address. Cancer Res. 38, 1479–1496. [PubMed] [Google Scholar]
- Delaney J. C.; Essigmann J. M. (2008) Biological properties of single chemical-DNA adducts: A twenty year perspective. Chem. Res. Toxicol. 21, 232–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A.; Harris C. C. (2008) Advances in chemical carcinogenesis: a historical review and prospective. Cancer Res. 68, 6863–6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemminki K. (1993) DNA adducts, mutations and cancer. Carcinogenesis 14, 2007–2012. [DOI] [PubMed] [Google Scholar]
- Poirier M. C. (1997) DNA adducts as exposure biomarkers and indicators of cancer risk. Environ. Health Perspect. 105, 907–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schut H. A.; Snyderwine E. G. (1999) DNA adducts of heterocyclic amine food mutagens: implications for mutagenesis and carcinogenesis. Carcinogenesis 20, 353–368. [DOI] [PubMed] [Google Scholar]
- Bartsch H.; Nair J. (2000) New DNA-based biomarkers for oxidative stress and cancer chemoprevention studies. Eur. J. Cancer 36, 1229–1234. [DOI] [PubMed] [Google Scholar]
- Kensler T. W.; Chen J. G.; Egner P. A.; Fahey J. W.; Jacobson L. P.; Stephenson K. K.; Ye L.; Coady J. L.; Wang J. B.; Wu Y.; Sun Y.; Zhang Q. N.; Zhang B. C.; Zhu Y. R.; Qian G. S.; Carmella S. G.; Hecht S. S.; Benning L.; Gange S. J.; Groopman J. D.; Talalay P. (2005) Effects of glucosinolate-rich broccoli sprouts on urinary levels of aflatoxin-DNA adducts and phenanthrene tetraols in a randomized clinical trial in He Zuo township, Qidong, People’s Republic of China. Cancer Epidemiol., Biomarkers Prev. 14, 2605–2613. [DOI] [PubMed] [Google Scholar]
- Sharma R. A.; Farmer P. B. (2004) Biological relevance of adduct detection to the chemoprevention of cancer. Clin. Cancer Res. 10, 4901–4912. [DOI] [PubMed] [Google Scholar]
- Hecht S. S. (2002) Human urinary carcinogen metabolites: biomarkers for investigating tobacco and cancer. Carcinogenesis 23, 907–922. [DOI] [PubMed] [Google Scholar]
- Hecht S. S. (2003) Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat. Rev. Cancer 3, 733–744. [DOI] [PubMed] [Google Scholar]
- Brambilla G.; Mattioli F.; Martelli A. (2010) Genotoxic and carcinogenic effects of gastrointestinal drugs. Mutagenesis 25, 315–326. [DOI] [PubMed] [Google Scholar]
- Hussain S. P.; Harris C. C. (2000) Molecular epidemiology and carcinogenesis: Endogenous and exogenous carcinogens. Mutat. Res. 462, 311–322. [DOI] [PubMed] [Google Scholar]
- Wogan G. N.; Hecht S. S.; Felton J. S.; Conney A. H.; Loeb L. A. (2004) Environmental and chemical carcinogenesis. Semin. Cancer Biol. 14, 473–486. [DOI] [PubMed] [Google Scholar]
- Grollman A. P. (2013) Aristolochic acid nephropathy: Harbinger of a global iatrogenic disease. Environ. Mol. Mutagen. 54, 1–7. [DOI] [PubMed] [Google Scholar]
- Arlt V. M.; Stiborova M.; vom B. J.; Simoes M. L.; Lord G. M.; Nortier J. L.; Hollstein M.; Phillips D. H.; Schmeiser H. H. (2007) Aristolochic acid mutagenesis: Molecular clues to the aetiology of Balkan endemic nephropathy-associated urothelial cancer. Carcinogenesis 28, 2253–2261. [DOI] [PubMed] [Google Scholar]
- Arlt V. M.; Stiborova M.; Schmeiser H. H. (2002) Aristolochic acid as a probable human cancer hazard in herbal remedies: A review. Mutagenesis 17, 265–277. [DOI] [PubMed] [Google Scholar]
- Grollman A. P.; Shibutani S.; Moriya M.; Miller F.; Wu L.; Moll U.; Suzuki N.; Fernandes A.; Rosenquist T.; Medverec Z.; Jakovina K.; Brdar B.; Slade N.; Turesky R. J.; Goodenough A. K.; Rieger R.; Vukelic M.; Jelakovic B. (2007) Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc. Natl. Acad. Sci. U.S.A. 104, 12129–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. H.; Dickman K. G.; Moriya M.; Zavadil J.; Sidorenko V. S.; Edwards K. L.; Gnatenko D. V.; Wu L.; Turesky R. J.; Wu X. R.; Pu Y. S.; Grollman A. P. (2012) Aristolochic acid-associated urothelial cancer in Taiwan. Proc. Natl. Acad. Sci. U.S.A. 109, 8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puisieux A.; Lim S.; Groopman J.; Ozturk M. (1991) Selective targeting of p53 gene mutational hotspots in human cancers by etiologically defined carcinogens. Cancer Res. 51, 6185–6189. [PubMed] [Google Scholar]
- Mace K.; Aguilar F.; Wang J. S.; Vautravers P.; Gomez-Lechon M.; Gonzalez F. J.; Groopman J.; Harris C. C.; Pfeifer A. M. (1997) Aflatoxin B1-induced DNA adduct formation and p53 mutations in CYP450-expressing human liver cell lines. Carcinogenesis 18, 1291–1297. [DOI] [PubMed] [Google Scholar]
- Ross R. K.; Yuan J. M.; Yu M. C.; Wogan G. N.; Qian G. S.; Tu J. T.; Groopman J. D.; Gao Y. T.; Henderson B. E. (1992) Urinary aflatoxin biomarkers and risk of hepatocellular carcinoma. Lancet 339, 943–946. [DOI] [PubMed] [Google Scholar]
- Kanaly R. A.; Hanaoka T.; Sugimura H.; Toda H.; Matsui S.; Matsuda T. (2006) Development of the adductome approach to detect DNA damage in humans. Antioxid. Redox Signaling 8, 993–1001. [DOI] [PubMed] [Google Scholar]
- Matsuda T.; Tao H.; Goto M.; Yamada H.; Suzuki M.; Wu Y.; Xiao N.; He Q.; Guo W.; Cai Z.; Kurabe N.; Ishino K.; Matsushima Y.; Shinmura K.; Konno H.; Maekawa M.; Wang Y.; Sugimura H. (2013) Lipid peroxidation-induced DNA adducts in human gastric mucosa. Carcinogenesis 34, 121–127. [DOI] [PubMed] [Google Scholar]
- Farmer P. B.; Singh R. (2008) Use of DNA adducts to identify human health risk from exposure to hazardous environmental pollutants: The increasing role of mass spectrometry in assessing biologically effective doses of genotoxic carcinogens. Mutat. Res. 659, 68–76. [DOI] [PubMed] [Google Scholar]
- Klaene J. J.; Sharma V. K.; Glick J.; Vouros P. (2012) The analysis of DNA adducts: The transition from 32P-postlabeling to mass spectrometry. Cancer Lett. 334, 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach A. C.; Gupta R. C. (1992) Human biomonitoring and the 32P-postlabeling assay. Carcinogenesis 13, 1053–1074. [DOI] [PubMed] [Google Scholar]
- Andrews C. L.; Vouros P.; Harsch A. (1999) Analysis of DNA adducts using high-performance separation techniques coupled to electrospray ionization mass spectrometry. J. Chromatogr. A 856, 515–526. [DOI] [PubMed] [Google Scholar]
- Koc H.; Swenberg J. A. (2002) Applications of mass spectrometry for quantitation of DNA adducts. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 778, 323–343. [DOI] [PubMed] [Google Scholar]
- Singh R.; Farmer P. B. (2006) Liquid chromatography-electrospray ionization-mass spectrometry: The future of DNA adduct detection. Carcinogenesis 27, 178–196. [DOI] [PubMed] [Google Scholar]
- Randerath K.; Reddy M. V.; Gupta R. C. (1981) 32P-labeling test for DNA damage. Proc. Natl. Acad. Sci U.S.A. 78, 6162–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. C.; Reddy M. V.; Randerath K. (1982) 32P-postlabeling analysis of non-radioactive aromatic carcinogen–DNA adducts. Carcinogenesis 3, 1081–1092. [DOI] [PubMed] [Google Scholar]
- Randerath K.; Randerath E.; Agrawal H. P.; Gupta R. C.; Schurda M. E.; Reddy M. V. (1985) Postlabeling methods for carcinogen-DNA adduct analysis. Environ. Health Perspect. 62, 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaska G.; al Juburi A. Z.; Kadlubar F. F. (1991) Smoking related carcinogen-DNA adducts in biopsy samples of human urinary bladder: Identification of N-(deoxyguanosin-8-yl)-4-aminobiphenyl as a major adduct. Proc. Natl. Acad. Sci. U.S.A. 88, 5350–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieler C. A.; Stiborova M.; Wiessler M.; Cosyns J. P.; van Ypersele De S. C.; Schmeiser H. H. (1997) 32P-post-labelling analysis of DNA adducts formed by aristolochic acid in tissues from patients with Chinese herbs nephropathy. Carcinogenesis 18, 1063–1067. [DOI] [PubMed] [Google Scholar]
- Chung F. L.; Nath R. G.; Nagao M.; Nishikawa A.; Zhou G. D.; Randerath K. (1999) Endogenous formation and significance of 1,N2-propanodeoxyguanosine adducts. Mutat. Res. 424, 71–81. [DOI] [PubMed] [Google Scholar]
- Cavalieri E.; Frenkel K.; Liehr J. G.; Rogan E.; Roy D. (2000) Estrogens as endogenous genotoxic agents--DNA adducts and mutations. J. Natl. Cancer Inst. Monogr. 75–93. [DOI] [PubMed] [Google Scholar]
- Phillips D. H. (2005) DNA adducts as markers of exposure and risk. Mutat. Res. 577, 284–292. [DOI] [PubMed] [Google Scholar]
- Nair U.; Bartsch H.; Nair J. (2007) Lipid peroxidation-induced DNA damage in cancer-prone inflammatory diseases: a review of published adduct types and levels in humans. Free Radical Biol. Med. 43, 1109–1120. [DOI] [PubMed] [Google Scholar]
- Gupta R. C.; Earley K. (1988) 32P-adduct assay: Comparative recoveries of structurally diverse DNA adducts in the various enhancement procedures. Carcinogenesis 9, 1687–1693. [DOI] [PubMed] [Google Scholar]
- Pfau W.; Lecoq S.; Hughes N. C.; Seidel A.; Platt K. L.; Grover P. L.; Phillips D. H. (1993) Separation of 32P-labelled nucleoside 3′,5′-bisphosphate adducts by HPLC. IARC Sci. Publ. 233–242. [PubMed] [Google Scholar]
- Jones N. J.; McGregor A. D.; Waters R. (1993) Detection of DNA adducts in human oral tissue: correlation of adduct levels with tobacco smoking and differential enhancement of adducts using the butanol extraction and nuclease P1 versions of 32P postlabeling. Cancer Res. 53, 1522–1528. [PubMed] [Google Scholar]
- Phillips D. H. (2002) Smoking-related DNA and protein adducts in human tissues. Carcinogenesis 23, 1979–2004. [DOI] [PubMed] [Google Scholar]
- Di Paolo O. A.; Teitel C. H.; Nowell S.; Coles B. F.; Kadlubar F. F. (2005) Expression of cytochromes P450 and glutathione S-transferases in human prostate, and the potential for activation of heterocyclic amine carcinogens via acetyl-coA-, PAPS- and ATP-dependent pathways. Int. J. Cancer 117, 8–13. [DOI] [PubMed] [Google Scholar]
- Talaska G.; Schamer M.; Skipper P.; Tannenbaum S.; Caporaso N.; Unruh L.; Kadlubar F. F.; Bartsch H.; Malaveille C.; Vineis P. (1991) Detection of carcinogen-DNA adducts in exfoliated urothelial cells of cigarette smokers: Association with smoking, hemoglobin adducts, and urinary mutagenicity. Cancer Epidemiol., Biomarkers Prev. 1, 61–66. [PubMed] [Google Scholar]
- Gorlewska-Roberts K.; Green B.; Fares M.; Ambrosone C. B.; Kadlubar F. F. (2002) Carcinogen-DNA adducts in human breast epithelial cells. Environ. Mol. Mutagen. 39, 184–192. [DOI] [PubMed] [Google Scholar]
- Arif J. M.; Dresler C.; Clapper M. L.; Gairola C. G.; Srinivasan C.; Lubet R. A.; Gupta R. C. (2006) Lung DNA adducts detected in human smokers are unrelated to typical polyaromatic carcinogens. Chem. Res. Toxicol. 19, 295–299. [DOI] [PubMed] [Google Scholar]
- Bartsch H.; Nair U.; Risch A.; Rojas M.; Wikman H.; Alexandrov K. (2000) Genetic polymorphism of CYP genes, alone or in combination, as a risk modifier of tobacco-related cancers. Cancer Epidemiol., Biomarkers Prev. 9, 3–28. [PubMed] [Google Scholar]
- Nath R. G.; Ocando J. E.; Guttenplan J. B.; Chung F. L. (1998) 1,N2-Propanodeoxyguanosine adducts: Potential new biomarkers of smoking-induced DNA damage in human oral tissue. Cancer Res. 58, 581–584. [PubMed] [Google Scholar]
- Phillips D. H.; Hewer A.; Martin C. N.; Garner R. C.; King M. M. (1988) Correlation of DNA adduct levels in human lung with cigarette smoking. Nature 336, 790–792. [DOI] [PubMed] [Google Scholar]
- Phillips D. H.; Arlt V. M. (2007) The 32P-postlabeling assay for DNA adducts. Nat. Protoc. 2, 2772–2781. [DOI] [PubMed] [Google Scholar]
- Phillips D. H. (2012) On the origins and development of the 32P-postlabelling assay for carcinogen-DNA adducts. Cancer Lett. 334, 5–9. [DOI] [PubMed] [Google Scholar]
- Rothman N.; Bhatnagar V. K.; Hayes R. B.; Zenser T. V.; Kashyap S. K.; Butler M. A.; Bell D. A.; Lakshmi V.; Jaeger M.; Kashyap R.; Hirvonen A.; Schulte P. A.; Dosemeci M.; Hsu F.; Parikh D. J.; Davis B. B.; Talaska G. (1996) The impact of interindividual variation in NAT2 activity on benzidine urinary metabolites and urothelial DNA adducts in exposed workers. Proc. Natl. Acad. Sci. U.S.A. 93, 5084–5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totsuka Y.; Fukutome K.; Takahashi M.; Takashi S.; Tada A.; Sugimura T.; Wakabayashi K. (1996) Presence of N2-(deoxyguanosin-8-yl)-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline (dG-C8-MeIQx) in human tissues. Carcinogenesis 17, 1029–1034. [DOI] [PubMed] [Google Scholar]
- Phillips D. H. (1997) Detection of DNA modifications by the 32P-postlabeling assay. Mutat. Res. 378, 1–12. [DOI] [PubMed] [Google Scholar]
- Wu K. Y.; Chiang S. Y.; Shih W. C.; Huang C. C.; Chen M. F.; Swenberg J. A. (2011) The application of mass spectrometry in molecular dosimetry: Ethylene oxide as an example. Mass Spectrom. Rev. 30, 733–756. [DOI] [PubMed] [Google Scholar]
- Giese R. W. (1997) Detection of DNA adducts by electron capture mass spectrometry. Chem. Res. Toxicol. 10, 255–270. [DOI] [PubMed] [Google Scholar]
- Dizdaroglu M.; Jaruga P.; Birincioglu M.; Rodriguez H. (2002) Free radical-induced damage to DNA: Mechanisms and measurement. Free Radical Biol. Med. 32, 1102–1115. [DOI] [PubMed] [Google Scholar]
- Bessette E. E.; Goodenough A. K.; Langouet S.; Yasa I.; Kozekov I. D.; Spivack S. D.; Turesky R. J. (2009) Screening for DNA adducts by data-dependent constant neutral loss-triple stage mass spectrometry with a linear quadrupole ion trap mass spectrometer. Anal. Chem. 81, 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou P. H.; Kageyama S.; Matsuda S.; Kanemoto K.; Sasada Y.; Oka M.; Shinmura K.; Mori H.; Kawai K.; Kasai H.; Sugimura H.; Matsuda T. (2010) Detection of lipid peroxidation-induced DNA adducts caused by 4-oxo-2(E)-nonenal and 4-oxo-2(E)-hexenal in human autopsy tissues. Chem. Res. Toxicol. 23, 1442–1448. [DOI] [PubMed] [Google Scholar]
- Gangl E. T.; Turesky R. J.; Vouros P. (1999) Determination of in vitro- and in vivo-formed DNA adducts of 2-amino-3-methylimidazo[4,5-f]quinoline by capillary liquid chromatography/microelectrospray mass spectrometry. Chem. Res. Toxicol. 12, 1019–1027. [DOI] [PubMed] [Google Scholar]
- Kanaly R. A.; Matsui S.; Hanaoka T.; Matsuda T. (2007) Application of the adductome approach to assess intertissue DNA damage variations in human lung and esophagus. Mutat. Res. 625, 83–93. [DOI] [PubMed] [Google Scholar]
- Kato K.; Yamamura E.; Kawanishi M.; Yagi T.; Matsuda T.; Sugiyama A.; Uno Y. (2011) Application of the DNA adductome approach to assess the DNA-damaging capability of in vitro micronucleus test-positive compounds. Mutat. Res. 721, 21–26. [DOI] [PubMed] [Google Scholar]
- Pietsch K. E.; van Midwoud P. M.; Villalta P. W.; Sturla S. J. (2012) Quantification of acylfulvene- and illudin S-DNA adducts in cells with variable bioactivation capacities. Chem. Res. Toxicol. 26, 146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rindgen D.; Turesky R. J.; Vouros P. (1995) Determination of in vitro formed DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine using capillary liquid chromatography/electrospray ionization/tandem mass spectrometry. Chem. Res. Toxicol. 8, 1005–1013. [DOI] [PubMed] [Google Scholar]
- Singh R.; Teichert F.; Seidel A.; Roach J.; Cordell R.; Cheng M. K.; Frank H.; Steward W. P.; Manson M. M.; Farmer P. B. (2010) Development of a targeted adductomic method for the determination of polycyclic aromatic hydrocarbon DNA adducts using online column-switching liquid chromatography/tandem mass spectrometry. Rapid Commun. Mass Spectrom. 24, 2329–2340. [DOI] [PubMed] [Google Scholar]
- Spilsberg B.; Rundberget T.; Johannessen L. E.; Kristoffersen A. B.; Holst-Jensen A.; Berdal K. G. (2010) Detection of food-derived damaged nucleosides with possible adverse effects on human health using a global adductomics approach. J. Agric. Food Chem. 58, 6370–6375. [DOI] [PubMed] [Google Scholar]
- Van den D. B.; Van Dongen W.; Lemiere F.; Esmans E. L. (2004) Implementation of data-dependent acquisitions in the study of melphalan DNA adducts by miniaturized liquid chromatography coupled to electrospray tandem mass spectrometry. Rapid Commun. Mass Spectrom. 18, 2001–2007. [DOI] [PubMed] [Google Scholar]
- Wolf S. M.; Vouros P. (1994) Application of capillary liquid chromatography coupled with tandem mass spectrometric methods to the rapid screening of adducts formed by the reaction of N-acetoxy-N-acetyl-2-aminofluorene with calf thymus DNA. Chem. Res. Toxicol. 7, 82–88. [DOI] [PubMed] [Google Scholar]
- Bransfield L. A.; Rennie A.; Visvanathan K.; Odwin S. A.; Kensler T. W.; Yager J. D.; Friesen M. D.; Groopman J. D. (2008) Formation of two novel estrogen guanine adducts and HPLC/MS detection of 4-hydroxyestradiol-N7-guanine in human urine. Chem. Res. Toxicol. 21, 1622–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner P. A.; Groopman J. D.; Wang J. S.; Kensler T. W.; Friesen M. D. (2006) Quantification of aflatoxin-B1-N7-guanine in human urine by high-performance liquid chromatography and isotope dilution tandem mass spectrometry. Chem. Res. Toxicol. 19, 1191–1195. [DOI] [PubMed] [Google Scholar]
- Hang B.; Chenna A.; Guliaev A. B.; Singer B. (2003) Miscoding properties of 1,N6-ethanoadenine, a DNA adduct derived from reaction with the antitumor agent 1,3-bis(2-chloroethyl)-1-nitrosourea. Mutat. Res. 531, 191–203. [DOI] [PubMed] [Google Scholar]
- Adams R. L. P., Knowler J. T., and Leader D. P. (1986) Degradation and modification of nucleic acids. In The Biochemistry of the Nucleic Acids, pp 87–119, Chapman and Hall, New York. [Google Scholar]
- Soglia J. R.; Turesky R. J.; Paehler A.; Vouros P. (2001) Quantification of the heterocyclic aromatic amine DNA adduct N-(deoxyguanosin-8-yl)-2-amino-3-methylimidazo[4,5-f]quinoline in livers of rats using capillary liquid chromatography/microelectrospray mass spectrometry: A dose-response study. Anal. Chem. 73, 2819–2827. [DOI] [PubMed] [Google Scholar]
- Inagaki S. (2010) Screening DNA adducts by LC-ESI-MS-MS: Application to screening new adducts formed from acrylamide. Chromatographia 72, 1043–1048. [Google Scholar]
- Wang P.; Fisher D.; Rao A.; Giese R. W. (2012) Nontargeted nucleotide analysis based on benzoylhistamine labeling-MALDI-TOF/TOF-MS: Discovery of putative 6-oxo-thymine in DNA. Anal. Chem. 84, 3811–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.; Gao J.; Li G.; Shimelis O.; Giese R. W. (2012) Nontargeted analysis of DNA adducts by mass-tag MS: Reaction of p-benzoquinone with DNA. Chem. Res. Toxicol. 25, 2737–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro-Perez J.; Plumb R.; Liang L.; Yang E. (2005) A high-throughput liquid chromatography/tandem mass spectrometry method for screening glutathione conjugates using exact mass neutral loss acquisition. Rapid Commun. Mass Spectrom. 19, 798–804. [DOI] [PubMed] [Google Scholar]
- Balbo S., Hecht S. S., Upadhyaya P., and Villalta P. W. (2014) Application of a high resolution mass spectrometry-based DNA adductomics approach for identification of DNA adducts in complex mixtures. Anal. Chem. [Online early access], DOI: 10.1021/ac403565m, Published Online: Jan 10, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu D.; Turesky R. J.; Tao Y.; Langouet S. A.; Nauwelaers G. C.; Yuan J. M.; Yee D.; Yu M. C. (2012) DNA adducts of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine and 4-aminobiphenyl are infrequently detected in human mammary tissue by liquid chromatography/tandem mass spectrometry. Carcinogenesis 33, 124–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht S. S.; Spratt T. E.; Trushin N. (1997) Absolute configuration of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol formed metabolically from 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 18, 1851–1854. [DOI] [PubMed] [Google Scholar]