SUMMARY
The noncoding Xist RNA triggers silencing of one of the two female X chromosomes during X inactivation in mammals. Gene silencing by Xist is restricted to a special developmental context in early embryos and specific hematopoietic precursors. Here, we show that Xist can initiate silencing in a lymphoma model. We identify the special AT-rich binding protein SATB1 as an essential silencing factor. Loss of SATB1 in tumor cells abrogates the silencing function of Xist. In lymphocytes Xist localizes along SATB1-organized chromatin and SATB1 and Xist influence each other’s pattern of localization. SATB1 and its homolog SATB2 are expressed during the initiation window for X inactivation in ES cells. Importantly, viral expression of SATB1 or SATB2 enables gene silencing by Xist in embryonic fibroblasts, which normally do not provide an initiation context. Thus, our data establish SATB1 as a crucial silencing factor contributing to the initiation of X inactivation.
INTRODUCTION
Mammals compensate the different number of X chromosomes between the sexes by X inactivation. One of the two female X chromosomes becomes transcriptionally silent in a developmentally regulated manner. The noncoding Xist RNA, which localizes to the inactive X chromosome (Xi), initiates chromosome-wide gene repression. Xist is required for the initiation of X inactivation in early embryonic cells (Marahrens et al., 1997; Penny et al., 1996), whereas in most differentiated cells the Xi is silenced in a stable manner which is independent of Xist (Brown and Willard, 1994; Csankovszki et al., 1999). Therefore, X inactivation is separated into an initiation and a maintenance phase. Several factors that act in maintaining the silent state of the Xi have been identified, including the histone variant macroH2A (Costanzi and Pehrson, 1998; Mermoud et al., 1999; Rasmussen et al., 2000), histone H4 hypoacetylation (Keohane et al., 1996), and DNA methylation (Sado et al., 2000, 2004). Recently, it has been shown that the SmcHD1 protein is required for maintenance of DNA methylation patterns and gene repression on the Xi (Blewitt et al., 2008).
At the initiation of X inactivation chromosome-wide silencing by Xist is initially reversible. In embryonic stem (ES) cells reactivation of genes can be observed when Xist expression is abolished (Wutz and Jaenisch, 2000). Gene silencing depends on the 5′-end of Xist which contains the repeat A sequence motif (Wutz et al., 2002). Mutation of repeat A abolishes the gene silencing function of Xist, but has no effect on Xist localization to the chromosome. Several factors that have been observed enriched on the Xi, such as Polycomb group complexes, are recruited by Xist in the absence of repeat A. This shows that these factors are not sufficient for gene repression, suggesting that an additional pathway is engaged in the initiation of chromosome-wide silencing (Plath et al., 2003; Schoeftner et al., 2006). No primary initiation factors have been identified to date.
The gene silencing function of Xist is restricted to cells of the early embryo (Savarese et al., 2006). In most differentiated cells, where the Xi is stably silenced, Xist expression can not initiate gene silencing (Sado et al., 2004; Savarese et al., 2006; Wutz and Jaenisch, 2000). We have previously observed that Xist expression can initiate gene silencing in lineage-restricted progenitor cells in the hematopoietic system of adult mice (Savarese et al., 2006). Thus, a special developmental context in the early embryo and the hematopoietic system exists in which chromosome-wide silencing can be established. Why Xist expression cannot establish silencing outside these developmental windows is unknown. One possibility is that expression of essential silencing factors is restricted to specific developmental contexts. We reasoned that perhaps we could find initiation factors by setting a condition where Xist could silence outside the known developmental windows.
RESULTS
Xist Initiates Ectopic X Inactivation in Lymphoma Cells and Blocks Tumor Growth
To assess whether an epigenetic context for Xist-mediated silencing exists in cancer cells we used an inducible Xist allele (TX), in which a tetracycline-inducible promoter is inserted upstream of the Xist transcription start (Savarese et al., 2006). Induction of Xist expression from the single X chromosome in TX/Y males causes X inactivation and consequently loss of X-linked genes leads to cell death in the context of T cell progenitors, particularly CD4+ CD8+ thymocytes (Savarese et al., 2006). We introduced the TX allele into a thymic lymphoma model, which is triggered by an oncogenic NPM-ALK fusion protein under the control of a CD4 promoter (Chiarle et al., 2003). The NPM-ALK protein results from the fusion of the anaplastic lymphoma kinase (ALK) to the nucleophosmin (NPM/B23) gene by a t(2;5) translocation found in human anaplastic large cell lymphoma (ALCL; Chiarle et al., 2008; Morris et al., 1994). We generated mice carrying the NPM-ALK transgene, which were either hemizygous TX/Y males or homozygous TX/TX females for the inducible Xist allele and homozygous for the Doxycycline-inducible transactivator R26rtTA/rtTA expressed from the ROSA26 locus (Savarese et al., 2006). Thymic lymphomas developed 6–12 weeks after birth and consisted of Thy 1.2+ B220− cells, which were mostly CD4+ CD8+ and in some cases CD4− CD8−. To measure the effect of Xist induction on tumor growth we injected 1 × 106 lymphoma cells subcutaneously into immunedeficient athymic nude mice using Matrigel as a carrier (Azuma et al., 2005). Untreated recipient mice developed lymphomas in all injection sites, whereas Xist induction by Doxycycline completely blocked tumor growth (Figures 1A–1C; see Figures S1D and S1E available online). TUNEL staining indicated that Xist induction triggered tumor cell death (Figure 1D). Comparable results were obtained with several independent primary lymphoma isolates including a female tumor with a homozygous TX allele, where Xist could be induced from both X chromosomes (Figure S2). To examine if Xist induction causes tumor regression, we administered Doxycycline after tumor establishment. When Doxycycline was administered 12 days after the graft, tumor remission was observed (Figure 1E). A control NPM-ALK X/Y R26rtTA/rtTA tumor that does not carry the TX allele was unaffected by Doxycycline treatment showing that tumor ablation was an effect of Xist induction (Figure S3).
Figure 1. Xist Initiates Ectopic X Inactivation in Lymphoma Cells and Blocks Tumorigenesis.
(A) Tumor growth was monitored in Doxycycline (Dox)-treated (red) and untreated (black) nude mice after subcutaneous injection of 2 × 106 NPM-ALK TX/Y R26rtTA/rtTA lymphoma cells (n = 10). Error bars represent standard deviation.
(B) Tumor weight on Day 30 (n = 10).
(C and D) Tumor morphology (C) and TUNEL analysis ([D], green) of histological tumor sections showing apoptotic cells in the Dox-treated mice. Nuclei are counterstained with DAPI (blue). Scale bar = 10 μm.
(E) Xist induction on day 12 after tumor graft (blue arrow) causes remission of established tumors (blue). Untreated tumors (black) and tumors treated with Dox from day 2 after tumor graft (red) are controls (n = 10). Error bars represent standard deviation.
(F) NPM-ALK TX/Y R26rtTA/rtTA lymphoma cell cultures on feeders on days 1 and 5 with and without Dox. Scale bar = 50 μm.
(G) RT-PCR showing silencing of the X-linked Pgk1 and Hprt genes after 48 hr of ectopic Xist induction in NPM-ALK TX/Y R26rtTA/rtTA lymphoma cells.
(H and I) Combined RNA FISH immunofluorescence analysis showing colocalization of H3K27me3 (H) and H2AK119ub1 (I) with Xist RNA. Percentage of total cells is given in the panels (n = 500). Scale bar = 5 μm.
We confirmed these results with a second tumor paradigm and injected 2 × 106 NPM-ALK TX/Y R26rtTA/rtTA lymphoma cells into the tail vein of Rag2−/− common-γ−/− immune compromised mice. After 21 days mice treated with Doxycycline were healthy and tumor-free, while untreated recipients showed massive infiltration of tumor cells in several organs (Figure S4).
To investigate if Xist induction caused ectopic X inactivation, we established several cell lines by culturing primary tumors in the presence of IL-7 on OP9-derived feeder cells. We plated 3 × 105 cells and measured cell growth in the presence or absence of Doxycycline. We observed that Xist induction from the single male X chromosome (TX/Y) or both female X chromosomes (TX/TX) caused a loss of tumor cells (Figure 1F and Figure S5A), whereas Doxycycline had no effect on control tumor cells without the TX allele. Doxycycline treated male NPM-ALK TX/Y R26rtTA/rtTA lymphoma cells showed Xist expression and a marked loss of the X-linked gene expression (Figure 1G). Xist clusters were detected in 98% of the cells induced with Doxycycline, whereas no Xist focus was detected without induction (Figure S5B). In over 90% of the cells Xist colocalized with trime-thylated histone H3 lysine 27 (H3K27me3) and histone H2A ubiquitinated on Lysine 119 (H2AK119ub1), which are two prominent markers of the Xi (Figures 1H and 1I). These data showed that Xist induction triggered ectopic X inactivation in lymphoma cells equivalent to embryonic cells.
Identification of SATB1 as a Silencing Factor for Xist
We reasoned that the lymphoma context might provide an opportunity for identifying initiation factors for Xist. For this we aimed at generating a tumor that is resistant to Xist-mediated killing. Tumor resistance is known to arise for virtually any therapeutic agent. Surprisingly, resistance to Xist-mediated killing was not observed in our tumor grafts despite considerable efforts. Therefore, we cultured lymphoma cells for 1 month in the absence of Doxycycline to trigger tumor cell evolution. Then 2 × 106 NPM-ALK TX/Y R26rtTA/rtTA cells were transplanted into the flank of nude mice and tumor development was studied in the presence of Doxycycline. Initially treated mice showed no signs of tumor growth in stark contrast to control mice that did not receive Doxycycline (Figure 2A). This is consistent with the large majority of tumor cells being ablated by Xist induction. However, after 21 days tumors started to appear at all injection sites in Doxycycline-treated mice (Figure 2A and Figure S6A). It is likely that these tumors originated from rare cells that acquired resistance to Xist-mediated killing during in vitro culture. In histological tumor sections Xist clusters colocalizing with H3K27me3 foci were observed (Figure 2B). The X-linked Pgk1 and Hprt genes were expressed at a level above or equal to that observed in untreated control tumors or thymic cells (Figure 2C). The presence of one X chromosome per tumor cell was confirmed by X chromosome painting ruling out the possibility that the resistance to Xist-mediated killing was due to rearrangement or gain of X chromosomes (Figure S6B). Furthermore, Xist induction by addition of Doxycycline did not affect proliferation of a cell line established from this tumor (Figure S6C). These results suggested that spontaneous resistance had arisen by loss of the pathways required for the silencing function of Xist.
Figure 2. Derivation of an Xist-Resistant Tumor for Identification of Silencing Factors.
(A) Tumor growth after transplantation of 2 × 106 cultured NPM-ALK TX/Y R26rtTA/rtTA lymphoma cells into nude mice. After 22 days (red arrow), Xist-resistant tumors appear in the Dox-treated group (red; n = 5). Error bars represent standard deviation.
(B) Xist RNA FISH (red) and H3K27me3 (green) in histological sections of the resistant tumor. Percentage of colocalization is given.
(C) Quantitative RT- PCR analysis showing that the X-linked Pgk1 and Hprt genes are still expressed in the resistant tumors when Xist is induced with Dox at a comparable level to normal thymus or parental tumor without Xist induction. Results were normalized to Gapdh. Error bars represent standard deviation (n = 3).
(D) Heatmap showing upregulated (red) and downregulated (green) genes after hierarchical clustering of gene expression profiles of Xist-resistant and Xist-responsive NPM-ALK TX/Y R26rtTA/rtTA lymphoma and normal male thymus. Probes for Xist and SATB1 are indicated (arrows) and gene groups are annotated (right).
To identify genes that were associated with resistance to Xist-mediated killing we performed genome-wide expression profiling of Xist-responsive and Xist-resistant tumors and of thymus (Figure 2D and Figure S7). Among the most significantly downregulated genes in resistant tumors (Table S1) we noted SATB1, a nuclear protein which functions in regulating chromatin structure and gene regulation in thymocytes (Dickinson et al., 1992; Alvarez et al., 2000; Cai et al., 2003). In thymus, expression of SATB1 is high in the CD4+ CD8+ double-positive T cell compartment, which we previously observed to possess the context for initiation of gene silencing by Xist (Savarese et al., 2006). This suggested a correlation of SATB1 expression and gene silencing by Xist.
SATB1 Is Essential for the Silencing Function of Xist in Thymocytes
To verify that SATB1 could be a limiting factor for the silencing function of Xist we analyzed expression in development. In thymus, high levels of SATB1 were found in the cortex, which is predominantly composed of CD4+ CD8+ thymocytes, whereas in the medulla fewer cells showed expression (Figures S8A and S8B). This is consistent with a loss of SATB1 expression upon further thymocyte differentiation to single positive T cells. To test that SATB1 expressing cells in the thymus provided the context for silencing by Xist, we induced Xist from the single male X chromosome in TX/Y R26rtTA/rtTA mice. We observed a rapid loss of SATB1 expressing cells upon Xist induction consistent with cell death caused by ectopic X inactivation (Figure S8C).
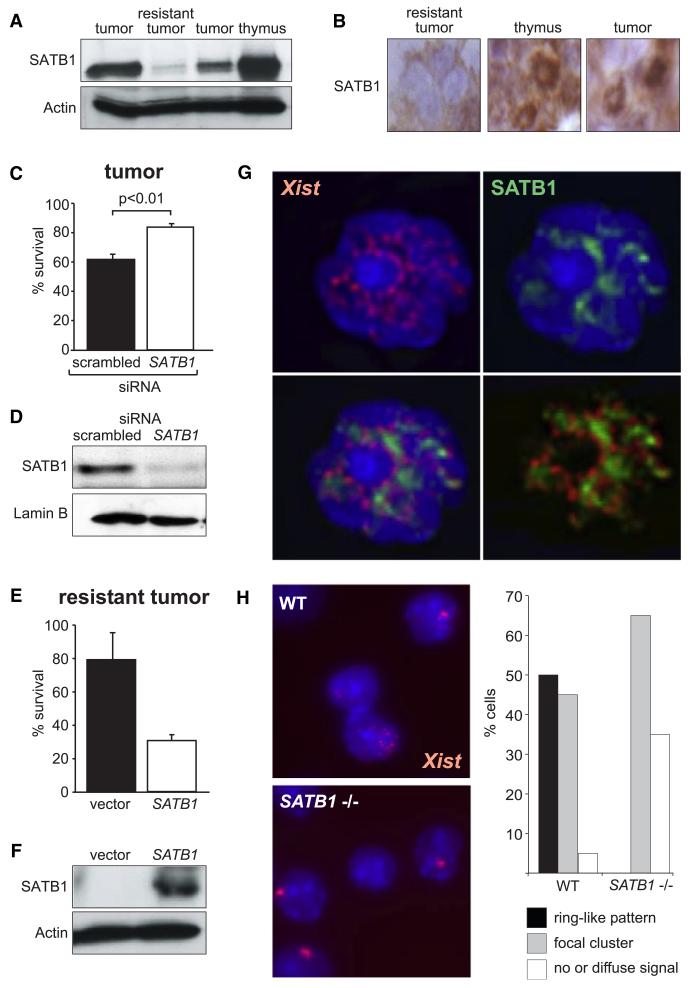
In NPM-ALK thymic lymphoma the normal thymic structure was lost and substituted by blasts, which were positive for the NPM-ALK protein and SATB1 (Figures S8A and S8B). Western analysis confirmed a loss of SATB1 protein in Xist-resistant tumors (Figure 3A). SATB1 immunohistochemistry in sections revealed a ring-like pattern in the thymus and Xist-responsive tumors, whereas Xist-resistant tumors showed no or minimal SATB1 expression (Figure 3B). None of the resistant tumors was positive for SATB1. To test if SATB1 was required for the gene silencing function of Xist in lymphoma cells we performed RNAi. RNAi in lymphoma cells was technically difficult and required the optimization of the protocol. For efficient reduction of SATB1 protein levels we used a double transfection protocol. We transfected NPM-ALK TX/Y R26rtTA/rtTA lymphoma cells with siRNA specific for SATB1 or control siRNA. After two days Xist expression was induced and a second siRNA transfection was performed 3 days thereafter. We measured the total cell numbers of triplicate cultures with and without Doxycycline induction after 5 days and calculated the percentage of surviving cells after Xist induction. Cells transfected with SATB1 siRNAs showed a significantly higher survival compared to controls (Figure 3C and Figure S8D). Western and immunofluorescence analysis showed reduced SATB1 expression in most cells treated with SATB1 siRNA, but not control siRNA, confirming the effect of the siRNA (Figure 3D and Figure S8E). SATB1 seemed to stabilize Xist RNA and thus after RNAi a reduction in Xist signal was observed (Figure S8E). We believe that Xist can localize onto SATB1 organized chromatin and is thereby stabilized leading to higher Xist levels. However, we also note that in the SATB1 siRNA-treated cells we observed efficient Xist induction.
Figure 3. SATB1 Is a Critical Silencing Factor for Xist in Lymphoma Cells.
(A) Western analysis of SATB1 expression in Xist-resistant and Xist-responsive tumor and normal thymus. Actin serves as loading control.
(B) Immunohistochemistry in sections reveals nuclear ring-like SATB1 structures in normal thymus and Xist-responsive tumor, whereas nuclear SATB1 staining is lost in resistant tumors.
(C) SATB1 RNAi in cultured NPM-ALK TX/Y R26rtTA/rtTA lymphoma cells significantly increases cell survival after Xist induction. Cell survival after 5 days of culture in presence of Dox relative to uninduced cultures is plotted. Scrambled siRNA treatment serves as control (n = 3). Error bars represent standard deviation.
(D) Western analysis confirming loss of SATB1 protein in SATB1 siRNA treated but not scrambled control siRNA treated cells. Lamin B serves as loading control.
(E) Virally expressed SATB1 restores Xist silencing function in Xist-resistant tumor cells. Cell survival after 5 days of cultures in presence of Dox relative to uninduced cultures is plotted. Empty vector-infected cells serve as control (n = 3).
(F) Western analysis confirming SATB1 protein expression in cells infected with SATB1 but not control virus. Actin serves as loading control.
(G) Xist RNA FISH (red) combined with SATB1 immunofluorescence (green) analysis showing Xist spreading along the SATB1 ring in TX/Y R26rtTA/rtTA thymic cells after 48 hr of Xist induction with Dox.
(H) Delocalization of Xist into a ring-like pattern is observed in thymocytes of wild-type (WT) but not SATB1 deficient female mice. Representative Xist RNA FISH images are shown. A statistical analysis of Xist patterns in SATB1 deficient and control thymocytes is given (n = 150).
To test if SATB1 is sufficient to restore Xist silencing in the resistant tumor we virally expressed SATB1 in a cultured Xist-resistant tumor cell line. SATB1 expressing tumor cells showed a strongly reduced survival upon Xist induction compared to Xist-resistant tumor cells infected with empty vector (Figures 3E and 3F). Taken together these data strongly support a crucial function for SATB1 in gene silencing by Xist in the lymphoid and tumor context.
Xist Localizes along Nuclear SATB1 Structures in Lymphoid Cells
To investigate a potential interaction we performed SATB1 immunofluorescence analysis combined with Xist RNA FISH. In thymocyte nuclei SATB1 is observed in a ring-like structure (Cai et al., 2003). We have previously observed that in CD4+ CD8+ T cells, which constitute the hematopoietic context for gene silencing in the thymus, Xist does not form a characteristic focus but appears in a diffuse pattern (Savarese et al., 2006). Therefore, we examined thymi from TX/Y R26rtTA/rtTA mice after induction of Xist for 24 and 48 hr. Xist formed a characteristic focus after 24 hr of induction. However, after 48 hr Xist spread along SATB1 in the majority of cells, whereby it did not colocalize with but rather outlined the SATB1 ring (Figure 3G and Figures S9A–S9E). In normal female thymocytes, we observed the expected SATB1 ring structure and in more than half of the cells Xist did not form a focus but localized along the SATB1 ring (Figure S9F). This localization pattern of Xist is therefore observed in a physiologically normal cell type. Furthermore, delocalization of Xist into a ring-like pattern was not observed in thymocytes isolated from SATB1-deficient mice, where more Xist foci were observed (Figure 3H). Thus, delocalization of Xist in thymocytes depends on SATB1.
Next we induced Xist expression in TX/TX R26rtTA/rtTA thymocytes from both female X chromosomes. We observed two Xist foci overlapped with two regions of intense SATB1 staining (Figure S9G). Such a focal SATB1 pattern was not observed before induction of Xist. This indicated that Xist expression from two X chromosomes had caused a redistribution of SATB1 from the ring. Thus, Xist and SATB1 apparently interacted and changed each other’s localization.
In NPM-ALK TY/Y R26rtTA/rtTA lymphoma cells a characteristic Xist focus was observed after 24 hr of induction whereas after 48 hr Xist spread along the SATB1 ring (Figures S9H and S10A). We further noted that the Polycomb group proteins Ring1B and Bmi1 were diffusely distributed in the nucleus and showed partial overlap with SATB1 in a ring-like pattern (Figure S10B). We suggest that the interaction between Xist and SATB1 in lymphoma cells and in normal thymocytes is potentially mediated by SATB1 organized chromatin. A direct physical attachment is unlikely, as SATB1 and Xist RNA showed little overlapping localization.
SATB1 Regulates Xist Silencing Function in Embryonic Cells
To analyze if SATB1 is relevant for the initiation of normal X inactivation we analyzed mouse embryonic stem (ES) cells. Western analysis showed SATB1 expression in ES cells, but 3 days after retinoic acid induced differentiation SATB1 expression became undetectable (Figure 4B). This expression precisely overlaps the window in which Xist is able to silence (Wutz and Jaenisch, 2000). Immunofluorescence analysis showed that SATB1 was diffusely distributed and surrounded a characteristic Xist focus when Xist expression was induced in TX/Y ES cells (Figure 4A) similar to the endogenous Xi in differentiating female ES cells (Figure S11A). Thus, SATB1 was expressed in the embryonic context for X inactivation and could contribute to the initiation of chromosome-wide silencing. However, disruption of SATB1 in mice is compatible with female development, suggesting that other factors can take over its function in X inactivation (Alvarez et al., 2000). To test if the close homolog SATB2 could provide such a redundant function, we analyzed its expression in ES cells. Similar to SATB1 we observed SATB2 expression in ES cells (Figure 4C). After 3 days of differentiation, SATB2 showed a switch to a lower molecular weight. The higher molecular weight likely corresponds to a SUMO-modified form that has been previously described as important for gene regulation (Dobreva et al., 2003). The shift in molecular weight at the end of the initiation window of X inactivation suggested that the higher molecular weight form of SATB2 could be important for Xist mediated silencing.
Figure 4. SATB1 Expression Defines the Silencing Function of Xist in Embryonic Cells.
(A) Combined RNA FISH immunofluorescence analysis of TX/Y R26rtTA/rtTA ES cells shows SATB1 (green) surrounding the characteristic Xist focus (red) after 24 hr of induction with Doxycycline. Scale bar = 5 μm.
(B and C) Western analysis of SATB1 (B) and SATB2 (C) protein during ES cell differentiation shows that SATB1 is lost and SATB2 shifts to a lower molecular weight on day 3, precisely when Xist loses its silencing capability. Asterisk indicates the high molecular weight form of SATB2. Actin serves as loading control.
(D) Repression of the puromycin (puro) marker, which is cointegrated with an Xist transgene on chromosome 11 in clone 36 ES cells, upon Xist induction is reduced by SATB1 or SATB2 RNAi compared to scrambled control. Quantification of puro expression after Xist induction relative to uninduced cells is given.
(E) Western analysis demonstrates strongly reduced SATB1 protein in ES cells treated with SATB1 siRNA but not control siRNA. Western analysis of SATB2 in ES cells treated with scrambles control, SATB2, and SATB1 siRNAs. The higher molecular weight form of SATB2 (asterisk) observed in ES cells is depleted by SATB2 RNAi. The lower molecular weight form of SATB2 is observed in brain. Lamin serves as loading control.
(F) Survival of immortalized fibroblasts established from TX/Y R26rtTA/rtTA embryos and infected with SATB1 and SATB2 expression vector or control empty vector (vector). The percent survival is calculated from the cell number with Xist induction relative to untreated cells (n = 6). Error bars represent standard deviation.
(G) Quantitative RT-PCR showing repression of the X-linked Hprt and Pgk1 genes upon Xist induction in MEFs infected with SATB1 and SATB2 but not empty vector controls. Expression relative to Gapdh is plotted (n = 6). Error bars represent standard deviation.
To further explore if SATB1 and SATB2 contribute to chromosome-wide silencing by Xist in ES cells we used RNAi. In clone 36 ES cells an inducible Xist transgene leads to repression of a puromycine selection marker, which provides a convenient readout for silencing (Wutz and Jaenisch, 2000). We transfected either control, SATB1, SATB2, or a combination of SATB1 and SATB2 siRNA into ES cells (Figure S11B). While there was little effect of control, SATB1, or SATB2 siRNAs on cell numbers, double RNAi of SATB1 and SATB2 led to a cell loss precluding further analysis (Figures S11C and S11D). Analysis of puro expression showed that silencing was efficient in control cells with 13% of puro signal remaining after Xist induction. The efficiency of silencing was reduced in cultures treated either with SATB1 or SATB2 siRNAs, with 26% and 20% of puro message remaining after Xist induction, respectively (Figure 4D). This indicated that an acute loss of SATB1 and SATB2 reduced the efficiency of gene silencing by Xist in ES cells. Furthermore, SATB2 RNAi depleted the high molecular weight form of SATB2 in ES cells confirming that the antiserum was specific (Figure 4E). SATB2 expressed in the brain corresponds to the lower molecular weight form, which we observed in differentiated ES cells after the initiation window. We next asked if SATB1 or SATB2 are limiting for Xist silencing function in embryonic cells. We previously reported that induction of Xist in embryonic fibroblasts (MEFs) does not cause gene silencing despite normal Xist RNA localization (Wutz and Jaenisch, 2000). Thus, MEFs lack essential initiation factors for Xist. To establish if either SATB1 or SATB2 would perform as primary initiation factors in the embryonic context, we virally expressed SATB1 in TX/Y MEFs and tested if gene silencing could be restored. In SATB1- and SATB2-infected MEFs cell numbers were significantly reduced upon induction of Xist expression from the single male X chromosome, whereas Xist had no effect on cells infected with a control virus (Figure 4F and Figure S12). Repression of the X-linked Pgk1 and Hprt genes showed that Xist induction had indeed led to ectopic X inactivation in SATB1- and SATB2-infected MEFs (Figure 4G). We conclude that expression of SATB1 or SATB2 substituted an essential primary initiation factor and allowed to reestablish the embryonic context for gene silencing by Xist in MEFs.
DISCUSSION
Here we identify SATB1 as a silencing factor for Xist. SATB1 is required and sufficient for gene silencing by Xist in thymic lymphoma cells. We find that SATB1 is expressed during the initiation window of X inactivation in ES cells and its expression is lost at the time point in differentiation when Xist can no longer initiate chromosome-wide silencing. Importantly, viral SATB1 expression enables gene silencing by Xist in mouse embryonic fibroblasts. Expression of SATB1 has also been observed in fetal brain (Alvarez et al., 2000) and in erythroid differentiation (Wen et al., 2005), which is consistent with our previous finding that Xist induction leads to ablation of erythroid cells in mice (Savarese et al., 2006). Mutation of SATB1 in mice leads to misregulation of multiple genes during T cell development and arrests development at the CD4+ CD8+ T cell stage (Alvarez et al., 2000). The development of female mice in the absence of SATB1 demonstrates that other factors can substitute SATB1 function in X inactivation. Thus, SATB1 contributes to X inactivation in embryonic cells together with other potentially as of yet unidentified factors.
We find that one such factor could be SATB2. We observe expression of SATB1 and SATB2 in ES cells. In western analysis SATB1 migrates at its expected molecular weight, whereas there is only a higher molecular weight band for SATB2. This higher molecular weight band is specifically eliminated by SATB2 RNAi. These data are also consistent with immunofluorescence staining showing SATB2 expression in ES cells, but not thymocytes, which are positive for SATB1 (data not shown). From this we conclude that there is a higher molecular weight form of SATB2 in ES cells. From bioinformatics and SATB2 transcript analysis we have not found evidence for additional SATB2 exons. From this we suggest that the higher molecular weight results from posttranslational modification. This is consistent with the shift on Day 2 of ES cell differentiation to the lower molecular weight form corresponding to the unmodified protein. Albeit, western analysis suggests that the lower molecular weight form in differentiated ES cells is more abundant than the higher molecular weight SATB2 in ES cells, we note that this might still be of functional significance since in ES cells SATB1 is also present. Disruption of SATB2 is also compatible with female development and causes skeletal abnormalities and perinatal lethality in mice (Dobreva et al., 2006; Britanova et al., 2006). SATB2 protein is expressed in the kidney (Dobreva et al., 2003) and the nervous system (Britanova et al., 2005; Dobreva et al., 2003), where we observe the lower molecular form of SATB2 which is not relevant to Xist silencing and which might have other functions. SATB2 expression is also reported in pre-B cells (Dobreva et al., 2003), which are efficiently ablated by Xist induction in mice (Savarese et al., 2006). The tissue distribution of SATB1 and SATB2 expression is, thus, consistent with a role in enabling chromosome silencing by Xist. So far, we have not identified additional proteins with a high sequence homology to SATB1 and SATB2 in the mouse genome. Albeit, more work is needed to fully address if other factors than SATB1 and SATB2 contribute to the initiation of X inactivation in the embryo, our data demonstrate a crucial contribution of SATB1 and SATB2 to chromosome-wide silencing by Xist and implicate a class of proteins with functions in chromatin organization in the mammalian dosage compensation mechanism.
Our finding that Xist spreads along the SATB1 ring in CD4+ CD8+ T cells suggests an interaction that links the localization of SATB1 and Xist RNA. This observation could be made in T cells where SATB1 shows a prominent ring- or cage-like staining pattern. In other cell types examined here, SATB1 shows a more diffuse distribution, which makes localization analysis less informative. We further made the intriguing observation that induction of Xist from both X chromosomes in TX/TX female thymocytes triggers a change in the localization pattern of SATB1. In these cells SATB1 is localized from a cage-like pattern toward the two Xist clusters. This mutual influence on localization suggests that the relative robustness of either the SATB1 cage structure or the chromosome territory organized by Xist determine the recruitment of SATB1 to the X chromosome or Xist onto SATB1 cage structure.
At the initiation of X inactivation in ES cells we observe SATB1 surrounding the Xist domain. Albeit, the precise mechanism of SATB1 in chromosome-wide silencing is not known at present, we suggest a regulatory role of SATB1 for genic chromatin of the X chromosome. SATB1 has been shown to organize transcriptionally active chromatin into chromatin loops (Galande et al., 2007; Cai et al., 2006). The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences (Clemson et al., 2006). Chaumeil et al. (2006) showed that genes are moved into the chromosome territory upon silencing in a manner that is dependent on repeat A of Xist. It is possible that Xist pulls genes into the repressive core compartment for silencing and SATB1 might act as an anchor in this chromosomal reorganization.
Although we cannot completely rule out that SATB1 acts indirectly in X inactivation via regulation of a target gene, we believe that this is unlikely based on the following evidence: (1) Gain- and loss-of-function experiments of SATB1 give the expected effect on Xist mediated silencing in tumor cells. (2) Expression profiles of Xist-resistant versus Xist-responsive tumors did not identify other candidate genes with a chromatin regulatory function or a plausible developmental expression pattern. (3) SATB1 expression in four different cell types enables gene silencing by Xist. Since SATB2 also contributes to gene silencing by Xist, we deem it extremely unlikely that a common SATB1 and SATB2 target gene is regulating silencing in all cell systems. (4) Our observation of mutual interaction between Xist and SATB1 localization in T cells supports a role of SATB1 in chromatin organization for the initiation of chromosome-wide silencing by Xist.
We have previously shown that the inactive X chromosome is maintained during the stages in T cell and B cell development when Xist can initiate silencing (Savarese et al., 2006). Since Xist is delocalized in pre-T cells and very low expression is observed in pre-B cells it is unlikely to contribute to maintaining X inactivation. Since SATB1 can only cause chromosome-wide silencing in combination with Xist, this fact strongly indicates that SATB1 is not essential for Xi maintenance in pre-T cells. Furthermore, the fact that the Xi is stably maintained in somatic cells which do not express SATB1 further argues against a general function of SATB1 in the maintenance of X inactivation. Thus, SATB1 is an initiation factor but not a maintenance factor for X inactivation.
SATB1 expression in thymic lymphoma suggests that the epigenetic context of progenitor cells enabling Xist gene silencing function is preserved in tumorigenesis. Progenitor-derived leukemia stem cells have also been observed to maintain progenitor identity and reactivate a self-renewal-associated program (Krivtsov et al., 2006). We find SATB1 expressed in a range of human lymphoma types including NPM-ALK positive and negative ALCL (Table S2 and Figure S13). Expression of SATB1 is further associated with more aggressive and poorly differentiated forms of breast cancer in humans (Han et al., 2008). These findings show that the Xist gene silencing pathway is also tied in with tumorigenesis. This idea is further consistent with the observation that XIST can initiate silencing in a human fibrosarcoma cell line, at least to some extent (Chow et al., 2007; Hall et al., 2002). Understanding the pathway enabling chromosomal silencing by Xist in progenitors, thus, might open new perspectives for cancer therapy.
EXPERIMENTAL PROCEDURES
Generation of NPM-ALK Tumors Carrying an Inducible Xist Allele, TX
NPM-ALK X/Y transgenic mice (Chiarle et al., 2003) were crossed with TX/TX R26rtTA/rtTA mice (Savarese et al., 2006) to obtain the desired genotypes NPM-ALK TX/Y R26rtTA/rtTA and NPM-ALK TX/TX R26rtTA/rtTA. Tumor cells isolated from thymic lymphoma were injected subcutaneously into both flanks of 6-week-old athymic Balb/c nu/nu mice. For each injection, 2 × 107 cells were suspended in 250 μl cold PBS and 250 μl of cold BD Matrigel Basement Membrane Matrix High Concentration (HC). For each experiment, 10 nude mice were injected. Xist expression in tumor grafts was induced by administration of Doxycycline via the drinking water (1 g/l Doxycycline and 100 g/l sucrose). Water bottles were protected from light and changed every second day. Tumor development at the site of injection was measured daily. Error bars throughout represent standard deviation unless stated otherwise. Tail vein injection model: single-cell suspensions were prepared from the thymic lymphoma NPM-ALK TX/Y R26rtTA/rtTA mice, and 1 × 106 lymphoma cells were injected into the tail vein of 10 Rag2−/− common-gamma chain cγ−/− mice. Five mice were administered Doxycycline and five were not treated. Pathology was analyzed after 21 days.
Cell Culture and Establishment of Lymphoma Cell Lines
ES cells and mouse embryonic fibroblasts were cultured as described previously (Wutz and Jaenisch, 2000). Xist expression was induced with 1 μg/ml of Doxycycline. Lymphoma tumor cell lines were established from primary NPM-ALK TX/TX R26rtTA/rtTA, NPM-ALK TX/Y R26rtTA/rtTA, and NPM-ALK X/Y R26rtTA/rtTA lymphoma. Cells were cultured on murine bone marrow stromal-derived OP9 feeders in IL-7 containing IMDM medium, as described (Hoflinger et al., 2004).
Histology, Immunohistochemistry, and TUNEL Assay
Tumors and normal organs were fixed in 4% neutral buffered formaldehyde at 4°C and embedded in paraffin. Hematoxilin and eosin (H&E) staining was performed on 3 μm sections. Immunohistochemistry was performed with antibodies for ALK (mouse anti-ALK Mab; Zymed), anti-Ki-67 (NCL-Ki67p; Novacostra), or SATB1 (ab49061 Rabbit polyclonal anti-SATB1; Abcam). TUNEL staining was performed by using the in situ cell death detection kit (Roche). Clinical specimens were handled according to institutional guidelines.
RNA Extraction, Reverse Transcription PCR Analysis, and Quantitative PCR Analysis
RNA was isolated using Trizol (Invitrogen, Life Technologies) and further purified using RNeasyTM columns (QIAGEN). Reverse transcription was primed with Oligo(dT)12–18 using the SuperScript II RNase H- RT Kit (Invitrogen). Quantitative PCR was performed for 40 cycles with SYBR Green on an Opticon 2 monitor fluorescence thermocycler (MJ Research, Waltham, MA). Relative concentrations were determined on basis of standard curves of cDNA and normalized for Gapdh. Melting curves were run to ensure amplification of a single product. Error bars represent standard deviation. Primers for Pgk1, Hprt, and Xist were previously published (Huynh and Lee, 2003). Primers for SATB1 were CCTGGATGAGCCCTTTGG and CTGCGTGGTGGAACATTATG and primers for mouse SATB2 were ACACCGACAACAGACCTC and GGGCTTGAGACACCTTGG. Human SATB2 primers were as described (Han et al., 2008).
Protein Extraction and Western Blotting
Whole cell extracts prepared with RIPA buffer and western analysis were performed as previously published (Schoeftner et al., 2006) using rabbit anti-SATB1 1:2000 (Dickinson et al., 1992) and SATB2 1:300 (ab34735 rabbit anti-SATB2; Abcam).
Immunofluorescence, RNA FISH, and X Chromosome Painting
ES cells were attached to poly-l-lysine coated coverslips or cytospun using a Cytospin 3 centrifuge (Thermo Shandon, USA). Differentiated cells were grown on Roboz slides (CellPoint Scientific, USA). Lymphoma cells were attached to adhesion slides (Marilienfeld, Germany). Immunostaining was performed as described previously (Leeb and Wutz, 2007). In brief, cells were fixed for 10 min in 4% PFA in PBS, permeabilized for 5 min in 0.1% Na-citrate/0.5% Triton X-100, and blocked for 30 min in PBS containing 5% BSA and 0.1% Tween 20. For H2AK119ub1 immunostaining, cells were pre-extracted in 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM Pipes (pH 6.8), and 0.5% Triton X-100 for 2 min before fixation, and washes after incubation with primary and secondary antibody were performed in KCM buffer (120 mM KCl, 20 mM NaCl, 10 mM Tris [pH 8.0], and 0.5 mM EDTA)/0.1% Tween 20. Antibodies and dilutions for Ring1B, Bmi1, H3K27me3, and secondary antibodies were used as previously published (Leeb and Wutz, 2007). SATB1 antibody (Dickinson et al., 1992) was used in 1:500 dilution. RNA FISH probes were generated by random priming (Stratagene) using Cy3-dCTP (GE Healthcare). After immunostaining, cells were fixed in 4% PFA in PBS for 10 min, dehydrated, hybridized, and washed as described previously (Leeb and Wutz, 2007). Vectashield (Vector Laboratories) was used as imaging medium. X chromosome painting was performed in tissue sections as published (Donadoni et al., 2004) using a commercial probe (Cambio). DAPI (4′,6′-diamidino-2-phenylindole) was used to stain the DNA. Combined Xist RNA and X chromosome painting was performed sequentially (Chaumeil et al., 2006) using a Cy3-labeled Xist probe and a biotin-labeled X paint (Cambio). Images were obtained at room temperature with a fluorescence microscope (Axioplan; Zeiss), a CCD camera (CoolSNAP fx; Photometrics), and MetaMorph image analysis software (Universal Imaging Corp.). Color levels were adjusted in Photoshop 7.0 (Adobe). All confocal pictures were obtained using a Zeiss LSM 510 Axiovert 200M. Deconvolution microscopy was carried out on a DeltaVision Spectris Restoration Microscopy System (Applied Precision) using a 100× 1.4 NA planachromat lens to collect a series of 0.2 μm z-sections and subsequent deconvolution using the proprietary SoftWorx algorithm (Applied Precision).
Expression Microarray and Bioinformatic Analysis
Total RNA was extracted using Trizol (Invitrogen) and whole genome expression profiles were generated using Affimetrix GeneChip® Mouse Genome 430 2.0 arrays (RZPD, Germany). Data were submitted to the GEO repository using the accession code GSE14585 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE14585). Microarray data were normalized using the Robust Multi-Array Analysis as implemented in Bioconductor (Gentleman et al., 2004; Irizarry et al., 2003). All analyses were performed with log2-transformed data. Hypothesis tests were performed using a modified t statistics with an empirical Bayes approach as implemented in the Bioconductor LIMMA package (Smyth, 2004). The p values were adjusted by the FDR method of Benjamini and Hochberg (1995). Unsupervised hierarchical clustering was performed using the Unweighted Pair Group Method with Arithmetic Mean (UPGMA) and the Pearson correlation coefficient distance measure. A batch effect was removed with a mixed model ANOVA, and all probe sets were ranked by their median absolute deviation. The 300 probe sets with the highest median absolute deviation were selected and z-transformed before clustering.
RNA Interference
NPM-ALK TX/Y R26rtTA/rtTA lymphoma cells were transfected twice with SATB1 siRNAs (Santa Cruz Biotechnology) or scrambled control siRNAs (Santa Cruz Biotechnology) following the recommendations of the supplier. Xist expression was induced by the addition of 1 μg/ml of Doxycycline 48 hr after the first transfection for 5 days. Viable cell number was measured using a CASY® Cell Counter (Schärfe System GmbH). Experiments were performed in triplicate, and error bars plotted represent the standard deviation. For RNA interference in ES cells, 2 × 104 cells were plated in a 6-well dish and incubated for 24 hr. Subsequently, cells were transfected using 75 μl/well of 5 μM siRNA (Santa Cruz Biotechnology) and 5 μl/well of Lipofectamlne 2000 (Invitrogen) in OPTI-MEM (GIBCO) without serum. A second round of transfection was performed after 3 days. Cells were then cultured for 48 hr in normal ES medium either with or without Dox and analyzed by northern analysis as described previously (Leeb and Wutz, 2007).
Cloning and Retrovirus Infections
The mouse SATB1 and human SATB2 cDNAs (RZPD, Germany) were cloned into the MSCV-CD2 and pBabe-puro vectors. For virus production, Plat-E cells were transfected with these vectors as described (Morita et al., 2000). The supernatant was collected and used for infection of primary or immortalized TX/Y R26rtTA/rtTA mouse embryonic fibroblasts. Infected cells were selected with Puromycin or sorted by FACS after staining with phycoeritrin (PE) antihuman CD2 (eBioscience) using a FACS Aria cell sorter (Becton Dickinson).
Supplementary Material
ACKNOWLEDGMENTS
We thank Gabi Stengl for FACS analysis, Pavel Pasierbek for help with miroscopy, Johannes Tkadletz for figure preparation, Andreas Bichl and Denise Imre for maintenance of the mouse colony, and Erwin F. Wagner, Denise Barlow, and Joseph Penninger for critically reading the mansucript. We thank Masaru Miyano for thymic cell preparations. This research was supported by a grant from the Vienna Science and Technology Fund (WWTF), by the IMP through Boehringer Ingelheim, and the Austrian Science Fund (FWF). The IMP is funded in part through Boehringer Ingelheim and C.H. is employed by Boehringer Ingelheim.
Footnotes
ACCESSION NUMBERS
Gene expression profiles have been deposited at GEO with the accession code GEO14585.
SUPPLEMENTAL DATA
Supplemental Data include thirteen figures and two tables and can be found with this article online at http://www.cell.com/developmental-cell/supplemental/S1534-5807(09)00096-3.
REFERENCES
- Alvarez JD, Yasui DH, Niida H, Joh T, Loh DY, Kohwi-Shigematsu T. The MAR-binding protein SATB1 orchestrates temporal and spatial expression of multiple genes during T-cell development. Genes Dev. 2000;14:521–535. [PMC free article] [PubMed] [Google Scholar]
- Azuma M, Hirao A, Takubo K, Hamaguchi I, Kitamura T, Suda T. A quantitative matrigel assay for assessing repopulating capacity of prostate stem cells. Biochem. Biophys. Res. Commun. 2005;338:1164–1170. doi: 10.1016/j.bbrc.2005.10.055. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series. 1995:289–300. [Google Scholar]
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat. Genet. 2008;40:663–669. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- Britanova O, Akopov S, Lukyanov S, Gruss P, Tarabykin V. Novel transcription factor Satb2 interacts with matrix attachment region DNA elements in a tissue-specific manner and demonstrates cell-type-dependent expression in the developing mouse CNS. Eur. J. Neurosci. 2005;21:658–668. doi: 10.1111/j.1460-9568.2005.03897.x. [DOI] [PubMed] [Google Scholar]
- Britanova O, Depew MJ, Schwark M, Thomas BL, Miletich I, Sharpe P, Tarabykin V. Satb2 haploinsufficiency phenocopies 2q32-q33 deletions, whereas loss suggests a fundamental role in the coordination of jaw development. Am. J. Hum. Genet. 2006;79:668–678. doi: 10.1086/508214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Cai S, Han H, Kowhi-Shigematsu T. Tissue-specific nuclear architecture and gene function regulated by SATB1. Nat. Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarle R, Gong JZ, Guasparri I, Pesci A, Cai J, Liu J, Simmons WJ, Dhall G, Howes J, Piva R, et al. NPM-ALK transgenic mice spontaneously develop T-cell lymphomas and plasma cell tumors. Blood. 2003;101:1919–1927. doi: 10.1182/blood-2002-05-1343. [DOI] [PubMed] [Google Scholar]
- Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat. Rev. Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- Chow JC, Hall LL, Baldry SE, Thorogood NP, Lawrence JB, Brown CJ. Inducible XIST-dependent X-chromosome inactivation in human somatic cells is reversible. Proc. Natl. Acad. Sci. USA. 2007;104:10104–10109. doi: 10.1073/pnas.0610946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl. Acad. Sci. USA. 2006;103:7688–7693. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- Dickinson LA, Joh T, Kowhi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- Dobreva G, Dambacher J, Grosschedl R. SUMO modification of a novel MAR-binding protein, SATB2, modulates immunoglobulin mu gene expression. Genes Dev. 2003;17:3048–3061. doi: 10.1101/gad.1153003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobreva G, Chahrour M, Dautzenberg M, Chirivella L, Kanzler B, Farinas I, Karsenty G, Grosschedl R. SATB2 is a multifunctional determinant of craniofacial patterning and osteoblast differentiation. Cell. 2006;125:971–986. doi: 10.1016/j.cell.2006.05.012. [DOI] [PubMed] [Google Scholar]
- Donadoni C, Corti S, Locatelli F, Papadimitriou D, Guglieri M, Strazzer S, Bossolasco P, Salani S, Comi GP. Improvement of combined FISH and immunofluorescence to trace the fate of somatic stem cells after transplantation. J. Histochem. Cytochem. 2004;52:1333–1339. doi: 10.1177/002215540405201009. [DOI] [PubMed] [Google Scholar]
- Galande S, Purbey PK, Notani D, Kumar PP. The third dimension of gene regulation: organization of dynamic chromatin loopscape by SATB1. Curr. Opin. Genet. Dev. 2007;17:408–414. doi: 10.1016/j.gde.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Byron M, Sakai K, Carrel L, Willard HF, Lawrence JB. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc. Natl. Acad. Sci. USA. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- Hoflinger S, Kesavan K, Fuxa M, Hutter C, Heavey B, Radtke F, Busslinger M. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J. Immunol. 2004;173:3935–3944. doi: 10.4049/jimmunol.173.6.3935. [DOI] [PubMed] [Google Scholar]
- Huynh KD, Lee JT. Inheritance of a pre-inactivated paternal X chromosome in early mouse embryos. Nature. 2003;426:857–862. doi: 10.1038/nature02222. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohane AM, O’Neill LP, Belyaev ND, Lavender JS, Turner BM. X-Inactivation and histone H4 acetylation in embryonic stem cells. Dev. Biol. 1996;180:618–630. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Twomey D, Feng Z, Stubbs MC, Wang Y, Faber J, Levine JE, Wang J, Hahn WC, Gilliland DG, et al. Transformation from committed progenitor to leukaemia stem cell initiated by MLL-AF9. Nature. 2006;442:818–822. doi: 10.1038/nature04980. [DOI] [PubMed] [Google Scholar]
- Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J. Cell Biol. 2007;178:219–229. doi: 10.1083/jcb.200612127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Mermoud JE, Costanzi C, Pehrson JR, Brockdorff N. Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J. Cell Biol. 1999;147:1399–1408. doi: 10.1083/jcb.147.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Rasmussen TP, Mastrangelo MA, Eden A, Pehrson JR, Jaenisch R. Dynamic relocalization of histone MacroH2A1 from centrosomes to inactive X chromosomes during X inactivation. J. Cell Biol. 2000;150:1189–1198. doi: 10.1083/jcb.150.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Fenner MH, Tan SS, Tam P, Shioda T, Li E. X inactivation in the mouse embryo deficient for Dnmt1: distinct effect of hypomethylation on imprinted and random X inactivation. Dev. Biol. 2000;225:294–303. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- Sado T, Okano M, Li E, Sasaki H. De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development. 2004;131:975–982. doi: 10.1242/dev.00995. [DOI] [PubMed] [Google Scholar]
- Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol. Cell. Biol. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Wen J, Huang S, Rogers H, Dickinson LA, Kohwi-Shigematsu T, Noguchi CT. SATB1 family protein expressed during early erythroid differentiation modifies globin gene expression. Blood. 2005;105:3330–3339. doi: 10.1182/blood-2004-08-2988. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.