Abstract
The catalytic moiety of Pseudomonas exotoxin A (domain III or PE3) inhibits protein synthesis by ADP-ribosylation of eukaryotic elongation factor 2 (eEF2). PE3 is widely used as a cytocidal payload in receptor-targeted protein toxin conjugates. We have designed and characterized catalytically inactive fragments of PE3 that are capable of structural complementation. We dissected PE3 at an extended loop and fused each fragment to one subunit of a heterospecific coiled coil. In vitro ADP-ribosylation and protein translation assays demonstrate that the resulting fusions — supplied exogenously as genetic elements or purified protein fragments — had no significant catalytic activity or effect on protein synthesis individually, but in combination catalyzed the ADP-ribosylation of eEF2 and inhibited protein synthesis. Although complementing PE3 fragments are less efficient catalytically than intact PE3 in cell-free systems, co-expression in live cells transfected with transgenes encoding the toxin fusions inhibits protein synthesis and causes cell death comparably as intact PE3. Complementation of split PE3 offers a direct extension of the immunotoxin approach to generate bispecific agents that may be useful to target complex phenotypes.
Keywords: exotoxin A, structural complementation, ADP-ribosylation, elongation factor 2, cytotoxicity
INTRODUCTION
Structural complementation, defined as the restoration of biological activity by noncovalent interaction of different inactive polypeptides, is a classical experimental tool for studying the structures and functions of oligomeric enzymes.1 For monomeric enzymes, structural complementation requires split fragments of the polypeptide chain, as in the case of the subtilisin-treated fragments (S-peptide and S-protein) of ribonuclease A.2 More recently, complementation has been used as a strategy for constructing reporter systems that respond conditionally to specific biophysical or biochemical signals. For example, split variants of fluorescent proteins,3–9 GAL4 transcriptional activator,10 β-galactosidase,11 dihydrofolate reductase,12 β-lactamase,13 luciferase,14 and DNA methylase15 have been fused to peptides and oligonucleotides to serve as reporters for library screening or biosensors for target-specific interactions. Target recognition is coupled to a biophysical signal or survival advantage that can be efficiently screened by high throughput methodologies.
Another recent use of complementation is to incorporate multiple cell-type specificities into functional proteins of biomedical interest. One example is afforded by engineered anthrax toxins harboring mutated subunits of the heptameric or octameric protective antigen.16,17 These subunits contain cleavage sites for cell-surface proteases — urokinase plasminogen activator and matrix metalloprotease — which render them incompetent for intoxication unless processed by cells that express both proteases. Here we report a complementing system designed from a monomeric toxin: the catalytic domain (domain III, or PE3) of Pseudomonas exotoxin A. PE3 is a structural and functional homolog of the catalytic domains of diphtheria toxin18,19 and cholix toxin.20 PE3 inhibits protein synthesis by ADP-ribosylating (with NAD+ as cofactor) a specific diphthamide residue in eukaryotic elongation factor 2 (eEF2).21 Intoxication induces death of the host cell through the activation of apoptotic pathways.22–24
From a biophysical point of view, covalent splitting of a monomeric protein significantly increases the total entropy of the split fragments. The magnitude of this increase depends on the extent to which the fragments retain the conformational constraints present in the original structure. In the case of subtilisin-treated ribonuclease A and certain schemes for split EGFP, this entropic penalty is not sufficient to prevent complementation. If the split fragments become significantly unfolded relative to the intact structure, however, a substantial net input in free energy may be required to drive complementation. Such a source of free energy could be furnished by fusion of the split fragments to an unrelated domain with strong affinity for heterodimerization. Thermodynamically, association of the latter domain limits the translational degrees of freedom in the split fragments, effectively destabilizing the fragments relative to the associated state and driving complementation.
We constructed a split toxin system by dissecting PE3 at an extended flexible loop and fusing each fragment to a heterospecific, antiparallel coiled coil. Individually, the fusion fragments are inactive. When both fragments are present, they spontaneously complement to yield a functional enzyme that inhibits protein translation and kills cells. Pseudomonas exotoxin A is widely used in targeted therapeutics such as immunotoxins for cancer and HIV.25–31 Structural complementation of split PE3 system offers a potential strategy to increase biological specificity by conditionally targeting two different molecular phenotypes in the same cell.
RESULTS
Design of a split ADP-ribosylating toxin
The C-terminal catalytic domain of the Pseudomonas aeruginosa exotoxin A (PE3; residues 400 to 613), inhibits protein synthesis by ADPribosylation of eEF2. Our goal was to split PE3 into two inactive fragments that would structurally complement to form functionally active enzyme.
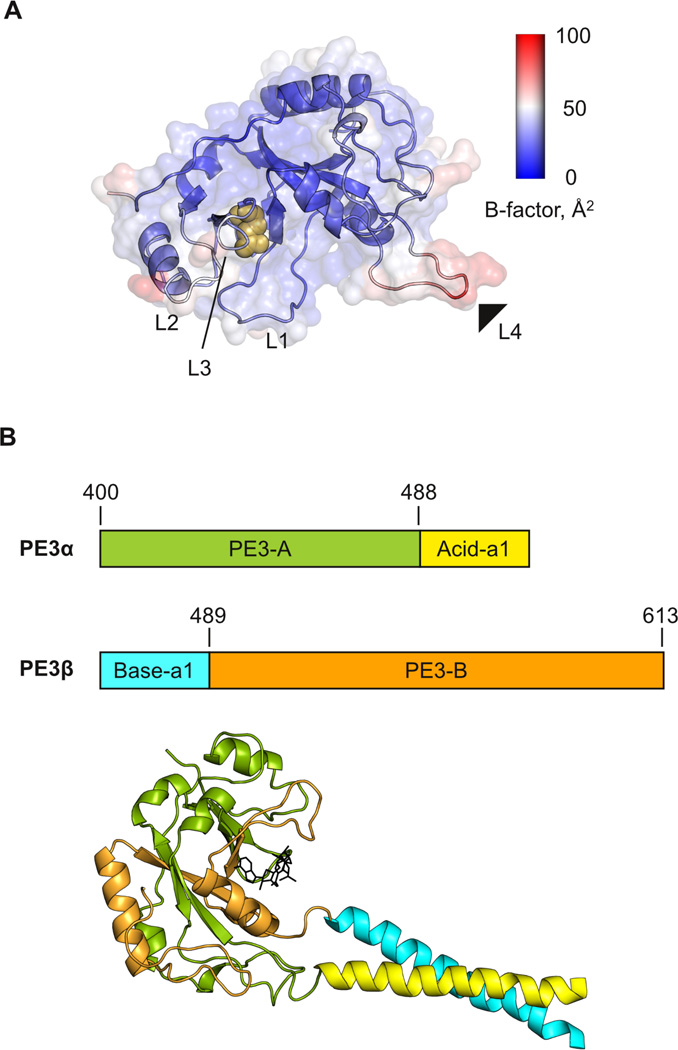
We followed a rational, biophysical approach by seeking an optimal dissection site that would minimize the thermodynamic (entropic) cost for reassembly. To do so, we screened the protein backbone for extended segments that are unfolded and mobile, using B-factors associated with the Cα atoms in the refined crystal structure of wildtype exotoxin A32 as a quantitative guide of dynamic fluctuations (Figures 1A and S1, Supplementary Data). Four distinct flexible loops were identified: L1 (residues 458 to 463), L2 (residues 517 to 522), L3 (residues 546 to 551), and L4 (residues 486 to 493) based on locally elevated B-factors. Since their Cα positions exhibit full occupancy, these segments represent dynamic and disordered loops rather than discrete stable conformations. Comparison of crystal structures of full-length exotoxin and PE3 bound to various NAD+ analogs revealed that the conformations of L1, L2, and L3 are coupled to residues in the catalytic cleft or ligand binding.32 These loops are also proximal to Glu553, an essential residue in the catalytic cleft.33 In contrast, L4 is distant from the catalytic site and also the most dynamic. It is also the most extended of the four loops and should provide the most spatial tolerance for fusion with a heterospecific domain. We therefore selected this loop for dissection of the PE3 monomer.
Figure 1. Design of split PE3.
A, Backbone trace of PE3 (residues 400 to 613) taken from the refined structure of the wildtype exotoxin A (PDB: 1IKQ), colored by B-factor. The four mobile loops identified are labeled L1 to L4. The site of dissection, Asp488/Ala489, is indicated by the solid triangle. The catalytically essential Glu553 residue is shown in gold. B, Schematic and homology model (templated to PDB co-ordinates from 1DMA and 1R48) of the fusion constructs PEα and PEβ generated by linking the split PE3 fragments to a heterodimeric coiled coil by Oakley and Kim 34. NAD+ is shown as black sticks.
We split PE3 genetically at L4 between Asp488 and Ala489 and fused each fragment [PE3-A (residues 400–488) and PE3-B (residues 489–613)] to each subunit of the antiparallel, heterodimeric coiled coil (Acid-a1 and Base-a1) devised by Oakley and Kim.34 The two fusion fragments are designated PE3α, consisting of PE3-A:Base-a1, and PE3β, consisting of Acida1: PE3-B (Figure 1B). No cysteine residue is present in the constructs. The antiparallel orientation of the coiled coil maintains correct polarity of the two fragments upon dimerization, as demonstrated with a split GFP by Regan and coworkers.4
Structural complementation of split PE3 restores ADPRT activity
PE3α and PE3β were initially expressed as His6-tagged constructs in E. coli. While we recovered PE3β by native purification, we could not isolate the PE3α fragment under native or denaturing conditions. Recloning PE3α as a C-terminal fusion to maltose binding protein (MBP) yielded protein when induced at 22°C. Co-expression of MBP-PEα and His6-PE3β yielded a complex that co-purified on Co-NTA agarose. To confirm specific, noncovalent association of the two fragments, the mixture was pulled down on amylose-agarose. The PEβ fragment (17 kDa), which lacked MBP, co-purified with MBP-tagged PEα (61 kDa) (Figure S2A, Supplementary Data). When fractionated by sizeexclusion chromatography, the coil-bearing MBP-PEα and His6-PE3β complex (3 µM) eluted as a monodisperse species with a retention volume similar to monomeric BSA (67 kDa; Figure S2B). While below the calculated molecular weight of the MBP/His6-tagged α+β heterodimer (60.3 + 19.2 = 79.5 kDa), the complex eluted well ahead of carbonic anhydrase (29 kDa) and cytochrome C (13 kDa). Cleavage of purified heterodimer with TEV protease yielded the α+β heterodimer free of all purification tags (including MBP).
To probe whether the heterodimeric coiled-coil is required for complementation of split PE3, we tested minimal constructs consisting only of PE3-A (residues 400–488) and PE3-B (residues 489–613) in pull-down experiments analogous to those described for PE3α and PE3β. Unlike their coil-bearing counterparts, the minimal constructs did not co-precipitate under the same conditions (c.f. Figures S2 and S3, Supplementary Data), indicating that assisted dimerization is required for structural complementation of split PE3.
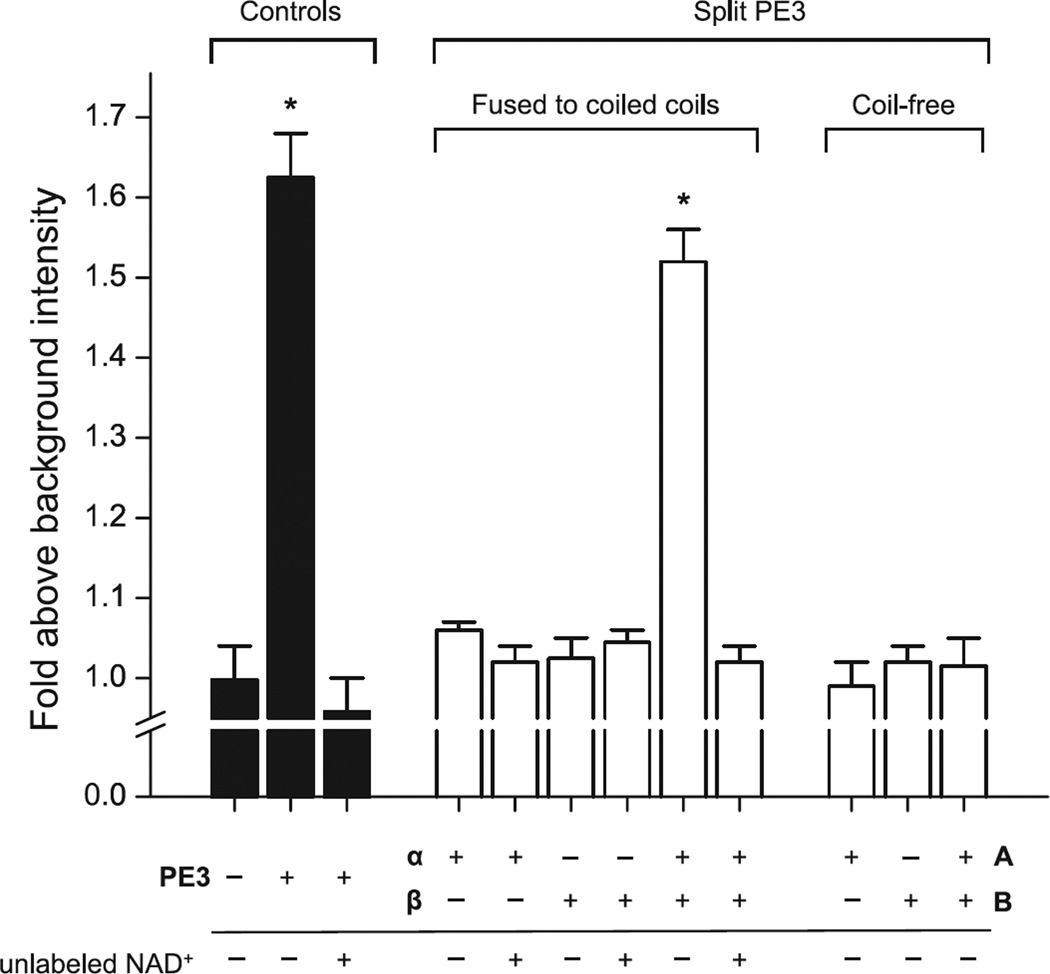
We measured the ADP-ribosyltransferase (ADPRT) activity of split PE3 by the incorporation of a fluorescently-labeled NAD+ analog (6-carboxyfluorescein-17-NAD+, 150 µM) to purified, recombinant yeast eEF2 (2 µM). ADPRT activity was first measured at a subunit concentration of 20 µM (Figure 2). Consistent with the pull-down data, coil-free PE3-A and PEB exhibited no ADPRT activity — individually or in combination — functionally confirming the absolute requirement of a heterodimerization domain for complementation of the PE3 fragments. For the coil-bearing fragments, neither PEα nor PEβ fragment was active individually, but together exhibited ADPRT activity, as did intact PE3 (positive control). Incorporation of fluorescence by the PEα+PEβ heterodimer and intact PE3 was abolished by excess unlabeled NAD+. Titration of the α+β heterodimer showed a potency in ADPRT activity of 16 ± 8 µM, compared with 4.9 ± 2.6 µM for intact PE3, a ~4-fold difference (Figure 3).
Figure 2. Structural complementation of split PE3 restores enzymatic activity.
ADP-ribosyltransferase activity was measured by incorporation of 6-carboxyfluorescein-17-NAD+ into yeast eEF2 as described in Materials and Methods. Background intensity refers to the fluorescence of a mock sample containing fluorescein-NAD+ alone. PE3 constructs were present at 20 µM (complex in the case of split PE3 fragments). The split PE3 fragments harboring a heterospecific coiled-coil are designated α and β, and their coiled-free counterparts A and B, following the nomenclature as described in the main text. Bars represent means ± SEM from duplicate measurements. Asterisks indicate statistically significant differences (p < 0.05) from background. Note the lack of ADPRT activity in coil-free PE3-A and PE3-B.
Figure 3. Comparative enzymatic activities of split and intact PE3.
ADPRT activity was measured by incorporation of 6-carboxyfluorescein-17-NAD+ into yeast eEF2 as described in Materials and Methods. Concentrations for split PE3 refer to the PEα:PEβ complex (harboring a heterodimeric coiled coil) or PE-A+PE-B (coil-free). The coil-free fragments do not heterodimerize as judged by pull-down experiments (Figure S2, Supplementary Data). Relative fluorescence incorporation was normalized against the fluorescence of a mock sample containing fluorescein-NAD+ alone. Symbols represent means ± SEM from duplicate measurements. Lines are best fits to the Hill equation.
Split PE3 inhibits protein synthesis following structural complementation
To evaluate the effect on protein synthesis by split PE3, we used rabbit reticulocyte lysate (RRL), a partially-purified, cell-free system containing all components required for protein synthesis from mRNA substrates. We treated RRL with 300 nM purified PE3α and/or PE3β, or intact PE3, for 30 min with or without NAD+ (50 µM). Luciferase mRNA was added to the reactions and protein synthesis was measured after 90 min by incorporation of [35S]-methionine into luciferase and detected by phosphorimagery of SDS-PAGE gels (Figure 4A). Consistent with the ADPRT results, whereas RRL pre-treated with PE3α and PE3β individually produced [35S]-luciferase, those exposed to both fragments (after TEV cleavage) or intact PE3 did not (Figure 4A; top panel). Inhibition of protein synthesis by the combined fragments was ADPRT-mediated, as confirmed by their dependence on NAD+ (bottom panel). Although the MBP/His6-tagged α+β heterodimer was manifestly dimeric before TEV cleavage, it did not inhibit protein synthesis, suggesting MBP prevented refolding of the catalytic domain or blocked the docking of eEF2 or NAD+.
Figure 4. Split PE3 inhibits protein synthesis following structural complementation in a dose-dependent manner.
Rabbit reticulocyte lysate (RRL) was treated with various purified constructs as indicated for 30 min. prior to the addition of 400 ng of luciferase mRNA. Residual translational activity was measured as [35S]-labeled luciferase or by luciferin chemiluminescence. A, Lysate were treated with the indicated protein constructs at 300 nM in the presence or absence of 50 µM NAD+. Densitometric measurements of [35S] incorporation for all samples, except for intact PE3 and the α+β heterodimer in the presence of NAD+, were randomly ±15%. B, RRL was titrated with intact PE3 (solid squares) or TEV-activated PE3 α+β heterodimer (open squares) in the presence of 50 µM NAD+. C, Titration with NAD+ in the presence of 300 nM intact PE3 (solid) or α+β heterodimer (open). Symbols represent means of four experiments ± SEM. Lines are best fits to the Hill equation.
To determine the dose-dependent activity of the PE3 α+β heterodimer, RRL was titrated with PE3 α+β heterodimer (activated by TEV cleavage) or intact PE3 in the presence of 50 µM NAD+, before addition of firefly luciferase mRNA. Residual translational activity was assessed by luciferin chemiluminescence (Figure 4B). A global fit of both data sets gave an apparent IC50 of 1.4 ± 0.4 nM for intact PE3 and 41 ± 11 nM (complex) for the α+β heterodimer, a ~40-fold difference. Utilization of NAD+ was assessed by titrating RRL with NAD+ in the presence of 300 nM of either toxin (Figure 4C). Under these conditions, the apparent IC50 values are 52 ± 12 nM and 1.1 ± 0.3 µM for intact PE3 and the α+β heterodimer (~22-fold difference), respectively. In summary, complementation of split PE3 substantially regains the specific enzymatic activity of intact PE3.
Inhibition of protein synthesis by split PE3 triggers cell death
We then investigated whether inhibition of protein synthesis could be achieved using tag-free constructs synthesized in situ. mRNA encoding PE3α, PE3β, and intact PE3 were separately prepared by in vitro transcription of pcDNA3.1-based plasmids. Purified mRNA was added to RRL to generate the corresponding protein in situ before the addition of luciferase mRNA. To account for the depletion of amino acid precursors from the first round of translation, we also included a control sample with mRNA encoding EGFP. Consistent with results using recombinant protein fragments, the combination of PE3α and PE3β mRNA inhibited translation similarly as intact PE3, while PE3α or PE3β individually had no significant effect beyond the EGFP control (Figure 5A).
Figure 5. Complementation of genetically encoded split PE3 inhibits protein synthesis in vitro and in live cells.
A, Rabbit reticulocyte lysate was treated with purified mRNA encoding the indicated constructs in the presence of 50 µM NAD+. Two different total mRNA doses (800 and 400 ng) were used. To account for the depletion of amino acid precursors prior to addition of luciferase mRNA, 800 ng of a control mRNA encoding for EGFP was also tested. Densitometric comparison of [35S] incorporation with a no-mRNA sample showed ~50% lower intensity for EGFP and other samples (except for those treated with intact PE3 and the α+β heterodimer, which are undetectable above background). B, HEK293 cells were transfected with expression plasmids encoding intact PE3 or various combinations of PEα and/or PEβ fragments. After 24 and 48 h, protein synthesis was measured by [3H]-leucine incorporation. Bars represent means ± SEM relative to vector control from triplicate experiments. Asterisks without brackets indicate statistically significant differences (p < 0.05) from vector control for the corresponding time point. Brackets indicate pairwise comparisons as shown. C, HEK293T cells stably expressing a destabilized EGFP (dEGFP) were transfected with plasmids as for the [3H]-leucine uptake assay. Cellular dEGFP was measured by flow cytometry at 24 h after transfection. Bars represent means ± SEM relative to vector control from four experiments.
To probe the effect on protein synthesis by the split PE3 fragments in live cells, the same pcDNA3.1 plasmids constitutively expressing PE3α, PE3β, or intact PE3 (500 ng total DNA) were transiently transfected into HEK293 cells. Protein synthesis was quantified by the incorporation of [3H]-leucine at 24 and 48 h after transfection (Figure 5B). Individually, PE3α and PE3β had no significant effect on [3H]-leucine incorporation over vector control. In contrast, at a total dose of 500 ng DNA, co-transfected PE3α and PE3β strongly inhibited protein synthesis by 24 h to an extent similar to intact PE3. To corroborate these observations, we examined the effect on protein synthesis using a destabilized EGFP (dEGFP) (Figure 5C) that is rapidly cleared by proteasomal degradation.35 In agreement with the [3H]-leucine data, cellular dEGFP fluorescence was indistinguishable from vector control at 24 h after transfection of PEα or PEβ. Co-transfection of both plasmids at a total dose of 500 ng DNA led to a significant loss of fluorescence similar to intact PE3. Neither treatment reduced dEGFP fluorescence to the same extent as cycloheximide, a chemical inhibitor of protein synthesis, reflecting a lag time between transfection and protein expression.
Cytotoxicity of split PE3 was measured by cellular metabolic status (reduction of XTT or resazurin) at 48 and 72 h after transfection (Figure 6). At a dose of 500 ng of total DNA, no significant difference in cell viability was observed after individual transfection of PE3α or PE3β relative to vector control. Co-transfection of PE3α and PE3β (500 ng total DNA) was significantly more toxic than either plasmid alone and to a similar extent as intact PE3 (Figure 6A). To estimate the difference in cytotoxic potency for the two toxins, we prepared serial 2-fold dilutions with vector DNA to maintain a constant quantity of total DNA (500 ng). A 7-fold difference was observed between the intact and α+β toxins at 48 h after reverse transfection (Figure 6B). Given the different gene sequences encoded by the two plasmids, the titrated doses of DNA cannot be directly interpreted as equivalent protein concentrations. The data should be interpreted as the expected difference in toxicity if the constructs were delivered genetically and their expression driven by the same promoter.
Figure 6. Co-transfection of split PE3 causes cell death.
HEK293 cells were transiently transfected with expression plasmids encoding the indicated constructs and incubated for 48 or 72 h. A constant amount of total DNA (500 ng per 105 cells) was used in each treatment. A, Cytotoxicity relative to intact PE3 was measured by XTT reduction. Bars represent means of viability relative to vector ± SEM from four experiments. Asterisks without brackets indicate statistically significant differences (p < 0.05) from vector control for the corresponding time point. Brackets indicate pairwise comparisons as shown. B. Relative dose dependence of cytotoxicity as measured by resazurin reduction. The dose of plasmids encoding each construct was diluted with vector as indicated while maintaining the total DNA at 500 ng. Symbols represent means of four experiments ± SEM. Lines are best fits to the Hill equation.
DISCUSSION
Our objective in this study was to develop a complementing cytotoxin using the catalytic domain of Pseudomonas exotoxin A (PE3). As proof-of-principle, we dissected PE3 and fused each fragment to one subunit of a heterospecific coiled coil (Figure 1). Individually, the fusion fragments, PE3α and PE3β, have no effect on protein translation in vitro — as purified proteins or genetic elements — or when transfected as plasmids into live cells. The two fragments associate noncovalently to form an α+β heterodimer (Figure 2) capable of ADP-ribosylating eEF2 (Figure 3) and inhibiting protein synthesis in a NAD+-dependent manner, with an IC50 of ~40 nM complex (Figure 4). Following transfection, PE3α and PE3β fragments inhibit protein synthesis only in combination (Figure 5) and are able to intoxicate HEK293 as well as intact PE3 (Figures 6). As an inhibitor of protein synthesis, the potency of the α+β heterodimer is approximately 40-fold lower than intact PE3 (Figure 4B). We observed that neither the α+β heterodimer nor intact PE3 caused complete cell death by chemical transfection due to the efficiency constraints of this method, as others have also reported.36,37 Since the cytocidal properties of PE3 are historically well-established 21,38–40 and used to great effect in therapeutic toxin conjugates,25–31 our objective here is to characterize the functional properties of complementing PE3 relative to intact PE3 under identical experimental conditions.
Perturbation to the catalytic activity in split PE3 relative to the intact protein
Observed differences in potency in terms of ADPRT activity (~4-fold), inhibition of protein synthesis (~40-fold) or NAD+ utilization (~22-fold difference) between intact and split PE3 reflect the oligomeric stability of the PE3 α+β heterodimer as well as its relative catalytic activity in the dimeric state. We found that structural complementation required thermodynamic stabilization by a dimerization domain. To drive heterodimerization, we have chosen the antiparallel coiled coils by Oakley and Kim, who showed that they are quantitatively dimeric at a complex concentration of 10 µM.34 We expect that other oligomerizing domains would also be tolerated, to the extent that their orientation does not strain the structure of the α+β heterodimer.
Overlaid on the thermal stability of the PE3 dimer is the intrinsic catalytic activity of the dimeric state itself (i.e. the activity if the coiled coil were covalently linked). To minimize the deficit in intrinsic activity of split PE3, we chose to dissect PE3 at L4 instead of the other disordered loops, in view of L4’s highly disordered conformation and the lack of coupling to the catalytic residues in the crystal structures of the PE3 apoenzyme (PDB: 1IKQ) and PE3-NAD+ holoenzyme (1DMA).32 Merill and coworkers have reported that L4 may be involved in conferring specificity for eEF2.41 However, in co-crystal structures involving eEF2,42,43 L4 remains completely disordered and does not make any close contact with eEF2, so the nature of the L4’s coupling to the catalytic mechanism is unclear. In addition, the site of dissection between Asp488 and Ala489 is at least 6.5 Å away from the closest eEF2 contact and points away from eEF2 into the solvent (Figure S4, Supplementary Data). The Asp488/Ala489 junction is or near the optimal site for dissecting PE3 in terms of minimizing perturbation on the enzyme’s conformation or blockage of the binding interface with eEF2. Our functional data in terms of ADPRT activity, inhibition of protein synthesis, and cytotoxicity all indicate that the our split PE3 design retains substantively the activity of intact PE3, and may serve in applications using PE3 or the homologous catalytic domain of diphtheria toxin.
Potential applications
Diphtheria toxin (DT), and by extension Pseudomonas exotoxin A, is famously known for their “one-molecule-can-kill-a-cell” potency.44 Both toxins are widely used in targeted therapeutics for various oncologic (particularly leukemic), infectious, and autoimmune diseases.25–31 Replacement of the native receptor-binding domains with other ligands, such as monoclonal antibodies, results in toxin fusions with redirected specificity for other cell-surface receptors.45,46 Currently, targeting is typically limited to single receptors such as CD22,47 the IL-2 48 and IL-13α2 receptors,49 viral envelope glycoproteins,50,51 and the EGFR family of receptor tyrosine kinases.52–56 Alternatively, targeting can be achieved with toxin transgenes whose expression is controlled by a tissue- or disease-specific promoter.57–62 Whichever the route, biological specificity is limited to cases where the phenotype is unique to the targeted cells, such CD20, a B cell-lineage restricted antigen, in non-Hodgkin’s lymphoma.63
In many diseases, a receptor or gene is overexpressed, but not unique, relative to normal cells. In these cases, monospecific targeting leads to unintended toxicity to normal cells. For example, because the HER2 receptor, a major marker in several cancers, is also found on myocardial tissues, targeting HER2 alone leads to cardiotoxicity in vivo.64,65 To increase biological specificity, several PE- and DT-based toxin conjugates with two ligands or antibodies directed at separate receptors on the target cells have been reported.66 Complementing PE3 represents an orthogonal strategy for bispecific targeting of a single cell. By directing the delivery or expression of each inactive fragment against a different molecular target, toxicity can be focused to a more refined biological phenotype. Complementation offers comparable cytotoxicity with potentially enhanced level of selectivity than bispecific ligands because toxicity is conditional on the presence of both fragments. Since complementation cannot occur in cells that present only one or the other target, off-target cells would be spared.
Our in vitro translation assays with purified PE3 fragments indicate that tagged α+β heterodimer does not inhibit protein synthesis even though it is dimeric. The heterodimer is activated by the removal of the MBP and His6 tags with TEV protease. Thus, the targeted delivery of a highly sequence-specific protease or incorporation of the cleavage site of a diseasespecific protease offers potential opportunities for additional specificities.
Our data also show that structural complementation occurs when the fragments are expressed in situ, supporting genetic delivery as an alternative approach to target phenotypes that are transcriptionally regulated.57,58 This is an advantage over bispecific toxins based on the anthrax protective antigen.16,17 In conclusion, complementing PE3 represents a potentially flexible platform for developing bispecific therapeutics by targeted transduction of the fragments or by delivery as transgenes under the control of specific promoters. Given the vast body of experience with exotoxin-based therapeutics,25–27 complementing PE3 may be readily adapted to the many exotoxin-based targeted therapies already in use.
MATERIALS AND METHODS
Molecular cloning
Synthetic genes were designed from sequences for Pseudomonas aeruginosa exotoxin A (Genbank: K01397.1) and the antiparallel heterodimeric coiled coil reported by Oakley and Kim.34 Inserts (PEα, PEβ, and PE3; Figure 1B) were cloned into the NheI/EcoRI sites of pcDNA3.1(+) for eukaryotic expression or T7-driven transcription in vitro. The same inserts were cloned into the NcoI/SacI sites of pET28b or the NcoI/NdeI sites of pET15b for bacterial expression. PEα was also cloned as a C-terminal fusion to maltose binding protein (MBP) in pMal-p2x (New England Biolab). All constructs for bacterial expression include additional sequences encoding His6 tags separated by cleavage sites for TEV protease (ENLYFQ↓G). Sequences were verified by Sanger sequencing.
Protein overexpression and purification
pET-based plasmids were transformed into BL21(DE3)Star E. coli (Invitrogen) and grown in LB Miller medium at 37°C. At OD600 ~ 0.6, cells were induced with IPTG to 0.5 mM. Induction was maintained for 4 to 5 h. Cells were harvested by centrifugation at 6,000 × g for 10 min. Cell pellets were resuspended in Buffer A (0.1 M TrisHCl, pH 7.5, 0.5 M NaCl, 10% w/v glycerol, 10 mM imidazole) with PMSF added at 0.1 mM, sonicated on ice, and cleared by centrifugation at 60,000 × g for 30 min. The supernatant was loaded onto Co-NTA agarose resin, washed exhaustively with Buffer A, and eluted in Buffer B (Buffer A with 150 mM imidazole). Fractions were checked by SDS-PAGE with Coomassie Blue staining. Protein concentration was determined by UV absorption at 280 nm based on a calculated extinction coefficient of 91,790 and 16,960 M−1 cm−1 for uncleaved PEα and PEβ fragments, respectively. Recombinant TEV protease (plasmid 8830; Addgene) was overexpressed in E. coli strain BL21(DE3)-RIL and purified on Co-NTA as described.67
Pull-down assay
Samples (10 µg) were incubated with 100 µL of amylose-agarose slurry (New England Biolab) for 1 h at room temperature. The resin was washed twice with 1 mL of Buffer C (20 mM TrisHCl, pH 7.4, 200 mM NaCl, 1 mM EDTA) and eluted in 150 µL of Buffer D (Buffer C plus 10 mM maltose). Aliquots (12 µL) were separated by SDS-PAGE and detected by Coomassie Blue staining.
Size-exclusion chromatography
Samples (300 µL) were loaded onto a Superdex 200 10/300 GL column and eluted under the control of an ÅKTAPurifier instrument (GE). The column was eluted isocratically with Buffer C at 0.5 mL/min and detected by UV absorption at 280 nm.
ADP-ribosylation assay
Yeast eEF2 was purified as previous described68 from S. cerevisiae in which the native chromosomal eEF2 has been replaced by a recombinant, C-terminally His6-tagged copy. eEF2 (2 µM; extinction coefficient of 74,300 M−1 cm−1 at 280 nm) was incubated with 150 µM 6-carboxyfluorescein-17-NAD+ (Trevigen) and with or without toxin construct and/or unlabeled NAD+ (750 µM) at 30°C for 2 h. These assay conditions are based on kinetic characterizations reported by Douglas and Collier.69 Unincorporated and eEF2-bound fluorescent label were separated by centrifugation through a desalting spin column (MWCO 7,000; Thermo Scientific). Fluorescence of the eluate was read in a black 96-well plate with a plate reader at 485/528 nm.
In vitro protein synthesis assay. Rabbit reticulocyte lysate (Promega) was treated with purified proteins or their corresponding mRNA (generated by in vitro transcription of the pcDNA-based plasmids) in the presence or absence of NAD+. Samples for SDS-PAGE analysis were incubated with a methionine-free amino acid mixture supplemented with [35S]-methionine (1175 Ci/mmol; MP Biomedical, Solon, OH). Following incubation at 30°C for 30 min, 400 ng of firefly luciferase mRNA were added. After an additional 90 min incubation, luciferase was assayed by chemiluminescence (unlabeled samples) using a commercial luciferin reagent (Promega) or by SDS-PAGE (35S-labeled samples). Gels were dried onto blotting paper and visualized by phosphorimagery using a Storm 860 instrument (GE Healthcare).
Cell culture and transfection
HEK293 cells were maintained in RPMI 1680 medium containing 10% heat-inactivated fetal bovine serum. Cells were seeded in 24- or 96-well plates in complete medium at 105 or 2 × 104 per well, respectively, 24 h before transfection. Cells in 24-well plates (500 µL) were transfected with 500 ng plasmid DNA per well using Fugene HD (Promega), according to the manufacturer’s directions. Cells in 96-well plates (100 µL) were transfected with 50 ng of DNA using a reverse transfection procedure.
Cell-based protein synthesis assay
At 24 or 48 h after transfection, medium was replaced with leucine-deficient medium supplemented with 1 µCi of [3H]-leucine/mL (Perkin Elmer, Billerica, MA) and incubated for 1 h. Plates were washed twice with 1 mL of PBS and counted in 0.5 mL scintillation fluid. Counts were normalized to cells transfected with pcDNA3.1 vector.
Alternatively, a destabilized-EGFP reporter plasmid was constructed by subcloning the CMV promoter from pcDNA3.1(+) between the BglII/KpnI sites of pd2EGFP-1 (Clontech). HEK293T cells stably expressing this construct were transiently transfected with PE3-related plasmids. After 24 h, cellular fluorescence was measured by flow cytometry at 488/510 nm. Data were gated to an untransfected sample and analyzed using FlowJo (Tree Star, Ashland, OR). A minimum of 10,000 events were counted for each sample.
Cell viability assay
At the indicated time points after transfection, cell viability was assessed by the metabolic reduction of XTT or resazurin. The XTT assay was performed in 24-well plates by adding 100 µL of activated XTT reagent (Biotium, Hayward, CA) to each well. After 6 h, background absorbance (675 nm) and absorbance of reduced XTT (475 nm) were measured and subtracted. For resazurin reduction, cells were reverse-transfected in 96-well black plates. Twenty µL of resazurin reagent (Cell-Titer Blue, Promega) was added to each well and incubated at 37°C for 2 h. Resorufin fluorescence was measured at 530/565 nm. All treatment data were normalized to cells transfected with pcDNA3.1 vector alone.
Data analysis
Statistics and least-square curve fitting was performed using Origin (Northampton, MA). Hypothesis testing was performed by two-sample t-tests or ANOVA with Bonferroni’s method for multiple comparisons. Statistical comparison is taken at p < 0.05. Error bars denote standard error of the mean (SEM) from replicate experiments as indicated.
Supplementary Material
ABBREVIATIONS
- PE3
Pseudomonas aeruginosa exotoxin A domain III (residue 400 to 613)
- ADPRT
ADP-ribosyltransferase
- eEF2
Eukaryotic elongation factor 2
- MBP
Maltose-binding protein
- RRL
Rabbit reticulocyte lysate
- DT
Diphtheria toxin
REFERENCES
- 1.Zabin I, Villarejo MR. Protein Complementation. Annu Rev Biochem. 1975;44:295–313. doi: 10.1146/annurev.bi.44.070175.001455. [DOI] [PubMed] [Google Scholar]
- 2.Richards FM, Vithayathil PJ. The preparation of subtilisn-modified ribonuclease and the separation of the peptide and protein components. J Biol Chem. 1959;234:1459–1465. [PubMed] [Google Scholar]
- 3.Demidov VV, Dokholyan NV, Witte-Hoffmann C, Chalasani P, Yiu HW, Ding F, Yu Y, Cantor CR, Broude NE. Fast complementation of split fluorescent protein triggered by DNA hybridization. Proc Natl Acad Sci U S A. 2006;103:2052–2056. doi: 10.1073/pnas.0511078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh I, Hamilton AD, Regan L. Antiparallel Leucine Zipper-Directed Protein Reassembly: Application to the Green Fluorescent Protein. J Am Chem Soc. 2000;122:5658–5659. [Google Scholar]
- 5.Magliery TJ, Wilson CG, Pan W, Mishler D, Ghosh I, Hamilton AD, Regan L. Detecting protein-protein interactions with a green fluorescent protein fragment reassembly trap: scope and mechanism. J Am Chem Soc. 2005;127:146–157. doi: 10.1021/ja046699g. [DOI] [PubMed] [Google Scholar]
- 6.Stains CI, Porter JR, Ooi AT, Segal DJ, Ghosh I. DNA sequenceenabled reassembly of the green fluorescent protein. J Am Chem Soc. 2005;127:10782–10783. doi: 10.1021/ja051969w. [DOI] [PubMed] [Google Scholar]
- 7.Cabantous S, Terwilliger TC, Waldo GS. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol. 2005;23:102–107. doi: 10.1038/nbt1044. [DOI] [PubMed] [Google Scholar]
- 8.Hu CD, Kerppola TK. Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat Biotechnol. 2003;21:539–545. doi: 10.1038/nbt816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerppola TK. Bimolecular Fluorescence Complementation (BiFC) Analysis as a Probe of Protein Interactions in Living Cells. Annual Review of Biophysics. 2008;37:465–487. doi: 10.1146/annurev.biophys.37.032807.125842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 11.Rossi F, Charlton CA, Blau HM. Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. Proc Natl Acad Sci U S A. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelletier JN, Campbell-Valois FX, Michnick SW. Oligomerization domain-directed reassembly of active dihydrofolate reductase from rationally designed fragments. Proc Natl Acad Sci U S A. 1998;95:12141–12146. doi: 10.1073/pnas.95.21.12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galarneau A, Primeau M, Trudeau LE, Michnick SW. Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat Biotechnol. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 14.Paulmurugan R, Gambhir SS. Monitoring protein-protein interactions using split synthetic renilla luciferase protein-fragment-assisted complementation. Anal Chem. 2003;75:1584–1589. doi: 10.1021/ac020731c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nomura W, Barbas CF. In Vivo Site-Specific DNA Methylation with a Designed Sequence-Enabled DNA Methylase. J Am Chem Soc. 2007;129:8676–8677. doi: 10.1021/ja0705588. [DOI] [PubMed] [Google Scholar]
- 16.Liu S, Redeye V, Kuremsky JG, Kuhnen M, Molinolo A, Bugge TH, Leppla SH. Intermolecular complementation achieves high-specificity tumor targeting by anthrax toxin. Nat Biotech. 2005;23:725–730. doi: 10.1038/nbt1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips DD, Fattah RJ, Crown D, Zhang Y, Liu S, Moayeri M, Fischer ER, Hansen BT, Ghirlando R, Nestorovich EM, Wein AN, Simons L, Leppla SH, Leysath CE. Engineering Anthrax Toxin Variants That Exclusively Form Octamers and Their Application to Targeting Tumors. J Biol Chem. 2013;288:9058–9065. doi: 10.1074/jbc.M113.452110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collier RJ, Cole HA. Diphtheria toxin subunit active in vitro. Science. 1969;164:1179–1181. doi: 10.1126/science.164.3884.1179. [DOI] [PubMed] [Google Scholar]
- 19.Collier RJ. Effect of diphtheria toxin on protein synthesis: inactivation of one of the transfer factors. J Mol Biol. 1967;25:83–98. doi: 10.1016/0022-2836(67)90280-x. [DOI] [PubMed] [Google Scholar]
- 20.Jørgensen R, Purdy AE, Fieldhouse RJ, Kimber MS, Bartlett DH, Merrill AR. Cholix Toxin, a Novel ADP-ribosylating Factor from Vibrio cholerae. J Biol Chem. 2008;283:10671–10678. doi: 10.1074/jbc.M710008200. [DOI] [PubMed] [Google Scholar]
- 21.Iglewski BH, Kabat D. NAD-dependent inhibition of protein synthesis by Pseudomonas aeruginosa toxin. Proc Natl Acad Sci U S A. 1975;72:2284–2288. doi: 10.1073/pnas.72.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du X, Youle RJ, FitzGerald DJ, Pastan I. Pseudomonas Exotoxin AMediated Apoptosis Is Bak Dependent and Preceded by the Degradation of Mcl-1. Mol Cell Biol. 2010;30:3444–3452. doi: 10.1128/MCB.00813-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenkins CE, Swiatoniowski A, Issekutz AC, Lin T-J. Pseudomonas aeruginosa Exotoxin A Induces Human Mast Cell Apoptosis by a Caspase-8 and-3-dependent Mechanism. J Biol Chem. 2004;279:37201–37207. doi: 10.1074/jbc.M405594200. [DOI] [PubMed] [Google Scholar]
- 24.Sharma AK, FitzGerald D. Pseudomonas exotoxin kills Drosophila S2 cells via apoptosis. Toxicon. 2010;56:1025–1034. doi: 10.1016/j.toxicon.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weldon JE, Pastan I. A guide to taming a toxin - recombinant immunotoxins constructed from Pseudomonas exotoxin A for the treatment of cancer. FEBS J. 2011;278:4683–4700. doi: 10.1111/j.1742-4658.2011.08182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolf P, Elsässer-Beile U. Pseudomonas exotoxin A: From virulence factor to anti-cancer agent. Int J Med Microbiol. 2009;299:161–176. doi: 10.1016/j.ijmm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Shapira A, Benhar I. Toxin-based therapeutic approaches. Toxins (Basel) 2010;2:2519–2583. doi: 10.3390/toxins2112519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antignani A, Fitzgerald D. Immunotoxins: the role of the toxin. Toxins (Basel) 2013;5:1486–1502. doi: 10.3390/toxins5081486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger EA, Pastan I. Immunotoxin complementation of HAART to deplete persisting HIV-infected cell reservoirs. PLoS Pathog. 2010;6:e1000803. doi: 10.1371/journal.ppat.1000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kreitman RJ. Immunotoxins for targeted cancer therapy. AAPS J. 2006;8:E532–E551. doi: 10.1208/aapsj080363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pastan I, Hassan R, Fitzgerald DJ, Kreitman RJ. Immunotoxin therapy of cancer. Nat Rev Cancer. 2006;6:559–565. doi: 10.1038/nrc1891. [DOI] [PubMed] [Google Scholar]
- 32.Wedekind JE, Trame CB, Dorywalska M, Koehl P, Raschke TM, McKee M, FitzGerald D, Collier RJ, McKay DB. Refined crystallographic structure of Pseudomonas aeruginosa exotoxin A and its implications for the molecular mechanism of toxicity. J Mol Biol. 2001;314:823–837. doi: 10.1006/jmbi.2001.5195. [DOI] [PubMed] [Google Scholar]
- 33.Douglas CM, Collier RJ. Exotoxin A of Pseudomonas aeruginosa: substitution of glutamic acid 553 with aspartic acid drastically reduces toxicity and enzymatic activity. J Bacteriol. 1987;169:4967–4971. doi: 10.1128/jb.169.11.4967-4971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oakley MG, Kim PS. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry. 1998;37:12603–12610. doi: 10.1021/bi981269m. [DOI] [PubMed] [Google Scholar]
- 35.Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC, Kain SR. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- 36.Hafkemeyer P, Brinkmann U, Gottesman MM, Pastan I. Apoptosis induced by Pseudomonas exotoxin: a sensitive and rapid marker for gene delivery in vivo. Hum Gene Ther. 1999;10:923–934. doi: 10.1089/10430349950018328. [DOI] [PubMed] [Google Scholar]
- 37.Glinka EM, Andryushchenko AS, Sapozhnikov AM, Zatsepina OV. Construction of the plasmid for expression of ETA-EGFP fusion protein under control of the cytomegalovirus promoter and its effects in HeLa cells. Plasmid. 2009;62:119–127. doi: 10.1016/j.plasmid.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 38.Pollack M. The role of exotoxin A in pseudomonas disease and immunity. Rev Infect Dis. 1983;5(Suppl 5):S979–S984. doi: 10.1093/clinids/5.supplement_5.s979. [DOI] [PubMed] [Google Scholar]
- 39.Liu PV. Extracellular Toxins of Pseudomonas aeruginosa. J Infect Dis. 1974;130:S94–S99. doi: 10.1093/infdis/130.supplement.s94. [DOI] [PubMed] [Google Scholar]
- 40.Kreitman RJ, Pastan I. Accumulation of a Recombinant Immunotoxin in a Tumor in Vivo: Fewer Than 1000 Molecules per Cell Are Sufficient for Complete Responses. Cancer Res. 1998;58:968–975. [PubMed] [Google Scholar]
- 41.Yates SP, Merrill AR. A Catalytic Loop within Pseudomonas aeruginosa Exotoxin A Modulates Its Transferase Activity. J Biol Chem. 2001;276:35029–35036. doi: 10.1074/jbc.M105002200. [DOI] [PubMed] [Google Scholar]
- 42.Jorgensen R, Wang Y, Visschedyk D, Merrill AR. The nature and character of the transition state for the ADP-ribosyltransferase reaction. EMBO Rep. 2008;9:802–809. doi: 10.1038/embor.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jorgensen R, Merrill AR, Yates SP, Marquez VE, Schwan AL, Boesen T, Andersen GR. Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature. 2005;436:979–984. doi: 10.1038/nature03871. [DOI] [PubMed] [Google Scholar]
- 44.Yamaizumi M, Mekada E, Uchida T, Okada Y. One molecule of diphtheria toxin fragment a introduced into a cell can kill the cell. Cell. 1978;15:245–250. doi: 10.1016/0092-8674(78)90099-5. [DOI] [PubMed] [Google Scholar]
- 45.Kondo T, FitzGerald D, Chaudhary VK, Adhya S, Pastan I. Activity of immunotoxins constructed with modified Pseudomonas exotoxin A lacking the cell recognition domain. J Biol Chem. 1988;263:9470–9475. [PubMed] [Google Scholar]
- 46.Pastan I, FitzGerald D. Pseudomonas exotoxin: chimeric toxins. J Biol Chem. 1989;264:15157–15160. [PubMed] [Google Scholar]
- 47.Kreitman RJ, Pastan I. Antibody Fusion Proteins: Anti-CD22 Recombinant Immunotoxin Moxetumomab Pasudotox. Clin Can Res. 2011;17:6398–6405. doi: 10.1158/1078-0432.CCR-11-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eklund JW, Kuzel TM. Denileukin diftitox: a concise clinical review. Expert Rev Anticancer Ther. 2005;5:33–38. doi: 10.1586/14737140.5.1.33. [DOI] [PubMed] [Google Scholar]
- 49.Kioi M, Seetharam S, Puri RK. Targeting IL-13Rα2-positive cancer with a novel recombinant immunotoxin composed of a single-chain antibody and mutated Pseudomonas exotoxin. Mol Cancer Ther. 2008;7:1579–1587. doi: 10.1158/1535-7163.MCT-07-2131. [DOI] [PubMed] [Google Scholar]
- 50.Van Oijen MGCT, Preijers FWMB. Rationale for the Use of Immunotoxins in the Treatment of HIV-infected Humans. J Drug Targeting. 1998;5:75–91. doi: 10.3109/10611869808995861. [DOI] [PubMed] [Google Scholar]
- 51.Chatterjee D, Chandran B, Berger EA. Selective killing of Kaposi's sarcoma-associated herpesvirus lytically infected cells with a recombinant immunotoxin targeting the viral gpK8.1A envelope glycoprotein. mAbs. 2012;4:233–242. doi: 10.4161/mabs.4.2.19262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cao Y, Marks JD, Huang Q, Rudnick SI, Xiong C, Hittelman WN, Wen X, Marks JW, Cheung LH, Boland K, Li C, Adams GP, Rosenblum MG. Single-Chain Antibody-Based Immunotoxins Targeting Her2/neu: Design Optimization and Impact of Affinity on Antitumor Efficacy and Off-Target Toxicity. Mol Cancer Ther. 2012;11:143–153. doi: 10.1158/1535-7163.MCT-11-0519. [DOI] [PubMed] [Google Scholar]
- 53.Huang Z-Q, Buchsbaum DJ. Monoclonal antibodies in the treatment of pancreatic cancer. Immunotherapy. 2009;1:223–239. doi: 10.2217/1750743X.1.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao Y, Marks JW, Liu Z, Cheung LH, Hittelman WN, Rosenblum MG. Design optimization and characterization of Her2/neu-targeted immunotoxins: comparative in vitro and in vivo efficacy studies. Oncogene. 2013 doi: 10.1038/onc.2012.612. 10.1038/onc.2012.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McCluskey AJ, Olive AJ, Starnbach MN, Collier RJ. Targeting HER2-positive cancer cells with receptor-redirected anthrax protective antigen. Mol Oncol. 2012 doi: 10.1016/j.molonc.2012.12.003. 10.1016/j.molonc.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mechaly A, McCluskey AJ, Collier RJ. Changing the receptor specificity of anthrax toxin. MBio. 2012;3:e00088-12. doi: 10.1128/mBio.00088-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Robson T, Hirst DG. Transcriptional Targeting in Cancer Gene Therapy. J Biomed Biotechnol. 2003;2003:110–137. doi: 10.1155/S1110724303209074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Papadakis ED, Nicklin SA, Baker AH, White SJ. Promoters and control elements: designing expression cassettes for gene therapy. Curr Gene Ther. 2004;4:89–113. doi: 10.2174/1566523044578077. [DOI] [PubMed] [Google Scholar]
- 59.Dong Z, Nor JE. Transcriptional targeting of tumor endothelial cells for gene therapy. Adv Drug Deliv Rev. 2009;61:542–553. doi: 10.1016/j.addr.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dorer DE, Nettelbeck DM. Targeting cancer by transcriptional control in cancer gene therapy and viral oncolysis. Adv Drug Deliv Rev. 2009;61:554–571. doi: 10.1016/j.addr.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 61.Lu Y. Transcriptionally regulated, prostate-targeted gene therapy for prostate cancer. Adv Drug Deliv Rev. 2009;61:572–588. doi: 10.1016/j.addr.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 62.Wu L, Johnson M, Sato M. Transcriptionally targeted gene therapy to detect and treat cancer. Trends Mol Med. 2003;9:421–429. doi: 10.1016/j.molmed.2003.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hauptrock B, Hess G. Rituximab in the treatment of non-Hodgkin's lymphoma. Biologics. 2008;2:619–633. doi: 10.2147/btt.s3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ewer MS, Ewer SM. Cardiotoxicity of anticancer treatments: what the cardiologist needs to know. Nat Rev Cardiol. 2010;7:564–575. doi: 10.1038/nrcardio.2010.121. [DOI] [PubMed] [Google Scholar]
- 65.Curigliano G, Mayer EL, Burstein HJ, Winer EP, Goldhirsch A. Cardiac Toxicity From Systemic Cancer Therapy: A Comprehensive Review. Prog Cardiovas Dis. 2010;53:94–104. doi: 10.1016/j.pcad.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 66.Kontermann R. Dual targeting strategies with bispecific antibodies. mAbs. 2012;4:182–197. doi: 10.4161/mabs.4.2.19000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 68.Jorgensen R, Carr-Schmid A, Ortiz PA, Kinzy TG, Andersen GR. Purification and crystallization of the yeast elongation factor eEF2. Acta Crystallogr D Biol Crystallogr. 2002;58:712–715. doi: 10.1107/s0907444902003001. [DOI] [PubMed] [Google Scholar]
- 69.Douglas CM, Collier RJ. Pseudomonas aeruginosa exotoxin A: alterations of biological and biochemical properties resulting from mutation of glutamic acid 553 to aspartic acid. Biochemistry. 1990;29:5043–5049. doi: 10.1021/bi00473a007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.