Abstract
The trace metal selenium is in demand for health supplements to human and animal nutrition. We studied the reduction of selenite (SeO3 −2) to red elemental selenium by Rhodopseudomonas palustris strain N. This strain was cultured in a medium containing SeO3 −2 and the particles obtained from cultures were analyzed using transmission electron microscopy (TEM), energy dispersive microanalysis (EDX) and X ray diffraction analysis (XRD). Our results showed the strain N could reduce SeO3 −2 to red elemental selenium. The diameters of particles were 80–200 nm. The bacteria exhibited significant tolerance to SeO3 −2 up to 8.0 m mol/L concentration with an EC50 value of 2.4 m mol/L. After 9 d of cultivation, the presence of SeO3 2− up to 1.0 m mol/L resulted in 99.9% reduction of selenite, whereas 82.0% (p<0.05), 31.7% (p<0.05) and 2.4% (p<0.05) reduction of SeO3 −2 was observed at 2.0, 4.0 and 8.0 m mol/L SeO3 2− concentrations, respectively. This study indicated that red elemental selenium was synthesized by green technology using Rhodopseudomonas palustris strain N. This strain also indicated a high tolerance to SeO3 −2. The finding of this work will contribute to the application of selenium to human health.
Introduction
Selenium is a trace element commonly found in materials of the earth's crust, and it is essential for humans and animals. In human beings, the nutritional functions of selenium are achieved by 25 selenoproteins that have selenocysteine at their active center [1], [2]. In at least three prospective studies [3]–[5], high selenium status has been associated with a low overall mortality. Selenium supplementation, even in apparently selenium-replete individuals, has pronounced immunostimulant effects, including an enhancement of proliferation of activated T-cells, increased cytotoxic lymphocyte-mediated tumor cytotoxicity, and natural killer cell activity [6]–[11].
Selenium enters food chains typically by the absorbance of plants from soil. The daily selenium intakes of human mainly come from plants and animals containing selenium [12], [13]. But, selenium deficiency in soil is a worldwide problem. The distribution of a low selenium belt showed a zonal area in each hemisphere (latitudes above 30 degrees), especially in Europe and North America [14], [15]. Therefore, it is necessary to enrich the uptake of selenium for health reasons. But, there is a relatively narrow margin between selenium intakes that result in deficiency or toxicity [16]. A form of Se with higher bioavailability and lower toxicity may have better prospects as a chemo-preventive agent. Amongst its various forms, the red amorphous selenium is with biological activity and effects similar to those of sodium selenite but with much lower acute toxicity [15], [17]. It was considered the most potent chemical form to the artificial selenium enrichment. Therefore, the red amorphous selenium has attracted much attention [18]–[22].
In natural environments, live cells, such as bacteria, fungi, yeasts and plants, are known to be capable of converting selenate and selenite to Se0 [23]–[28]. Among these, bacteria are preferred for biosynthesis due to their extracellular particle production ability, short generation time, ease of culturing, downstream processing and manipulation [29]. Rhodopseudomonas palustris is a purple, non-sulphur, photosynthetic bacterium. It can grow not only aerobically in the dark but also anaerobically in the light [30] and has tolerance to a variety of metal and transition metal oxyanions, including selenium [31], [32]. This resistance is attributed to the capacity of the organisms to reduce Se oxyanions to their elemental ground state [29], [33], [34].
The objective of the present study was to investigate the ability of Rhodopseudomonas palustris strain N to reduce selenite to its elemental state. It will provide an eco-friendly and potentially economically viable ‘green’ synthesis route towards synthesis of red elemental selenium and contribute to the application of selenium for human health.
Materials and Methods
Ethics statement
The collection of sludge was permitted by Taiyuan sewage plant, Shanxi Province, China.
Growth of bacteria
Rhodopseudomonas palustris strain N was employed in these experiments. This strain was originally isolated from sludge in Taiyuan sewage plant (112°33′E 37°52′N) and synchronously stored in the China General Microbiological Culture Collection [35]. Before the experiment, the strain was cultured in 500 mL bottles of full medium described by Ormerod [36] with the exception that DL-malate was omitted and 1.8 g sodium acetate was added. Bottles were closed by a rubber plug and the gas phase was exchanged with Ar by 5 cycles consisting of 1 atm of vacuum followed by the addition of Ar to a pressure of 1.5 atm. The cultures were incubated at 30 °C in the presence of incandescent light (1500 Lux) at gentle stirring (80 r/min) with magnetic stirrer. When it reached the end of the exponential growth phase (about 72 h after cultured), the cells were used to inoculate new culture flasks. Three transfers were done before the cultures were used for experiments.
Transformation of selenite (SeO3 2−) to Se0
The microbial transformation of SeO3 2− was studied under anaerobic conditions as mentioned above. Rhodopseudomonas palustris strain N was cultured under the same condition above, except that medium was supplemented with 1.0 m mol/L sodium selenite and the strain was incubated at 30 °C in the presence of incandescent light (1500 Lux) for 8 days. The volume of culture used for inoculation was calculated so that the starting cell concentration corresponded to an absorbance at 680 nm of 0.21 with a 10-mm path length.
Scanning electron microscopy (SEM)
The strains were fixed in 2.5% glutaraldehyde for 48 h, rinsed three times in phosphate buffer (0.2 mol/L), and dehydrated in an ethanol series (30%, 50%, 70%, 90%, and 100%). Ethanol was displaced by isoamyl acetate, and the cells were dried using an EMS 850 critical point drying apparatus. Next, the samples were mounted on microscope slides (approximately 2.5 cm×2.5 cm) and sputter-coated with gold to a thickness of approximately 20 nm. Finally, they were observed under a JSM-6360LV scanning electron microscope (JEOL, Tokyo, Japan) operating at 30 kV.
Transmission electron microscopy (TEM) and energy dispersive microanalysis (EDX)
The harvested cells cultured in the medium containing selenite were centrifuged at 12,000×g for 10 min. The precipitated pellet was collected and washed with normal saline. Cells were resuspended in 2.5% glutaraldehyde and fixed for 2 h, washed with phosphate buffer (0.2 m mol/L, pH 7.0), and embedded in low-melting-point agarose. Agar blocks (approximately 1 by 1 by 1 mm) were fixed in 1% OsO4 in running water for 60 min, dehydrated with ethanol and acetone, and embedded in Epon-Araldite. Sections cut from the Epon-Araldite preparation were contrasted with uranyl acetate and lead citrate as described by Hess [37]. For energy-dispersive X-ray (EDX) analysis, whole cells were applied to scanning electron microscopy grids, dried at room temperature, and coated with 5 nm of carbon before measurements were obtained. The samples were examined using a JEM-1011 scanning electron microscope (JEOL, Tokyo, Japan) with an accelerating voltage of 80 kV. The EDX analysis was performed with a JEM-2010 transmission electron microscope (JEOL, Tokyo, Japan) equipped with a Link-Inca microanalysis system.
X ray diffraction analysis
The bacterial culture was centrifuged at 3,000×g and 4°C for 20 min after the reduction phase was completed. The pellet was discarded, and the supernatant containing Se0 in the form of Se-NPs was centrifuged at 100,000×g and 4°C for 30 min. The supernatant was discarded, and the pellet with the selenium-containing particles was resuspended in 50 m mol/L Tris-HCl (pH 7.5). The suspension was washed twice in the same buffer by repeating the two centrifugation steps. The pellet was freeze-dried on Thermo Dryer (Thermo, Savant ModulyoD-2300, USA) and stored in lyophilized powdered form until used for further characterization. The finely powdered sample was analyzed by a D8 Advance (Bruker, Germany) X-ray diffractometer using CuKα radiation (λ = 1.5406 A) in the range of 5°≤2θ≤80° at 40 kV. In order to calculate the particle size (D) of the sample, the Scherrer's relationship (D = 0.9 λ/βcosθ) has been used [38], where λ is the wavelength of X-ray, β is the broadening of the diffraction line measured half of its maximum intensity in radians and θ is the Bragg's diffraction angle. The particle size of the sample was estimated from the line width of the (101) XRD peak.
Resistance to SeO3 2− Stress
Bacteria culture
The effect of SeO3 2− on bacterial growth was determined by culturing the cells (the starting cell concentration corresponded to an absorbance at 680 nm of 0.21 with a 10-mm path length) in 500 mL bottle containing 500 mL of medium modification of described by Ormerod [36] supplemented with 1.0, 2.0, 4.0, and 8.0 m mol/L of Na2SeO3. The bottles were incubated at 30°C in the presence of incandescent light (1500 Lux). The growth in each bottle was sampled by at 0–5 days after cultured. The fermentation was used for bacterial biomass determination (calculated by cell concentration and protein content according to Kessi et al. [31]).
Protein content determination
The protein content of cells was determined by using a modification of the method of Lowry et al. [39]. 1 mL cultures were disrupted by ultrasonic treatment with 160 W for 15 min, at 4°C. A cell suspension of 200 µL was centrifuged at 15,000×g for 15 min, at 4°C and resuspended in 125 µL NaOH (0.1 mol/L). The resulting sample was frozen and stored. The samples were incubated in boiling water for 10 min. Then 875 µL of a solution containing 0.025% copper sulfate, 0.050% sodium tartrate, and 2.5% sodium carbonate was added, and the sample was incubated at room temperature for 10 min. After the addition of 250 µL of Folin-Ciocalteu solution (diluted 1/6 with H2O) and incubation for an additional 3 h, the absorbance at 750 nm of the copper-protein complex was determined by using a blank containing all of the reagents except protein. Selenium-containing samples were centrifuged at 15,000×g for 10 min before measurements were obtained. The optical density value was recorded using a microplate reader (M5, Spectra Max, www.spectramax.com). All measurements were done in triplicate. Bovine serum albumin was used as a standard. Growth ratio was calculated with a growth curve and the medium exposure concentration (EC50) [40], [41] was calculated using the Probit Analysis of Regression (Origin 8.0).
Reduction of SeO3 2−
Bacteria were cultured as described in ‘bacteria culture’ for 9 days. The fermentation product was centrifuged at 15,000×g for 15 min, at 4°C. The concentration of selenium in the supernatant fraction was determined following by the method reported by Zhang et al. [42]. The contents of the obtained supernatant was estimated by using LH-2A hydride generation (General research institute for nonferrous metals, Beijing, China) and AA-6300 atomic absorption spectrometer (Shimadzu, Tokyo, Japan) in an air-acetylene flame at 196.1 nm wavelength. Reagent channel: 2.0% KBH4/0.5% NaOH; Acid channel: 10% HCl; Speed of supported gas (N2): 200 mL/min. The samples were determined three times.
Reduction of SeO3 2− in a cell-free spent medium
A 250 µL portion of 0.1 mol/L SeO3 2− was added to 25 mL of cell-free medium obtained from a 1.0 m mol/L SeO3 2− culture in which reduction was complete and culture without SeO3 2−. The solution was thoroughly mixed and incubated at 30°C in the presence of incandescent light (1500 Lux) and observed after 120 h cultured.
Statistical analysis
All data presented are the mean values of three independent sets of experiments. Each value was presented as means ± standard deviation (S.D.). Statistical analysis was carried out by one-way ANOVA using the Tukey's posthoc test to evaluate whether the means were significantly different. The level of significance was set at P<0.05. Statistical computations were performed with SPSS 16.0 for Windows (SPSS Inc.).
Results
Transformation of SeO3 2− to Se0
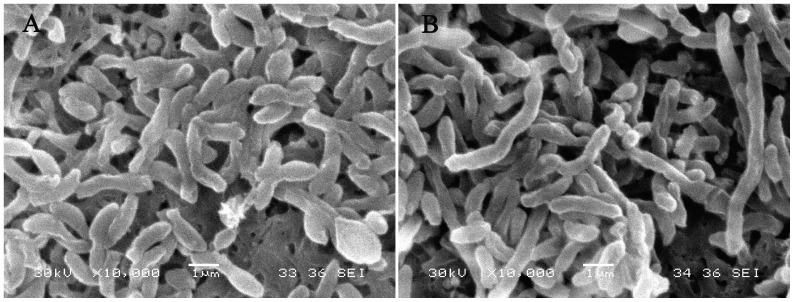
Rhodopseudomonas palustris strain N was cultured in medium containing SeO3 2− under optimized growth conditions. At the start of culture, the culture with and without SeO3 2− had the same color (Fig. 1A). In the control group, the strain began to proliferation at the 1st day after inoculated and shown a red color at the 2nd day. But in the treated group, it began to grow and showed an orange color at the 2nd day (Fig. 1B). At the 8th day, the control group turned dark reddish brown (this color was mainly due to strain in the presence of chlorophyll a and carotenoids [43]) and the treated group exhibited a red color (Fig. 1C). Moreover, cells of strain N exhibited elongation when exposed to 2.0 m mol/L SeO3 2− (Fig. 2B) contrasting to the cells in the control group under scanning electron microscope (Fig. 2A).
Figure 1. Culture of Rhodopseudomonas palustris strain N in medium containing selenite.
A, at the start of cultivation; B, at the 2nd day of cultivation; C, at the 8th day of cultivation. Control: strain cultured in a medium without selenite; Treatment: strain cultured in medium containing selenite.
Figure 2. Cell morphology of Rhodopseudomonas palustris strain N cultured without selenite (A) and with 2.0 m mol/L selenite (B) under scanning electron microscopy.
TEM and EDX analysis
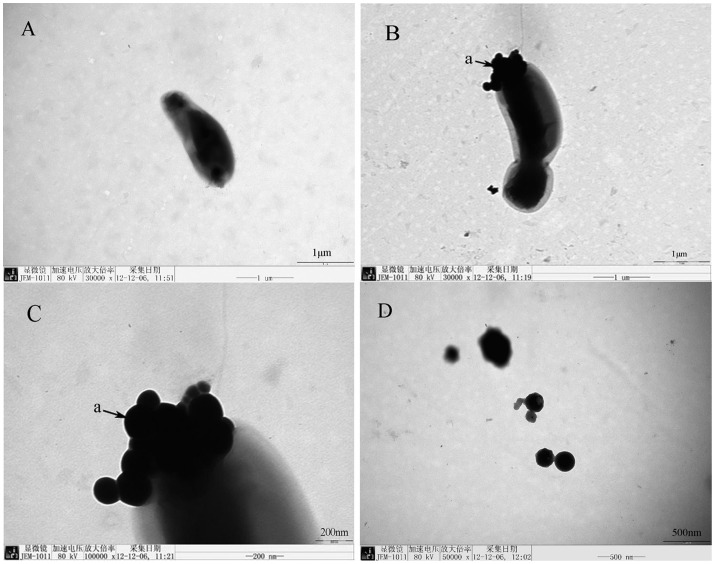
An electron micrograph of cells and selenium-containing particles obtained from the medium of a culture amended with 1.0 m mol/L SeO3 2− is shown in Fig. 3. In the control group, the strain surface was smooth (Fig. 3A). In the treated group, there were particles on the surface of stain (Fig. 3B), the shape of the particles was approximately spheric with a diameter of 80–200 nm (Fig. 3C). The particles free in the medium were observed also (Fig. 3D).
Figure 3. Transmission electron micrograph of Rhodopseudomonas palustris strain N.
A, cells cultured without selenite; B–C, cells grown in the presence of selenite are showing electron-dense particles formed by the strain N; D, particles free in the medium, a: particles.
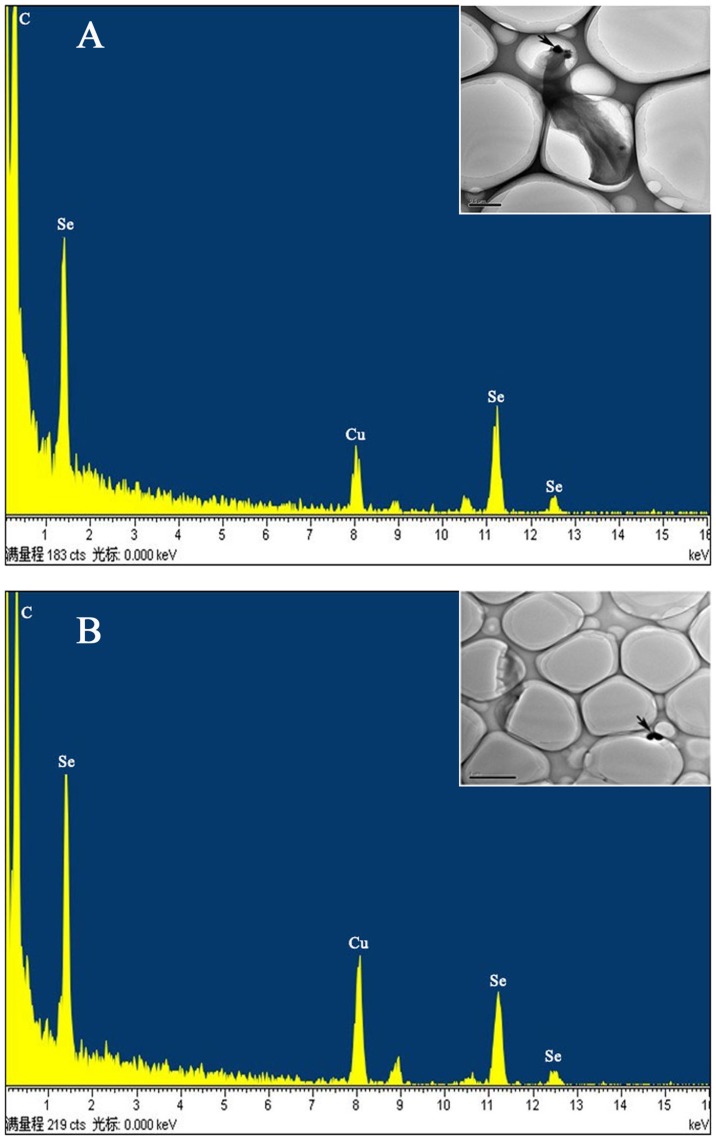
When preparations of obtained cells (Fig. 4A) and selenium-containing particles (Fig. 4B) were analyzed by using EDX analysis, the electrondense particles produced specific selenium absorption peaks at 1.37 keV (peak SeLa), 11.22 keV (peak SeKa), and 12.49 keV (peak SeKb). This result indicated that SeO3 2− was reduced to red elemental selenium by Rhodopseudomonas palustris strain N.
Figure 4.
(A) Particles on the cell membrane. Energy levels (in kiloelectron volts) are indicated on the x axis. The emission lines for selenium are at 1.37 keV (peak SeLa), 11.22 keV (peak SeKa), and 12.49 keV (peak SeKb). (B) Particles in the culture medium. Energy levels (in kiloelectron volts) are indicated on the x axis. The emission lines for selenium are at 1.37 keV (peak SeLa), 11.22 keV (peak SeKa), and 12.49 keV (peak SeKb).
X ray diffraction analysis
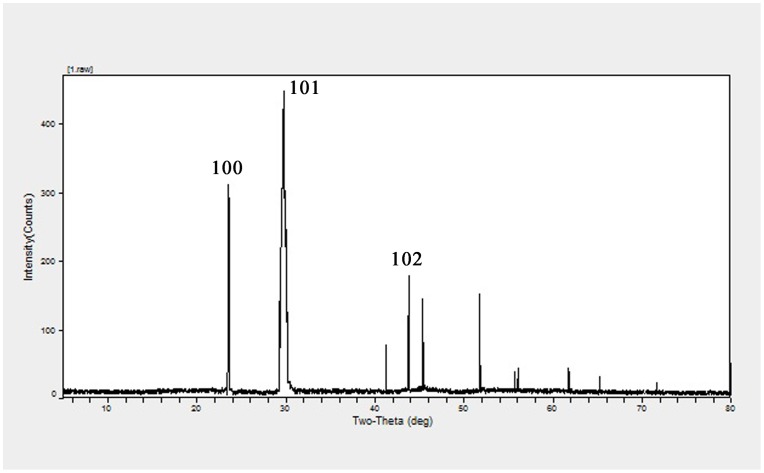
The XRD pattern obtained for the extracellular red elemental selenium with three intense peaks in the whole spectrum of 2θ values ranging from 5 to 80 is shown in Fig. 5. The diffractions at 23.599°, 29.777° and 43.778° can be indexed to the (100), (101) and (102) planes of the face-centered cubic (fcc) red elemental selenium, respectively. The full-width-at-half-maximum (FWHM) values measured for 101 planes of reflection were used to calculate the size of the particles. The calculated average size of the red elemental selenium particles was determined to be 165 nm.
Figure 5. XRD pattern of selenium-containing particles formed by Rhodopseudomonas palustris strain N.
The characteristic strong diffraction peak located at 29.777° is ascribed to the (101) facets of the face-centered cubic elemental Se0 structure.
Resistance to SeO3 2− Stress
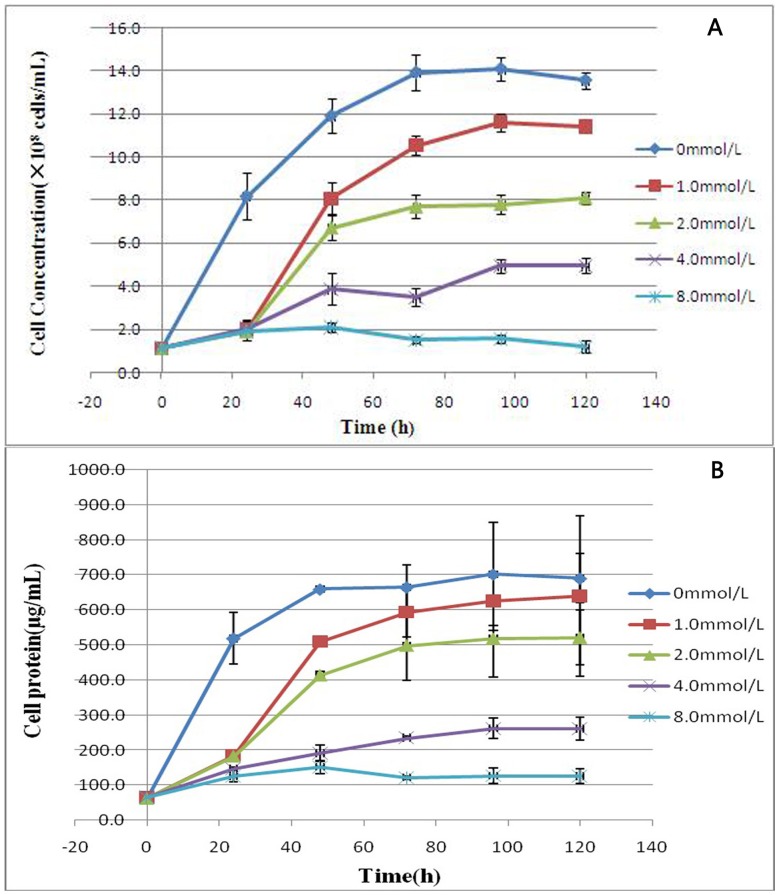
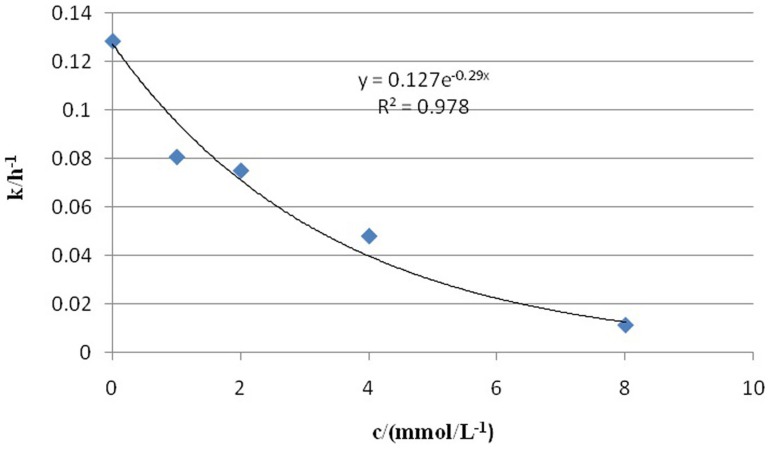
The results in Fig. 6 showed the SeO3 2− tolerance of Rhodopseudomonas palustris strain N at increasing concentrations of Na2SeO3. The cells were viable in the medium containing SeO3 2− and reached the end of log growth at 72 h after cultured. Growth kinetics data obtained at different SeO3 2− concentrations (1.0 to 8.0 m mol/L) showed that increasing the SeO3 2− concentration reduced the maximum attainable cell concentration (Fig. 6A) and protein content (Fig. 6B). The change trends of cell concentration were similar to the protein content of strain. The inhibition of the growth of bacteria increased with the increasing of SeO3 2− concentration in the medium. After 120 h, the presence of SeO3 2− up to 1 m mol/L resulted in 7.3% growth inhibition of the protein content of the control, whereas 24.5% (p<0.05), 62.1% (p<0.05) and 81.7% (p<0.05) growth inhibition was observed at 2.0, 4.0 and 8.0 m mol/L SeO3 2− concentrations, respectively. Based on the extent of growth inhibition (Fig. 6B), the effective concentration (EC50) of SeO3 2− was determined to be 2.4 m mol/L (k = 0.127exp(-0.29c), R2 = 0.978) (Fig. 7).
Figure 6. Time course of anaerobic phototrophic growth of Rhodopseudomonas palustris strain N in the presence of different selenite concentrations.
(A) Cell concentration of the strain. (B) Protein content of the strain. Symbols: ⧫, control cells (no selenite); ▪, 1.0 m mol/L; ▴, 2.0 m mol/L; ×, 4.0 m mol/L; *, 8.0 m mol/L. Selenite was added at time zero. Each curve shows means based on the results of three experiments.
Figure 7. Influence of sodium selenite on the average growth rate of cells.
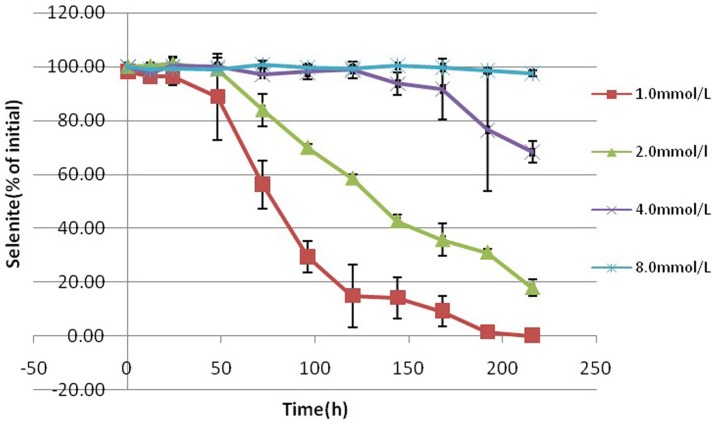
The decrease in the SeO3 2− concentration during growth is shown in Fig. 8. This decrease was steep and complete at SeO3 2− concentrations of 1.0 m mol/L. In the culture amended with 2.0 m mol/L SeO3 2− reduction showed a similar slope as the slope in culture amended with 1.0 m mol/L SeO3 2−, while the reduction kinetics of cultures containing 4.0 and 8.0 m mol/L SeO3 2− were different from them. After 9 d, the presence of SeO3 2− up to 1.0 m mol/L resulted in 99.9% reduction of SeO3 2−, whereas 82.0% (p<0.05), 31.7% (p<0.05) and 2.4% (p<0.05) reduction of SeO3 2− was observed at 2.0, 4.0 and 8.0 m mol/L SeO3 2− concentrations, respectively. Furthermore, the start of the reactions was further delayed with increasing concentrations of SeO3 2−.
Figure 8. Time course of selenite reduction by Rhodopseudomonas palustris strain N during anaerobic phototrophic growth in the presence of different selenite concentrations.
Symbols: ▪, 1.0 m mol/L; ▴, 2.0 m mol/L; ×, 4.0 m mol/L; *, 8.0 m mol/L. Selenite was added at time zero. Each curve shows means based on the results of three experiments.
The location of elemental red selenium formation in grown cells
Under transmission electric microscope, high-electron-density particles having a regular geometrical shape were present in the cytoplasm of grown cells with SeO3 2− (Fig. 9). In addition, SeO3 2− was added to cell-free medium obtained from a 1.0 m mol/L SeO3 2− culture in which reduction was complete. The color of the solution did not changed at 30°C in the presence of incandescent light (1500 Lux) for 120 h. These results indicated that red elemental selenium formed in the cytoplasm of Rhodopseudomonas palustris strain N cells grown in the presence of SeO3 2−.
Figure 9. Transmission electron micrograph of the cells cultured without selenite (A) and exposed to 1.0 m mol/L selenite (B).
Arrows indicate the paticles.
Discussion
Se0 is well known for its photoelectric, semiconductor, free-radical scavenging, anti-oxidative and anti-cancer properties [44]. The production of Se0 can be achieved through chemical and biological methods. Chemical detoxification of metals is proven to be very expensive and often results in secondary effects on the environment. The synthesis of Se0 using microorganisms has been suggested as a possible green method. In this study, Rhodopseudomonas palustris strain N was cultured in a medium containing SeO3 2− and particles obtained from the medium were proven to be Se0 by EDX and XRD analysis. The maximum concentration of SeO3 2− culture in which was complete of this strain was more than 1.0 m mol, and it was similar to Bacillus megaterium (≥0.25 m mol) [22] and Rhodospirillum rubrum (≥1.0 m mol) [31]. This study provided a new bacterial stain resource for the reduction of SeO3 2− to Se0.
In recent years, several different bacteria have been reported for the biological synthesis of Se0, such as Thauera selenatis [45] and Rhizobium selenitireducens strain B1 [23], [46], [47], Escherichia coli [48], Clostridium pasteurianum [49] and Bacillus selenitireducens [50]. In our study, Rhodopseudomonas palustris strain N also showed this ability. Moreover, the present Rhodopseudomonas palustris strain N belongs to the photosynthetic bacteria. The photosynthetic bacteria have more digestible bacterial cell wall, and are rich in protein, carotenoids, biological cofactors, and vitamins [51]. They were already shown to be suitable amendments to health foods for humans and animals. Therefore, reduction of SeO3 2− to Se0 by Rhodopseudomonas palustris strain N provides a potential application to artificially enrich food with selenium for human health.
Microorganisms have been shown to be particularly resistant to SeO3 2− [31], [32]. This resistance is attributed to the capacity of the organisms to reduce Se oxyanions to their elemental ground state. Rhodopseudomonas palustris strain N was resistant to the toxic effects of SeO3 2− and the minimum inhibitory concentration (the lowest amount of an antimicrobial agent that will completely inhibit the growth of an organism) for SeO3 2− for this organism exceeds 8 m mol/L. It was equal or greater than the MIC of other strains. For example, the MIC of Bacillus subtilis was 1.0 m mol/L [52], the MIC of E. coli W1485 was 5. 8 m mol /L [53], the MIC of Bradyrhizobium japonicum was 6.0–12.0 m mol/L [54], and the MIC of Rhodobacter spheroids was 2.9–4.6 m mol/L [55].
Both electron micrographs showing intact cells after SeO3 2− reduction (Fig. 3B) and growth kinetics showing that the cell concentration and protein concentration in the stationary phase is rather constant (Fig. 6) suggest that cells were not severely damaged in the presence of SeO3 2− but rather viable. On the other hand, large amounts of selenium-containing particles present in the culture medium after SeO3 2− reduction (Fig. 10), indicating that Rhodopseudomonas palustris strain N is able to efficiently transport elemental selenium out of the cell. For collection of elemental selenium, this metabolism method was easier than others [56].
Figure 10. Electron micrograph of selenium-containing particles obtained after bacterial reduction in a culture amended with 1.0 m mol/L selenite.
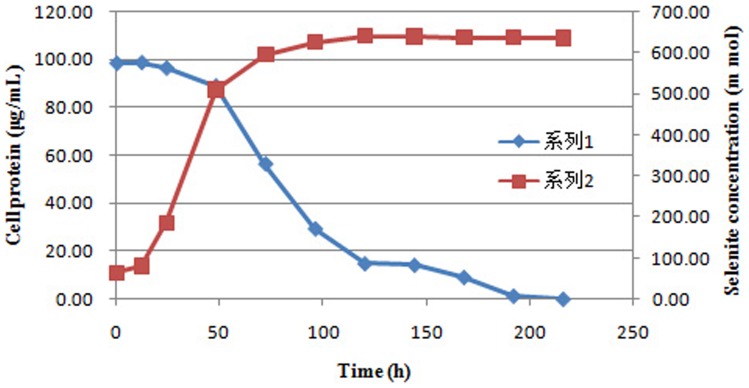
In our study, SeO3 2− reduction was closely related to the growth kinetics of cultures. It occurred at the end of the exponential phase and the stationary phase of bacterial growth (Fig. 11). This finding is similar to the results of Kessi et al. [31], who reported that the production of volatile selenium compounds occurred during the stationary phases. It means that the reduction is independent of strain growth. Moreover, this result suggests that reduction reaction is controlled by the stationary phase of the cultures.
Figure 11. Time course of growth and selenite reduction by Rhodopseudomonas palustris strain N.
Symbols: ▪, cell protein; ⧫, selenite concentration.
In conclusion, we found that Rhodopseudomonas palustris strain N was capable of converting SeO3 2− to Se0. And this study provided a new microorganism resource and a potentially economically valuable ‘green’ synthesis route towards the synthesis of red elemental selenium, therefore contributing to the application of selenium for human health.
Acknowledgments
The authors are grateful to Professors Hans-Uwe Dahms and Enmin Zou for critical comments on this manuscript and language editing.
Funding Statement
This work was supported by the Shanxi Provincial Key Technologies Research and Development Program (20130321033-03 and 20100311050) and Postgraduate Innovation Project of Shanxi Province (011352901019). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, et al. (2003) Characterization of mammalian selenoproteomes. Science 300: 1439–1443. [DOI] [PubMed] [Google Scholar]
- 2. Rayman MP (2012) Selenium and human health. Lancet 379: 1256–1268. [DOI] [PubMed] [Google Scholar]
- 3. Akbaraly NT, Arnaud J, Hininger-Favier I, Gourlet V, Roussel AM, et al. (2005) Selenium and mortality in the elderly: results from the EVA study. Clin Chem 51: 2117–2123. [DOI] [PubMed] [Google Scholar]
- 4. Bleys J, Navas-Acien A, Guallar E (2008) Serum selenium levels and all-cause, cancer, and cardiovascular mortality among US adults. Arch Intern Med 168: 404–410. [DOI] [PubMed] [Google Scholar]
- 5. Ray AL, Semba RD, Walston J, Ferrucci L, Cappola AR, et al. (2006) Low serum selenium and total carotenoids predict mortality among older women living in the community: the women's health and aging studies. J Nutr 136: 172–176. [DOI] [PubMed] [Google Scholar]
- 6. Broome CS, McArdle F, Kyle JA, Andrews F, Lowe NM, et al. (2004) An increase in selenium intake improves immune function and poliovirus handling in adults with marginal selenium status. Am J Clin Nutr 80: 154–162. [DOI] [PubMed] [Google Scholar]
- 7. Hawkes WC, Kelley DS, Taylor PC (2001) The effects of dietary selenium on the immune system in healthy men. Biol Trace Elem Res 81: 189–213. [DOI] [PubMed] [Google Scholar]
- 8. Hoffmann FW, Hashimoto AC, Shafer LA, Dow S, Berry MJ, et al. (2010) Dietary selenium modulates activation and differentiation of CD4+ T cells in mice through a mechanism involving cellular free thiols. J Nutr 140: 1155–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kiremidjian-Schumacher L, Roy M, Glickman R, Schneider K, Rothstein S, et al. (2000) Selenium and immunocompetence in patients with head and neck cancer. Biol Trace Elem Res 73: 97–111. [DOI] [PubMed] [Google Scholar]
- 10. Rayman MP (2000) The importance of selenium to human health. Lancet 356: 233–241. [DOI] [PubMed] [Google Scholar]
- 11. Wood SM, Beckham C, Yosioka A, Darban H, Watson RR (2000) β-Carotene and selenium supplementation enhances immuneresponse in aged humans. Integr Med 2: 85–92. [DOI] [PubMed] [Google Scholar]
- 12. Rayman MP (2008) Food-chain selenium and human health: emphasis on intake. Br J Nutr 100: 254–268. [DOI] [PubMed] [Google Scholar]
- 13.Reilly C (2006) Selenium in food and health. New York (NY, USA): Springer.
- 14. Johnson CC, Fordyce FM, Rayman MP (2010) Conference on ‘Over- and undernutrition: challenges and approaches’ Symposium on ‘Geographical and geological influences on nutrition’ factors controlling the distribution of selenium in the environment and their impact on health and nutrition. Proc Nutr Soc 69: 119–132. [DOI] [PubMed] [Google Scholar]
- 15. Wang H, Zhang J, Yu H (2007) Elemental selenium at nano size possesses lower toxicity without compromising the fundamental effect on selenoenzymes: comparison with selenomethionine in mice. Free Radic Biol Med 42: 1524–1533. [DOI] [PubMed] [Google Scholar]
- 16.Fordyce FM (2005) Selenium deficiency and toxicity in the environment. In: Selinus O, Alloway B, Centeno JA, Finkelman RB, Fuge R, et al. (eds.). Essentials of medical geology. Elsevier Academic Press, Amsterdam, Holland. 373–416 pp.
- 17. Zhang J, Wang X, Xu T (2007) Elemental selenium at nano size (Nano-Se) as a potential chemopreventive agent with reduced risk of selenium toxicity: comparison with Se-methylselenocysteine in mice. Toxicol Sci 101: 22–31. [DOI] [PubMed] [Google Scholar]
- 18. Domokos-Szabolcsy E, Marton L, Sztrik A, Babka B, Prokisch J, et al. (2012) Accumulation of red elemental selenium nanoparticles and their biological effects in Nicotinia tabacum . Plant Growth Regul 68: 525–531. [Google Scholar]
- 19. Dwivedi S, AlKhedhairy AA, Ahamed M, Musarrat J (2013) Biomimetic synthesis of selenium nanospheres by bacterial strain JS-11 and its role as a biosensor for nanotoxicity assessment: A novel Se-bioassay. Plos One 8(3): e57404 doi:10.1371/journal.pone.0057404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunter WJ, Kuykendall LD (2007) Reduction of selenite to elemental red selenium by Rhizobium sp. strain B1. Curr Microbiol 55: 344–349. [DOI] [PubMed] [Google Scholar]
- 21. Hunter WJ, Manter DK (2009) Reduction of selenite to elemental red selenium by Pseudomonas sp. strain CA5. Curr Microbiol 58: 493–498. [DOI] [PubMed] [Google Scholar]
- 22. Mishra RR, Prajapati S, Das J, Dangar TK, Das N, et al. (2011) Reduction of selenite to red elemental selenium by moderately halotolerant Bacillus megaterium strains isolated from Bhitarkanika mangrove soil and characterization of reduced product. Chemosphere 84: 1231–1237. [DOI] [PubMed] [Google Scholar]
- 23. Hunter WJ, Manter DK (2008) Bio-reduction of selenite to elemental red selenium by Tetrathiobacter kashmirensis . Curr Microbiol 57: 83–88. [DOI] [PubMed] [Google Scholar]
- 24. Narasingarao P, Haggblom MM (2007) Identification of anaerobic selenate respiring bacteria from aquatic sediments. Appl Environ Microbiol 73: 3519–3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bansal V, Ramanathan R, Bhargava SK (2011) Fungus-mediated biological approaches towards ‘Green’ synthesis of oxide nanomaterials. Aust J Chem 64: 279–293. [Google Scholar]
- 26. Bansal V, Bharde A, Ramanathan R, Bhargava SK (2012) Inorganic materials using ‘unusual’ microorganisms. Adv Colloid Interf Sci 179–182: 150–168. [DOI] [PubMed] [Google Scholar]
- 27. Oremland RS, Hollibaugh JT, Maest AS, Presser TS, Miller L, et al. (1989) Selenate reduction to elemental selenium by anaerobic bacteria in sediments and culture: biogeochemical significance of a novel, sulfate-independent respiration. Appl Environ Microbiol 55: 2333–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oremland RS, Steinberg NA, Maest AS, Miller LG, Hollibaugh JT (1990) Measurement of in situ rates of selenate removal by dissimilatory bacterial reduction in sediments. Environ Sci Technol 24: 1157–1164. [Google Scholar]
- 29. Ramanathan R, Field MR, O'Mullane AP, Smooker PM, Bhargava SK, et al. (2013) Aqueous phase synthesis of copper nanoparticles: a link between heavy metal resistance and nanoparticle synthesis ability in bacterial systems. Nanoscale 5: 2300–2306. [DOI] [PubMed] [Google Scholar]
- 30. Giotta L, Agostiano A, Italiano F, Milano F, Trotta M (2006) Heavy metal ion influence on the photosynthetic growth of Rhodobacter sphaeroides . Chemosphere 62: 1490–1499. [DOI] [PubMed] [Google Scholar]
- 31. Kessi J, Ramuz M, Wehrli E, Spycher M, Bachofen R (1999) Reduction of selenite and detoxification of elemental selenium by phototrophic bacterium Rhodospirillum rubrum . Appl Environ Microbiol 65: 4734–4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kessi J (2006) Enzymatic systems proposed to be involved in the dissimilatory reduction of selenite in the purple non-sulfur bacteria Rhodospirillum rubrum and Rhodobacter capsulatus . Microbiology 152: 731–743. [DOI] [PubMed] [Google Scholar]
- 33. Moore MD, Kaplan S (1992) Identification of intrinsic high-level resistance to rare-earth oxides and oxyanions in members of the class Proteobacteria: characterization of tellurite, selenite, and rhodium sesquioxide reduction in Rhodobacter sphaeroides . J Bacteriol 174: 1505–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ramanathan R, O'Mullane AP, Parikh RY, Smooker PM, Bhargava SK, et al. (2011) Bacterial kinetics-Controlled shape-Directed biosynthesis of silver nanoplates using Morganella psychrotolerans . Langmuir 27: 714–719. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Z, Yang S, Zhao C (1992) Study on isolation and identification of Rhodopseudonas palustris . J Shanxi Univ (Nat Sci) 15: 379–385. [Google Scholar]
- 36. Ormerod JG, Ormerod KS, Gest H (1961) Light dependent utilization of organic compounds and photoproduction of molecular hydrogen by photosynthetic bacteria; relationships with nitrogen metabolism. Arch Biochem Biophys 94: 449–463. [DOI] [PubMed] [Google Scholar]
- 37. Hess WM (1966) Fixation and staining of fungus hyphae and host plant root tissues for electron microscopy. Stain Technol 41: 27–35. [DOI] [PubMed] [Google Scholar]
- 38. Patterson AL (1939) The Scherrer formula for X-ray particle size determination. Phys Rev 699: 978–982. [Google Scholar]
- 39. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193: 265–275. [PubMed] [Google Scholar]
- 40. Lü CJ, Zhang YY, Zhao CG, Guo SW, Yang SP, et al. (2012) Arsenic resistance mechanisms in Rhodopseudomonas palustris under anaerobic and light conditions. Acta Scientiae Circumstantiae 32: 2375–2383. [Google Scholar]
- 41. Rider CV, LeBlanc GA (2005) An integrated addition and interaction model for assessing toxicity of chemical mixtures. Toxicol Sci 87: 520–528. [DOI] [PubMed] [Google Scholar]
- 42. Zhang Y, Frankenberger WT, Moore JN (1999) Measurement of selenite in sediment extracts by using hydride generation atomic absorption spectrometry. Sci Total Environ 229: 183–193. [Google Scholar]
- 43. Getha K, Vikineswary S, Chong VC (1998) Isolation and growth of the phototrophic bacterium Rhodopseudomonas palustris strain B1 in sago-starch-processing wastewater. World J Microb Biot 14: 505–511. [Google Scholar]
- 44. Zhang J, Zhang S, Xu J, Chen H (2004) A new method for the synthesis of selenium nanoparticles and the application to construction of H2O2 biosensor. Chinese Chem Lett 15: 1345–1348. [Google Scholar]
- 45. Bledsoe TL, Cantafio AW, Macy JM (1999) Fermented whey - an inexpensive feed source for a laboratory-scale selenium-bioremediation reactor system inoculated with Thauera selenatis . Appl Microbiol Biotechnol 51: 682–685. [DOI] [PubMed] [Google Scholar]
- 46. Euzeby J (2008) Validation List no. 121. List of new names and new combinations previously effectively, but not validly, published. Int J Syst Evol Microbiol 58: 1057. [DOI] [PubMed] [Google Scholar]
- 47. Hunter WJ, Kuykendall LD, Manter DK (2007) Rhizobium selenireducens sp. nov.: a selenite reducing a-Proteobacteria isolated from a bioreactor. Curr Microbiol 55: 455–460. [DOI] [PubMed] [Google Scholar]
- 48. Avazeri C, Turner RJ, Pommier J, Weiner JH, Giordano G, et al. (1997) Tellurite and selenate reductase activity of nitrate reductases from Escherichia coli: correlation with tellurite resistance. Microbiology 143: 1181–1189. [DOI] [PubMed] [Google Scholar]
- 49. Yanke LJ, Bryant RD, Laishley EJ (1995) Hydrogenase (I) of Clostridium pasteurianum functions as a novel selenite reductase. Anaerobe 1: 61–67. [DOI] [PubMed] [Google Scholar]
- 50. Afkar E, Lisak J, Saltikov C, Basu P, Oremland RS, et al. (2003) The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol Lett 226: 107–112. [DOI] [PubMed] [Google Scholar]
- 51. Kobayashi M, Kurata S (1978) The mass culture and cell utilization of photosynthetic bacteria. Process Biochem 13: 27–30. [Google Scholar]
- 52. Garbisu C, Carlson D, Adamkiewicz M, Yee BC, Wong JH, et al. (1999) Morphological and biochemical responses of Bacillus subtilis to selenite stress. Biofactor 10: 311–319. [DOI] [PubMed] [Google Scholar]
- 53. Schmidt MG, Konetzka WA (1986) Glutathion overproduction by selenite-resistant Escherichia coli . Can J Microbiol 32: 825–827. [DOI] [PubMed] [Google Scholar]
- 54. Kinkle BK, Sadowsky MJ, Johnstone K, Koskinen WC (1994) Tellurium and selenium resistance in Rhizobia and its potential use for direct isolation of Rhizobium meliloti from soil. Appl Environ Microb 60: 1674–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. William JH, Kuykendall LD (2007) An Azospira oryzae (syn Dechlorosoma suillum) strain that reduces selenate and selenite to elemental red selenium. Curr Microbiol 54: 376–381. [DOI] [PubMed] [Google Scholar]
- 56. Bebien M, Chauvin JP, Adriano JM, Grosse S, Vermeglio A (2001) Effect of selenite on growth and protein synthesis in the phototrophic bacterium Rhodobacter sphaeroides . Appl Environ Microbiol 67: 4440–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]