Abstract
Mediator is an essential, broadly utilized eukaryotic transcriptional co-activator. How and what it communicates from activators to RNA polymerase II (RNAPII) remains an open question. Here we performed genome-wide location profiling of Saccharomyces cerevisiae Mediator subunits. Mediator is not found at core promoters but rather occupies the upstream activating sequence (UAS), upstream of the pre-initiation complex. In the absence of Kin28 (CDK7) kinase activity, or in cells where the RNAPII C-terminal domain (CTD) is mutated to replace Ser5 with alanines, however, Mediator accumulates at core promoters together with RNAPII. We propose that Mediator is quickly released from promoters upon Ser5 phosphorylation by Kin28 (CDK7), which also allows for RNAPII to escape from the promoter.
INTRODUCTION
Regulation of transcription by RNA polymerase II (RNAPII) by sequence-specific transcription factors (TF) requires co-activator proteins. Co-activators are usually large protein complexes carrying one or more enzymatic activities1. First identified in budding yeast (yeast hereafter) via both genetic and biochemical approaches, Mediator is one of the most widely studied co-activator complexes (recently reviewed in2–4). Conserved throughout eukaryotes, Mediator is thought to be an essential component for the expression of most if not all genes, at least in yeast.
The 25 (yeast) to 30 (human) proteins that comprise Mediator are organized into four distinct modules, but -given its size and complexity-Mediator can easily be envisioned to be highly multifunctional (reviewed in2,3). The Tail module interacts with sequence-specific TF that recruit Mediator to DNA. The Head and Middle modules make several interactions with the RNAPII and the general transcription machinery. Finally, the Kinase module, connected to the rest of Mediator via the Middle module, contains a cyclin-dependent kinase (CDK) (CDK8 (Srb10)) that has been shown to have both positive and negative regulatory roles in gene expression. Biochemical and structural evidence suggests that the Kinase module and RNAPII interact with Mediator in a mutually exclusive manner5–8 but so far, genome-wide location profiling of Mediator subunits has failed to detect differences in location of the Kinase module relative to the rest of Mediator9,10. Mediator purified from mammalian cells missing the Kinase module generally contains an additional subunit called MED26. Interestingly, both CDK8 and MED26 have been shown to stimulate transcriptional elongation in different systems (reviewed in11).
The best described function for Mediator, both in yeast and mammalian cells, is to promote pre-initiation complex (PIC) assembly12–20, although this has not been previously investigated in vivo at the genomic scale. In addition, human Mediator has been shown to stimulate the release from promoter-proximal pausing15, most likely by recruiting various elongation factors to the paused polymerase21,22. Mediator also participates in enhancer-promoter gene looping in mammalian cells and in promoter-terminator looping in yeast (reviewed in4). Although less understood, Mediator interacts with nucleosomes, histone tails and chromatin regulators (reviewed in3). Finally, evidence also suggests a role for Mediator in mRNA processing23–25.
Despite two decades of intense research, several basic questions remain unanswered about Mediator function. Surprisingly, the genomic location of Mediator, while well understood in mammalian cells, remains a topic of intense debate in yeast. Indeed, Mediator has been proposed to bind to just about any kind of genomic regions, from UAS, promoters and even coding regions (ORFs)9,10,26–28. Also controversial is the notion that Mediator may be recruited to specific genes rather than acting globally (reviewed in3). In order to address these issues, we performed a thorough investigation of Mediator genomic location in yeast and propose a model for how Mediator associates with genes in vivo. Our data provide explanation for divergent observations among previous studies. Furthermore, we identified a transient state of Mediator and RNAPII occupancy at promoters that is controlled by Kin28 (CDK7) phosphorylation on Ser5 of the CTD.
RESULTS
Mediator associates with UAS elements rather than promoters
To determine where in the yeast genome Mediator binds, we performed extensive genome-wide location profiling of Mediator subunits. After testing several antibodies against various Mediator subunits, we realized that the best signal to noise ratio is achieved using epitope-tagged proteins. Indeed, all the polyclonal antibodies against Mediator tested generated not only poor enrichment, but also extensive enrichment in ORFs of highly expressed genes (See Supplementary Fig. 1a,b). Although we cannot completely rule out the possibility that Mediator occupies ORFs, several lines of evidence argue against such a conclusion. First, the enrichment on ORFs was also observed using IgG control ChIP experiments (Supplementary Fig. 1a,b). Second, the enrichment on ORFs largely disappeared when ChIPs were performed using epitope-tagged subunits and normalized against non-tagged ChIP controls (Supplementary Fig. 1c) or Mock IP samples using control IgGs (Supplementary Fig. 1d). Finally, we failed to show that Mediator moves along a gene with RNAPII using the GAL1pr-YLR454W elongation assay developed by Mason and Struhl 29 (unpublished data). Thus, our experiments fail to support the model where yeast Mediator travels with RNAPII during elongation9,10 (See Discussion). Recent work from the Rine, van Oudenaarden and Iyer groups30,31 similarly reported artifactual ChIP enrichments in highly transcribed coding regions. While the use of no tag control ChIPs was not efficient at eliminating this systematic error in their hands, we found that using our ChIP protocol (which uses magnetic beads instead of agarose beads) and normalising ChIP samples with no tag controls reliably eliminated most of the signal in coding regions of highly transcribed genes (See Supplementary Fig. 1c, bottom panel). All Mediator ChIP experiments described in this study were therefore performed using strains with Myc-tagged Mediator subunits [Head: MED19 (Rox3); Tail: MED15 (Gal11), MED16 (Sin4); and Kinase: CDK8 (Srb10), CycC (Srb11)]. All experiments have been hybridized against ChIP samples performed in isogenic non-tagged strains, as we have done previously (See32 for an example).
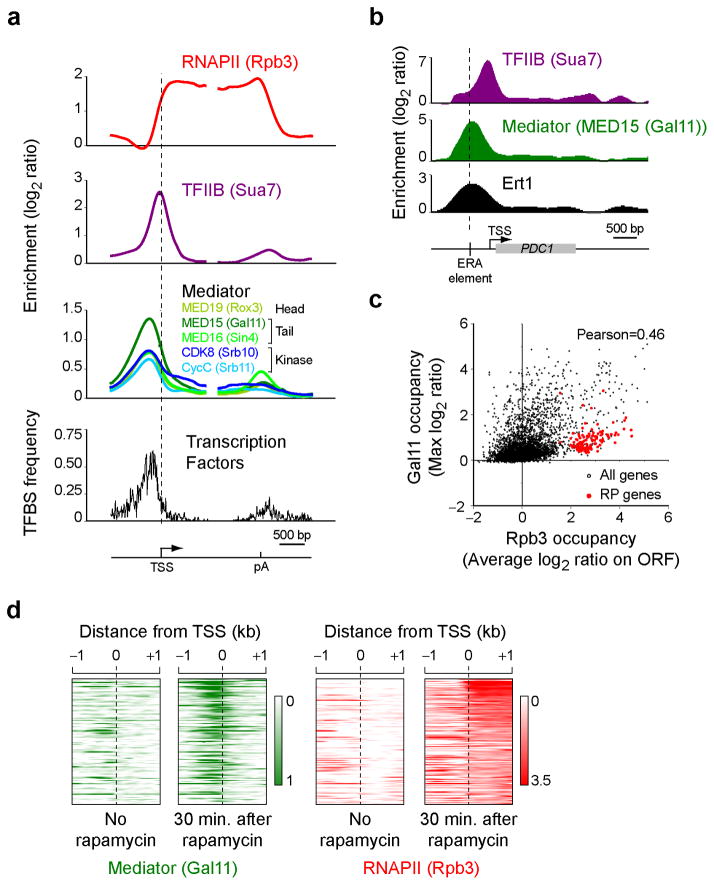
All Mediator subunits tested showed very similar binding profiles (Fig. 1a, green and blue traces; see also Supplementary Fig. 2a for a heat map representation). Interestingly, Mediator was not detected on core promoters (highlighted here by the presence of TFIIB (encoded by the SUA7 gene) in purple) but rather occupies a region further upstream. This location is heavily enriched for TF binding sites (Fig. 1a, black trace) demonstrating that Mediator actually occupies the upstream activating sequence (UAS). A specific example is shown in Figure 1B where the transcription factor Ert133 and Mediator co-occupy the cis-acting element (ERA)34 upstream of the PDC1 gene.
Figure 1.
Mediator occupies the UAS. (a) The enrichment of RNAPII (Rpb3, red), TFIIB (Sua7, purple) and Mediator subunits (different shades of green and blue) is shown over a metagene made of all the yeast genes that are more than 1 kb-long and have an Rpb3 average ORF occupancy >1 (n=299). The frequency of gene-specific transcription factor binding sites (TFBS) is shown in black. Genes were split in half and aligned on their transcription start site (TSS) and polyadenylation (pA) signal site. A heat map representation of the same data is shown in Supplementary Fig. 2a. (b) The enrichment of TFIIB (Sua7, purple), Mediator (MED15 (Gal11), green) and Ert1 is shown over the PDC1 gene, a gene regulated by Ert1. (c) A scatter plot of Mediator (Gal11) occupancy upstream of genes (measured as the maximum log2 enrichment level observed in the first kilobase upstream of each gene) versus RNAPII (Rpb3) occupancy in the coding region (measured as the average log2 enrichment level across the ORF). (d) A heat map representing Mediator (Gal11, green) and RNAPII (Rpb3, red) occupancy around the TSS of rapamycin induced genes (n=123; see Online Methods for details) before and 30 min after addition of rapamycin.
Two of the Mediator subunits tested show genomic binding sites in addition to UAS regions. First, CDK8 (Srb10) showed a modest occupancy on highly transcribed regions (See dark blue trace in Fig. 1a), perhaps speaking to a possible role in transcription elongation as described in mammalian cells35 (reviewed in11), although we can not rule out the possibility that this enrichment represents an artifact of the ChIP assay (see above and30,31). A second exception is observed with MED16 (Sin4), which, in addition to binding to UAS regions, binds in the 3′end of genes. This may be a consequence of 5′-to-3′ gene looping and perhaps relates to the recently described role of Mediator in that phenomenon24.
Mediator promotes PIC assembly
Mediator occupancy correlates weakly but positively (Pearson=0.46) with RNAPII (Rpb3) occupancy (Fig. 1c). Quite strikingly, ribosomal protein (RP) genes appear as an exception to this trend as Mediator was barely detectable at these very highly transcribed genes (See red dots in Fig. 1c and Supplementary Fig. 2b–d). While this observation might be used to argue that Mediator is not operating at all genes, further analyses suggested that this rather reflects different dynamics in the association of Mediator with different classes of genes (See below). The correlation between RNAPII and Mediator occupancy suggests a rather global role for Mediator. To further test this model, we examined whether Mediator occupancy is dynamically reorganized upon remodelling of gene expression after a 30 minute treatment with rapamycin, an inhibitor of the TOR pathway that triggers the induction of several nutrient response genes36. Upon treatment with rapamycin, we observed an increased in Mediator occupancy upstream of rapamycin-induced genes (Fig. 1d) consistent with a global role for Mediator in gene expression.
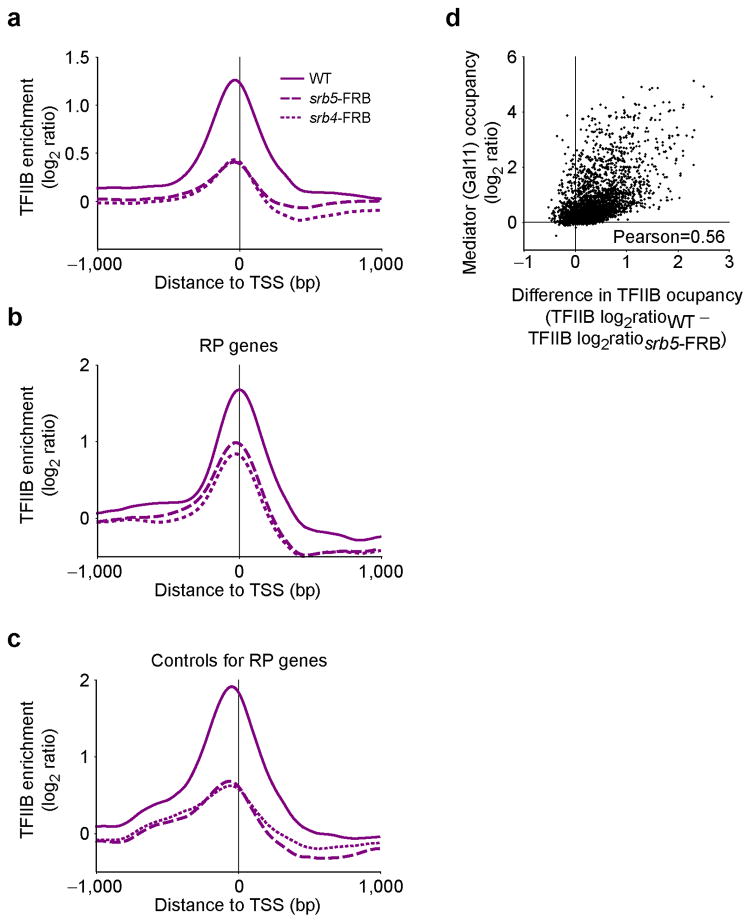
The main proposed function for Mediator is to promote PIC assembly (See Introduction). This, however, was never directly tested in vivo at the genomic scale. We therefore took advantage of the recently developed anchor-away (AA) system37 to test for the effect of depleting Mediator components from the nucleus on PIC assembly. SRB4 and SRB5 were tagged with the FRB domain of human mTOR in the appropriate AA strain and TFIIB occupancy was assayed by ChIP-chip 90 minutes after rapamycin addition. Note that these strains are immune to rapamycin stress as they are defective for TOR1 and FPR1. As shown in Figure 2a, TFIIB promoter occupancy was reduced upon depletion of either MED17 (Srb4) or MED18 (Srb5) from the nucleus. Importantly, TFIIB occupancy was also reduced at RP genes (Fig. 2b,c), although to a smaller extent, despite Mediator being difficult to detect at these genes (Supplementary Fig. 2b–d). Overall, the effect of Mediator depletion on PIC assembly correlates with Mediator occupancy (Fig. 2d; Pearson=0.56). Altogether, these data suggest that Mediator broadly promotes PIC assembly in vivo, although perhaps not to the same extent at all genes.
Figure 2.
Mediator is involved in PIC assembly in vivo. (a–c) Average TFIIB (purple) enrichment around the TSS in either wild type cells (WT, solid traces) or in cells where Srb5 (srb5-FRB, dashed traces) or Srb4 (srb4-FRB, dotted traces) were depleted from the nucleus using the anchor-away system for all genes with an Rpb3 average ORF occupancy >1 in WT cells (n=299) (a), ribosomal protein (RP) genes (n=76) (b) and a control group for the RP genes shown in b (n=132) (c). These genes are transcribed at similar level as the RP genes from b. (d) A scatter plot of Mediator occupancy (Gal11) versus the difference in TFIIB occupancy between WT and Srb5-depleted cells.
Transient association of Mediator with the PIC
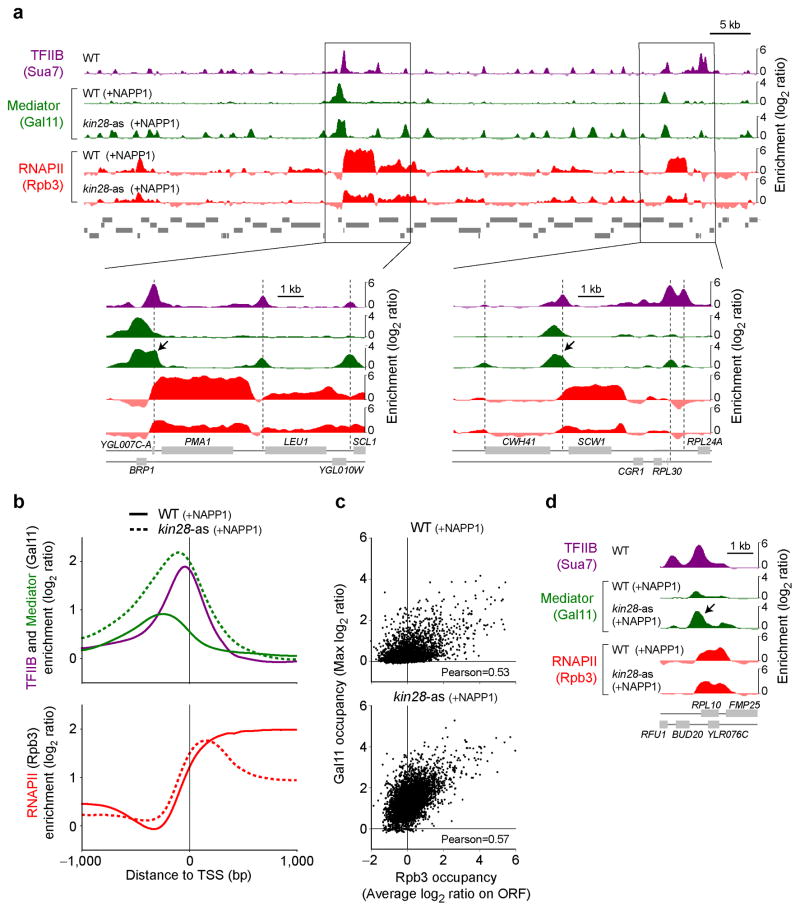
Genes encoding many Mediator subunits were originally identified as suppressors of partial truncation of the RNAPII C-terminal domain (CTD) 38,39. Later, the association of Mediator with the CTD was shown to be disrupted by CTD phosphorylation in vitro40,41. Finally, Mediator’s ability to act as a co-activator in vitro requires the CTD5,39,42. For all these reasons, Mediator function is thought to be intimately linked to the CTD and its phosphorylation state, though the nature of this relationship has remained unclear. We therefore looked at Mediator occupancy in CTD kinase mutants. With the exception of the TFIIH-associated Kin28 (CDK7) (hereafter called Kin28), no CTD kinase mutant substantially affected Mediator occupancy (Supplementary Fig. 3a–c). Following abrogation of Kin28 kinase activity with the inhibitor NAPP1 in kin28-analog sensitive (as) cells, however, overall Mediator occupancy was widely increased at several genomic locations (Fig. 3a). Quite strikingly, this increase in Mediator binding does not mainly occur at UAS regions. Instead, in the absence of Kin28 activity, binding of Mediator was observed at promoters, where it is normally not detected (Fig. 3, see arrows for specific examples). A simple interpretation of our data, which is also completely consistent with the in vitro data from the literature40,41, is that Kin28 mediates the dissociation of Mediator from core promoters, and presumably the PIC. This result has important implications as it shows that while Mediator is not a stable component of the PIC in vivo, it nevertheless colocalizes with it when Kin28 kinase activity is abrogated, and this appears unique to Kin28 among the transcriptional CDKs. What accounts for the apparent increase in Mediator residence time at promoters? One possible contributor to increased Mediator signal at promoters upon Kin28 inhibition could be the presence of a long-lived open complex consequent to defective RNAPII promoter escape defect described below (additionally, ssDNA in an open PIC may crosslink more efficiently than dsDNA43). Regardless, our result raises the intriguing question of why Mediator becomes detectable over core promoters.
Figure 3.
Mediator accumulates at core promoters in the absence of Kin28 kinase activity (see also Supplementary Fig. 3). (a) Mediator (Gal11, green) and RNAPII (Rpb3, red) occupancy in wild type (WT) and kin28 ATP analog-sensitive (kin28-as) cells, both treated with NAPP1 (+NAPP1), along a segment of chromosome VII. TFIIB (purple) from WT cells is shown as a placeholder for core promoters. Zooms into regions around the PMA1 and SCW1 genes are shown at the bottom. Vertical dashed line indicates PIC positions and arrows point toward accumulation of Mediator in core promoter regions. (b) Average Mediator (Gal11, green) and RNAPII (Rpb3, red) occupancy in wild type (WT, solid traces) and kin28-as (dotted traces) cells, both treated with NAPP1, around the TSS of all genes with an Rpb3 average ORF occupancy >1 in WT cells (n=266). TFIIB (purple) is shown as a placeholder for core promoters. (c) Scatter plot of Mediator (Gal11) occupancy versus RNAPII (Rpb3) occupancy in wild type (WT, top) and kin28-as (kin28-as, bottom) cells, both treated with NAPP1. Gal11 occupancy is defined as the maximum Gal11 enrichment observed in the first kilobase upstream from the TSS for each gene. Rpb3 occupancy is defined as the average occupancy across the entire ORF. (d) Mediator (Gal11, green) and RNAPII (Rpb3, red) occupancy in wild type (WT) and kin28-as cells, both treated with NAPP1, around the RPL10 gene. TFIIB (purple) is shown as a placeholder for core promoters. The arrow indicates the accumulation of Mediator in the promoter of RPL10 in kin28-as cells.
We set out to test the model that Mediator-PIC interactions have now been trapped due to loss of Kin28 activity, which hypothetically would be required to dissolve them. Even more importantly, this model immediately posits a mechanism for Kin28’s role in RNAPII promoter clearance. We found that accumulation of Mediator upon Kin28 inhibition was accompanied by a shift in RNAPII towards the core promoter, leading to RNAPII accumulation in the 5′ end of genes, at the expense of the 3′ region, as documented by us and others before (REFs44–46, Fig. 3a,b and Supplementary Fig. 3d). This result raises the possibility that Kin28 function in promoter escape by RNAPII could be Mediator-dependent. Noteworthy is the fact that, like Mediator, RNAPII was not normally detected exactly coincidentally with TFIIB (a marker of the core promoter or TSS). Instead, average RNAPII occupancy rised further downstream (See Figs.1a and 3). Taken together, these data suggest that RNAPII and Mediator association in PICs is transient, and also in both cases, the release from the PIC is stimulated by Kin28-dependent phosphorylation.
Interestingly, Mediator occupancy upstream of genes is more reliably detected in kin28-as cells than it is in WT cells (Fig. 3a,b and Supplementary Fig. 3d). Indeed, when the activity of Kin28 was abrogated, Mediator, now trapped at promoters, became readily detectable even at genes where it is otherwise difficult to detect (Fig. 3c and Supplementary Fig. 3d), such as the RP genes (Fig. 3d). This has important implications as it provides further support for a global role for Mediator in transcription. Indeed, our data are consistent with a model where Mediator associates with all genes but is more difficult to detect at some of them (the RP genes being extreme cases) due to the transient nature of its association with promoters and most likely also with UAS regions.
P-Ser5 mediates Mediator release and RNAPII escape
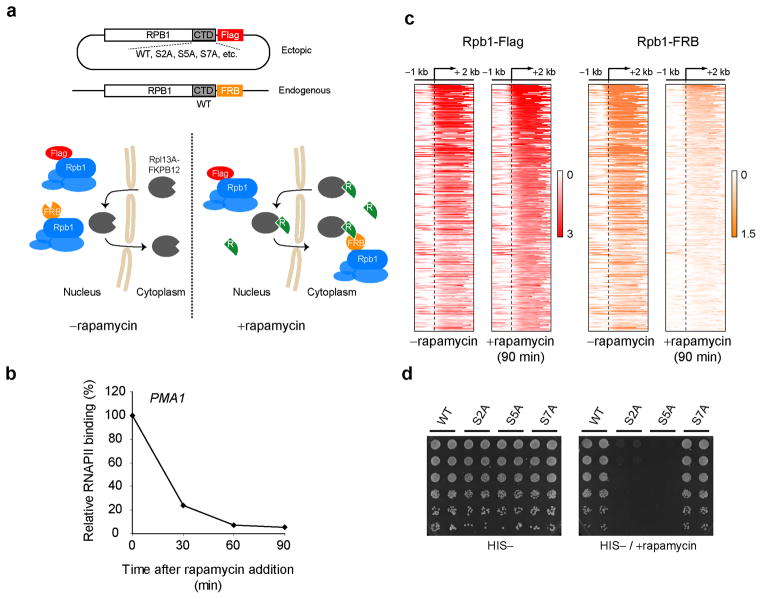
While the experiments described above uncover a role for Kin28 in Mediator release and RNAPII escape, the functional target of Kin28 kinase activity has not been addressed. An obvious candidate that would fit with prior Mediator literature would be the RNAPII CTD. However, Kin28 also has been shown to have non-CTD substrates41,47. Even if the CTD is presumed the relevant Kin28 target, two different serines in the CTD heptapeptide sequence (Ser5 and Ser7) are substrates for Kin28 (reviewed in48). CTD kinase mutants, despite their utility, have serious limitations in dissecting CTD function. Complementation systems have been developed, using α-amanitin-resistant Rpb1 mutants49,50 or Rpb1 conditional knockout in DT40 chicken cells51, to assay for various phenotypes of CTD mutants but these systems require several hours or days to switch from the WT to the mutant allele, which can lead to secondary and toxicity effects. Here we took advantage of the recently developed anchor-away system37 to rapidly deplete the endogenous Rpb1 protein from the nucleus. By expressing various mutant alleles (such as CTD mutants) that are immune to nuclear depletion, we can assay for different phenotypes associated with these mutations (Fig. 4a). We call this system “Complementation After Nuclear Depletion” (CAND). As shown in Figure 4b, Rpb1 was acutely depleted from the highly expressed PMA1 gene in as short as 60 minutes after addition of rapamycin in this system. Importantly, ChIP-chip for an exogenous RPB1 allele, here tagged with a Flag epitope, was unaffected by the addition of rapamycin though the FRB-tagged endogenous RPB1 allele was completely wiped out from genes (Fig. 4c). Consequently, the exogenous allele can functionally complement the endogenous allele. As expected, WT or non-lethal mutations such as S7A could rescue growth in the presence of rapamycin while lethal mutations such as S2A or S5A could not (Fig. 4d).
Figure 4.
The Complementation After Nuclear Depletion (CAND) system to study CTD function in vivo. (a) A schematic diagram of the experimental CAND system. The endogenous RPB1 gene is epitope tagged with the FRB (orange) domain of human mTOR in the anchor-away-ready strain. An additional RPB1 allele, carrying the desired CTD mutation is expressed ectopically from a CEN-HIS3 plasmid and under the RPB1 promoter, therefore ensuring physiological expression levels. This ectopic Rpb1 if Flag-tagged (red). In the absence of rapamycin (−rapamycin panel), both RPB1 alleles co-exist in the nucleus and the cell is kept healthy thanks to the presence of the WT (endogenous) protein in the nucleus. Upon addition of rapamycin (+rapamycin panel), the WT (endogenous) protein is rapidly sequestrated in the cytoplasm while the mutant protein stays in the nucleus. Various assays can be applied to these cells shortly (60–90 minutes) after addition of rapamycin. (b) Relative RNAPII (Rpb3) binding on the PMA1 gene following rapamycin addition. (c) Heat map representation of the occupancy of the endogenous (FRB-tagged, orange) and an ectopically expressed (Flag-tagged, red) Rpb1 proteins over the genes with an average Rpb1 (Flag) ORF occupancy >1 (n=345) in the absence of rapamycin and 90 minutes after addition of rapamycin. (d) Spotting assay of strains expressing RPB1 alleles carrying different CTD mutations. After growth in HIS-liquid media, cultures were washed, resuspended at equal optical density, serial diluted (5 fold series) and spotted on HIS- plates containing (right) or not (left) rapamycin.
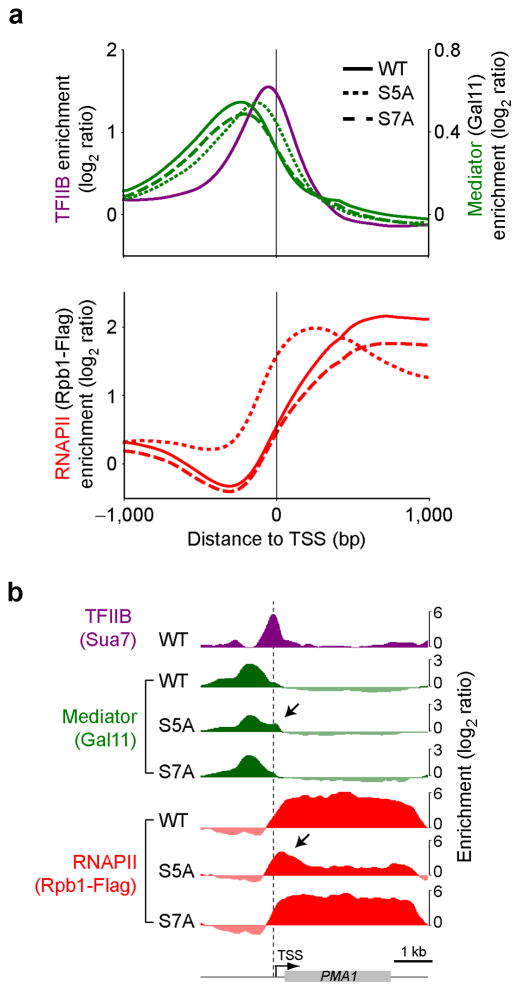
Using the CAND system, we tested whether specific CTD mutations could recapitulate the Mediator and RNAPII accumulation phenotypes observed in kin28-as cells. Because Kin28 is known to phosphorylate CTD Ser5 and Ser7, we tested both S5A and S7A mutants separately, together with a wild type (WT) allele as control. Importantly, replacing the endogenous Rpb1 with a WT allele generated wild type RNAPII and Mediator profiles (Fig. 5). Interestingly, complementing Rpb1 depletion with an S5A mutant led to RNAPII (dotted red) and Mediator (dotted green) ChIP profiles that are similar to those from the kin28-as mutant. Indeed, after depletion of the endogenous Rpb1, the S5A mutant caused Mediator to shift towards promoters and RNAPII to pile up in the 5′ end of genes (See black arrows in specific examples shown in Fig. 5b). This phenotype was not observed when complementing with the S7A mutant (dashed traces in Fig. 5a,b). Taken together, these observations are entirely consistent with the model that Kin28-dependent phosphorylation of the CTD leads to release of Mediator and RNAPII from core promoters or TSS, leaving Mediator’s most stable chromatin association upstream towards presumptive activator recruitment sites, and RNAPII’s being downstream within transcription units.
Figure 5.
Mediator occupancy shifts towards core promoters in the absence of CTD Ser5 phosphorylation. (a) Average Mediator (Gal11, green) and RNAPII (Rpb1-Flag, red) occupancy on all genes with Rpb1-Flag average ORF occupancy >1 in the absence of rapamycin (n=345) in cells expressing wild type (WT, solid traces), S5A (dotted traces) and S7A (dashed traces) Rpb1 proteins, 90 minutes after nuclear depletion of the endogenous Rpb1 protein by CAND. TFIIB (purple) from WT cells is shown as a placeholder for core promoters. (b) Genome browser view of a region around the PMA1 gene. The vertical dashed line indicates PIC position and black arrows point toward Mediator accumulation at the promoter and RNAPII pile up in the 5′ end in the S5A mutant.
DISCUSSION
We report an extensive investigation of Mediator occupancy across the yeast genome. The location of Mediator along yeast genes has a long history of debate. It has been reported to associate (or not to associate) with UAS, promoters and even transcribed regions9,10,26–28. Our examination of Mediator occupancy using a number of systems (antibodies versus tagged proteins), approaches (normalization to input versus to non-tagged or Mock controls) and subunits (a total of 14 subunits were tested), in addition to the use of different CTD kinase mutants, allows us to more fully understand the nature of previous discrepancies. We showed that Mediator stably associates with UAS regions but only transiently associates with core promoters (See Fig. 6 for a schematic representation). The prior lack of clarity about Mediator being at core promoters or not very likely has something to do with the resolution of the ChIP assays and the kinetics of steps in the transcription cycle. The distance between the UAS and the core promoter can be as small as 60 base pairs and rarely exceeds 300 base pairs in yeast. Given the resolution of ChIP, a signal from a binding event at the UAS would “spill over” the promoter to a considerable extent. When using a single PCR amplicon, this has certainly led some people to conclude that Mediator occupies core promoters. As for binding of Mediator in the coding regions, much might relate to non-specific enrichment of highly transcribed regions in ChIP assays, as these regions are enriched using IgG control ChIPs or anti-Myc ChIP samples from non-tagged strains. Consequently, most of this signal disappears when the signal from these controls are factored into the analyses. Previous ChIP-chip experiments reporting binding of Mediator in ORFs9,10 were using input DNA as the baseline (as opposed to control ChIPs), perhaps explaining the discrepancies with other studies including the current work. Another contentious issue about Mediator relates to whether it is ubiquitously used by all genes or rather specifically recruited by specific TFs at some but not all genes26–28. Here, the fact that Mediator is not detected as easily at some genes than others in wild type cells, coupled to the fact that it binds at variable distances from the TSS from gene to gene, most likely contributed to divergent views. Because Mediator occupies promoters in a manner that correlates well with RNAPII in Kin28 mutant cells (conditions where Mediator is proposed to be trapped at promoters), and because it affects PIC assembly at all genes, our data are consistent with the original view of Mediator as a general co-activator28,52–54. Our data also suggest that the variability in the ability to detect Mediator in wild type cells has to do with the dynamics at which it goes from the nucleoplasm, to the UAS, to the promoter and to the nucleoplasm again, rather than reflecting a gene-specific function.
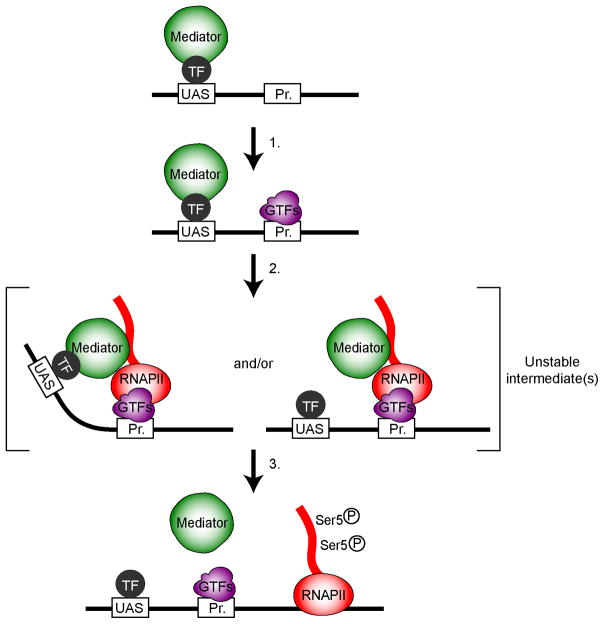
Figure 6.
A schematic representation of the dynamic association of Mediator with yeast genes. Mediator (green) is recruited to the Upstream Activating Sequence (UAS) by sequence-specific transcription factors (TF, dark grey). In step 1, Mediator stimulates pre-initiation complex (PIC, purple) assembly. RNAPII (red) and Mediator are the last components to join the PIC, creating a very unstable (transient) fully assembled PIC (step 2). Note that our data do not allow determining whether promoter-bound Mediator preserves its contact with the UAS. Phosphorylation of the CTD on Ser5 by Kin28 promotes the eviction of Mediator and the concomitant or subsequent release of RNAPII from the promoter area (step 3).
Our data are consistent with a well-conserved Mediator mechanism between yeast and mammals. In mammalian cells, Mediator is mostly detected in enhancer regions55–58, reminiscent to the UAS binding described here. In addition to enhancers, a somewhat smaller Mediator peak is often observed at the core promoter in mammalian cells. While this is generally considered as the consequence of the well described enhancer-promoter looping, our data at least suggest that some of this promoter signal may be a consequence of a transient association of mammalian Mediator with promoters. Finally, our data are also consistent with previous reports showing that the Kinase module has a binding profile similar to that or core Mediator subunits9,10,55.
The short life of Mediator at core promoters is mediated by Kin28-dependent phosphorylation of the RNAPII CTD on Ser5. Indeed, phosphorylation of Ser5 promotes the eviction of Mediator, in agreement with previous in vitro studies40,41. In addition, Ser5 phosphorylation promotes the escape of RNAPII from the promoter. Our results therefore provide compelling in vivo support for diverse in vitro and structural studies regarding possible functions of TFIIH and Ser5 in promoter clearance40,41,59,60. We and others previously reported a pile up of RNAPII in promoter regions in a Kin28 mutant44–46 but the current work is the first formal demonstration that this is mediated by Ser5 phosphorylation, as opposed to Ser7 or some other Kin28 substrate. Indeed, Kin28 has at least two substrates in addition to the CTD. Interestingly, these two substrates are components of Mediator, namely MED14 (Rgr1) and MED441,47. Abrogating phosphorylation of these two substrates, however, had no effect on Mediator or RNAPII occupancy (unpublished data).
In this study, we also describe a simple and powerful system to dissect the function of the RNAPII CTD in transcription and transcription-coupled processes. This assay, which we called CAND for “Complementation After Nuclear Depletion” consists of complementing the nuclear depletion of the endogenous Rpb1 protein with various Rpb1 alleles that are immune to the depletion. This allows replacing WT Rpb1 with any mutant (including lethal mutants) in as little as 60 minutes. CAND can be used to test growth properties of mutants, but can also be combined to other assays such as RNA-Seq or, as we did here, ChIP assays. CAND-ChIP will be a powerful method to test for the molecular function of the CTD in vivo.
ONLINE METHODS
Yeast strains and plasmids
Genotypes for the yeast strains used in this study are listed in Supplementary Table 1. The 9Myc (klTRP1) and 18Myc (klURA3) tags on Mediator subunits (Gal11, Rox3, Sin4, MED6, MED8, Srb10 and Srb11) were inserted by homologous recombination of PCR products amplified from the p3536 and p3747 plasmids (a gift from Richard Young), respectively. The strain yFR1544 (Srb4-FRB-kanMX6) was generated by homologous recombination of a PCR cassette obtained from plasmid pFA6a-FRB-kanMX6 (Euroscarf) with the appropriate primer pairs into the anchor-away-ready strain yFR1321 (HHY168, Euroscarf).
Details about the construction of the plasmid expressing RPB1-3Flag without the CTD under its endogenous promoter (pRS313-RPB1-CTDless-3Flag-HIS3) are available upon request. To construct the final RPB1-3Flag-expressing plasmids, various CTD derivatives [either WT or where all serines at position 2, 5 or 7 have been replaced by alanines (CTDS2A, CTDS5A, CTDS7A)] were synthesized as minigenes by IDT and cloned into the pRS313-RPB1-CTDless-3Flag-HIS3 plasmid. The sequence of each minigene was optimized in order to facilitate synthesis. All sequences are available upon request.
Growth conditions
Yeast cells were grown to an OD600 of 0.6–0.8 at 30 °C following standard procedures. For inhibition of Kin28 activity experiments (Fig. 3), ATP analog-sensitive kin28-as strains were pre-cultured in yeast nitrogen base (YNB) medium lacking uracil before inoculation in yeast extract-peptone-dextrose (YPD) medium. kin28-as strains and their controls were treated with 6μM of NAPP1 (Tocris Bioscience) for 15 min prior to crosslinking. Strains expressing the RPB1 CTDwt or CTDmutant plasmids were cultured in YNB medium lacking histidine (HIS). For the anchor-away experiments (Figs. 2, 4 and 5), all strains were treated with 1 μg/mL of rapamycin (Bio Basic Inc.) for 90 min before crosslinking. For the rapamycin-induced genes ChIP experiment (Fig. 1d), cells were treated with 200 ng/ml of rapamycin for 30 min before crosslinking.
Serial-dilution growth assay
Cells were grown to saturation in −HIS medium at 30 °C, washed and resuspended to an OD600 of 1.0 in water. Cells were then subjected to fivefold serial dilutions and spotted onto −HIS, with or without rapamycin (1 μg/mL). Plates were incubated at 30 °C and inspected daily.
ChIP
ChIP experiments were performed at least in duplicates as previously described32 with minor modifications. In brief, yeast cultures were grown in 50 mL of the appropriate medium (see above) to an OD600 of 0.6–0.8 before cross-linking with 1% formaldehyde for 30 min and quenched with 125mM glycine. The following amounts of antibody per immunoprecipitation were used: Rpb3 (W0012 from Neoclone, 3μL), TFIIB (a gift from Richard Young, 2μL), Myc (9E10, 10μL), Flag (Sigma F3165, 5μg), FRB (mTor, human FRB domain from Enzo Life Sciences ALX-215-065, 5μL), rabbit IgG (Millipore 12-370, 2μg), Srb4 (MED17) (Abcam ab63812, 5μL) and rabbit polyclonal antibodies against various Mediator subunits (from Richard Young, 10μL). All comercial antibodies have been validated for ChIP (see manufacturers’ web sites). The TFIIB and Mediator subunits antibodies have been described before61,62. Rabbit antibodies were coupled to Dynabeads coated with Protein G (Life Technologies) and mouse antibodies were coupled to Pan Mouse IgG antibodies (Life Technologies).
ChIP-qPCR
To assess the relative RNAPII (Rpb3) binding on the PMA1 gene following different time points after rapamycin addition (Fig. 4b), ChIP DNA was analyzed by quantitative real-time PCR (qPCR) using SYBR green. Primers directed against the ORF of the PMA1 gene (Fwd: TCTTCTGTGTTTTGGGTGGTT, Rev: TCTTTGCATAGCAGCCATGA) were used. For each time point “n”, the enrichment [relative to time 0 (t0)] was calculated using the ΔCt method as follow: 1/(2^[Ctn−Ctt0]).
ChIP-chip
For epitope-tagged proteins, the ChIP DNA was hybridized in competition with a control ChIP DNA prepared from an isogenic untagged strain. For ChIPs performed using rabbit polyclonal antibodies, the ChIP DNA was hybridized in competition with input DNA. The microarrays were custom designed by Agilent Technologies and contain a total of about 180,000 Tm-adjusted 60-mer probes covering the entire yeast genome with virtually no gaps between probes.
ChIP-chip data analysis
The ChIP-chip data were normalized using the Limma Loess method and replicates were combined as described previously63. The data was subjected to one round of smoothing using a Gaussian sliding window with a standard deviation of 100bp to generate data points in 10bp intervals as described before64.
Correction of systematic errors in ChIP-chip data
As indicated above, ChIP-chip performed using epitope tagged proteins were hybridized against ChIP samples from isogenic non-tagged strains. This reliably removes systematic errors, notably the enrichment in highly expressed ORFs, as shown in Supplementary Figure 1c. For ChIP experiments performed with polyclonal antibodies (TFIIB, FRB and various Mediator subunits shown in Supplementary Figure 1a), however, the ChIP DNA was hybridized against input DNA. In order to reduce the systematic error in these experiments, the log2 ratio from a Mock ChIP-chip experiment performed using rabbit IgG was subtracted from the log2 ratio of these ChIP-chip experiments. As shown in Supplementary Figure 1d, this eliminates most of the systematic error enrichment observed in highly transcribed ORFs.
Aggregate profiles
Aggregate profiles (as shown in Fig. 1a, 2a–c, 3b, 5a, etc.) were generated using the Versatile Aggregate Profiler (VAP), a stand alone program for which a manuscript is currently under review and is based on methods used in our previous work32,44,65–67. In brief, for Figure 1, genes were virtually cut in the middle and the first half aligned on the TSS while the second half was aligned on the polyA (pA) site. The TSS and pA sites were deduced using the UTR sizes as determined in Xu and collaborators68. Genes for which the 5′ and 3′ UTRs have not been previously determined were therefore not included in these analyses. The aligned data were averaged over 10bp bins (100 bins upstream from the TSS, 100 bins downstream from the TSS, 100 bins upstream of the pA and 100 bins downstream of the pA). For Figures 2a–c, 3b and 5a, genes were aligned on their TSS only and averaged over 10bp bins (100 bins upstream of the TSS and 100 bins downstream of the TSS). In these analyses, only genes that are at least 1 kilobase-long were retained since including shorter genes creates noise due to the resolution of the ChIP assay (signal from the 5′ end “contaminating” the 3′ end and vice versa).
Transcription factor binding sites frequency
The transcription factor binding sites (TFBS) frequency was generated using the UCSC transRegCode genome browser track representing data from Harbison et al69. These data are associated with a quality score and information about whether or not there is ChIP evidence for binding (as opposed to solely computational prediction). Using this information, we filtered the TFBS to keep only those with a score above 300 and having ChIP evidence for DNA binding. Essentially, this keeps only high quality binding site predictions and removes a large number of predictions that are likely false positives. The frequency of these TFBS was computed relative to the TSS and pA sites in Figure 1a.
Heat map representation of ChIP-chip data
To generate heat maps (as shown in Figs. 1d and 4c), genes were aligned on their TSS and the intensity was parsed into 10bp bins. The heat map images were generated using ThreeView70.
Supplementary Material
Acknowledgments
We thank K. Struhl (Harvard Medical School) for discussing unpublished data, P. Collin (Robert laboratory) for assistance with the CTD mutant constructs, as well as R. Young (Whitehead Institute for Biomedical Research), S. Hahn (Fred Hutchinson Cancer Research Center) and L. Myers (Dartmouth-Hitchcock Medical Center) for sharing strains and antibodies. We are also grateful to C. Kaplan and N. Francis for their critical reading of the manuscript. This work was funded by a Canadian Institutes for Health Research grant (MOP-82891) to F.R.
Footnotes
Accession codes. Raw and normalized ChIP-chip data are available at Gene Expression Omnibus (GEO) database under accession number GSE55402.
AUTHOR CONTRIBUTIONS
C.J. and F.R. designed the project and wrote the manuscript. C.J. performed most of the experiments. F.R. performed most of the analyses.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 2.Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit Rev Biochem Mol Biol. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansari SA, Morse RH. Mechanisms of Mediator complex action in transcriptional activation. Cell Mol Life Sci. 2013;70:2743–56. doi: 10.1007/s00018-013-1265-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlsten JO, Zhu X, Gustafsson CM. The multitalented Mediator complex. Trends Biochem Sci. 2013;38:531–7. doi: 10.1016/j.tibs.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Myers LC, et al. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1998;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spahr H, et al. Mediator influences Schizosaccharomyces pombe RNA polymerase II-dependent transcription in vitro. J Biol Chem. 2003;278:51301–6. doi: 10.1074/jbc.M306750200. [DOI] [PubMed] [Google Scholar]
- 7.Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes & development. 2009;23:439–51. doi: 10.1101/gad.1767009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 2002;16:1339–44. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrau JC, et al. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Molecular cell. 2006;22:179–92. doi: 10.1016/j.molcel.2006.03.023. [DOI] [PubMed] [Google Scholar]
- 10.Zhu X, et al. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Molecular cell. 2006;22:169–78. doi: 10.1016/j.molcel.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 11.Conaway RC, Conaway JW. The Mediator complex and transcription elongation. Biochim Biophys Acta. 2013;1829:69–75. doi: 10.1016/j.bbagrm.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuras L, Struhl K. Binding of TBP to promoters in vivo is stimulated by activators and requires Pol II holoenzyme. Nature. 1999;399:609–13. doi: 10.1038/21239. [DOI] [PubMed] [Google Scholar]
- 13.Cantin GT, Stevens JL, Berk AJ. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc Natl Acad Sci U S A. 2003;100:12003–8. doi: 10.1073/pnas.2035253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–43. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G, et al. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Molecular cell. 2005;17:683–94. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 16.Baek HJ, Kang YK, Roeder RG. Human Mediator enhances basal transcription by facilitating recruitment of transcription factor IIB during preinitiation complex assembly. J Biol Chem. 2006;281:15172–81. doi: 10.1074/jbc.M601983200. [DOI] [PubMed] [Google Scholar]
- 17.He Q, Battistella L, Morse RH. Mediator requirement downstream of chromatin remodeling during transcriptional activation of CHA1 in yeast. J Biol Chem. 2008;283:5276–86. doi: 10.1074/jbc.M708266200. [DOI] [PubMed] [Google Scholar]
- 18.Rana R, Surapureddi S, Kam W, Ferguson S, Goldstein JA. Med25 is required for RNA polymerase II recruitment to specific promoters, thus regulating xenobiotic and lipid metabolism in human liver. Mol Cell Biol. 2011;31:466–81. doi: 10.1128/MCB.00847-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin JJ, et al. Mediator coordinates PIC assembly with recruitment of CHD1. Genes Dev. 2011;25:2198–209. doi: 10.1101/gad.17554711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen XF, et al. Mediator and SAGA have distinct roles in Pol II preinitiation complex assembly and function. Cell Rep. 2012;2:1061–7. doi: 10.1016/j.celrep.2012.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takahashi H, et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galbraith MD, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell. 2013;153:1327–39. doi: 10.1016/j.cell.2013.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukundan B, Ansari A. Novel role for mediator complex subunit Srb5/Med18 in termination of transcription. J Biol Chem. 2011;286:37053–7. doi: 10.1074/jbc.C111.295915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukundan B, Ansari A. Srb5/Med18-mediated termination of transcription is dependent on gene looping. J Biol Chem. 2013;288:11384–94. doi: 10.1074/jbc.M112.446773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, et al. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 2012;45:459–69. doi: 10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan X, Chou DM, Struhl K. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol. 2006;13:117–20. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 27.Fan X, Struhl K. Where does mediator bind in vivo? PLoS One. 2009;4:e5029. doi: 10.1371/journal.pone.0005029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ansari SA, He Q, Morse RH. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci U S A. 2009;106:16734–9. doi: 10.1073/pnas.0905103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strasser K, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–8. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 30.Teytelman L, Thurtle DM, Rine J, van Oudenaarden A. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci U S A. 2013;110:18602–7. doi: 10.1073/pnas.1316064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park D, Lee Y, Bhupindersingh G, Iyer VR. Widespread Misinterpretable ChIP-seq Bias in Yeast. PLoS One. 2013;8:e83506. doi: 10.1371/journal.pone.0083506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drouin S, et al. DSIF and RNA Polymerase II CTD Phosphorylation Coordinate the Recruitment of Rpd3S to Actively Transcribed Genes. PLoS genetics. 2010;6:e1001173. doi: 10.1371/journal.pgen.1001173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soontorngun N, et al. Genome-wide location analysis reveals an important overlap between the targets of the yeast transcriptional regulators Rds2 and Adr1. Biochem Biophys Res Commun. 2012;423:632–7. doi: 10.1016/j.bbrc.2012.05.151. [DOI] [PubMed] [Google Scholar]
- 34.Liesen T, Hollenberg CP, Heinisch JJ. ERA, a novel cis-acting element required for autoregulation and ethanol repression of PDC1 transcription in Saccharomyces cerevisiae. Mol Microbiol. 1996;21:621–32. doi: 10.1111/j.1365-2958.1996.tb02570.x. [DOI] [PubMed] [Google Scholar]
- 35.Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nature structural & molecular biology. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loewith R, Hall MN. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics. 2011;189:1177–201. doi: 10.1534/genetics.111.133363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haruki H, Nishikawa J, Laemmli UK. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell. 2008;31:925–32. doi: 10.1016/j.molcel.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 38.Nonet ML, Young RA. Intragenic and Extragenic Suppressors of Mutations in the Heptapeptide Repeat Domain of Saccharomyces Cerevisiae RNA Polymerase II. Genetics. 1989;123:715–24. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–75. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 40.Sogaard TM, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem. 2007;282:14113–20. doi: 10.1074/jbc.M701345200. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y, et al. Two Cyclin-Dependent Kinases Promote RNA Polymerase II Transcription and Formation of the Scaffold Complex. Molecular and Cellular Biology. 2004;24:1721–1735. doi: 10.1128/MCB.24.4.1721-1735.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 43.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bataille AR, et al. A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell. 2012;45:158–70. doi: 10.1016/j.molcel.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 45.Kim H, et al. Gene-specific RNA polymerase II phosphorylation and the CTD code. Nature structural & molecular biology. 2010;17:1279–1286. doi: 10.1038/nsmb.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tietjen JR, et al. Chemical-genomic dissection of the CTD code. Nat Struct Mol Biol. 2010;17:1154–61. doi: 10.1038/nsmb.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guidi BW, et al. Mutual targeting of mediator and the TFIIH kinase Kin28. J Biol Chem. 2004;279:29114–20. doi: 10.1074/jbc.M404426200. [DOI] [PubMed] [Google Scholar]
- 48.Jeronimo C, Bataille AR, Robert F. The writers, readers, and functions of the RNA polymerase II C-terminal domain code. Chem Rev. 2013;113:8491–522. doi: 10.1021/cr4001397. [DOI] [PubMed] [Google Scholar]
- 49.McCracken S, et al. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & development. 1997;11:3306–18. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fong N, Bird G, Vigneron M, Bentley DL. A 10 residue motif at the C-terminus of the RNA pol II CTD is required for transcription, splicing and 3′ end processing. Embo J. 2003;22:4274–82. doi: 10.1093/emboj/cdg396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hsin JP, Sheth A, Manley JL. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 2011;334:683–6. doi: 10.1126/science.1206034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson CM, Young Ra. General requirement for RNA polymerase II holoenzymes in vivo. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:4587–90. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holstege FCP, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–28. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 54.Takagi Y, Kornberg RD. Mediator as a general transcription factor. The Journal of biological chemistry. 2006;281:80–9. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 55.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–5. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svejstrup JQ, et al. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci U S A. 1997;94:6075–8. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson PJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD. Structure of the mediator head module bound to the carboxy-terminal domain of RNA polymerase II. Proc Natl Acad Sci U S A. 2012;109:17931–5. doi: 10.1073/pnas.1215241109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–9. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 62.Liao SM, et al. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–6. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 63.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science (New York, NY) 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 64.Guillemette B, et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS biology. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szilard RK, et al. Systematic identification of fragile sites via genome-wide location analysis of gamma-H2AX. Nature structural & molecular biology. 2010;17:299–305. doi: 10.1038/nsmb.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rufiange A, Jacques P-É, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Molecular cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 67.Hardy S, et al. The euchromatic and heterochromatic landscapes are shaped by antagonizing effects of transcription on H2A.Z deposition. PLoS genetics. 2009;5:e1000687. doi: 10.1371/journal.pgen.1000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu Z, et al. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–7. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.