Abstract
The U1 small nuclear (sn)RNA participates in splicing of pre-mRNAs by recognizing and binding to 5′ splice sites at exon/intron boundaries. U1 snRNAs associate with 5′ splice sites in the form of ribonucleoprotein particles (snRNPs) that are comprised of the U1 snRNA and 10 core components, including U1A, U1-70K, U1C and the ‘Smith antigen’, or Sm, heptamer. The U1 snRNA is highly conserved across a wide range of taxa; however, a number of reports have identified the presence of expressed U1-like snRNAs in multiple species, including humans. While numerous U1-like molecules have been shown to be expressed, it is unclear whether these variant snRNAs have the capacity to form snRNPs and participate in splicing. The purpose of the present study was to further characterize biochemically the ability of previously identified human U1-like variants to form snRNPs and bind to U1 snRNP proteins. A bioinformatics analysis provided support for the existence of multiple expressed variants. In vitro gel shift assays, competition assays, and (IPs) precipitations revealed that the variants formed high molecular weight assemblies to varying degrees and associated with core U1 snRNP proteins to a lesser extent than the canonical U1 snRNA. Together, these data suggest that the hU1 snRNA variants analyzed here are unable to efficiently bind U1 snRNP proteins. The current work provides additional biochemical insights into the ability of the variants to assemble into snRNPs.
Keywords: Splicing, Spliceosome, U1A protein, U1-70K protein, Sm proteins
1. Introduction
Eukaryotes enhance protein diversity from a limited number of protein-coding genes via alternative processing of pre-mRNAs, including the use of alternative transcription start sites (Sandelin, Carninci et al. 2007), alternative splicing (Kornblihtt, Schor et al. 2013) and variable 3′ cleavage and polyadenylation sites (Proudfoot 2011). In the case of alternative splicing, it has been estimated that as many as 92-94% of human transcripts undergo some form of alternative splicing (Wang, Sandberg et al. 2008), which is mediated by a high molecular weight complex known as the spliceosome. The spliceosome is comprised of five small nuclear (sn)RNAs (U1, U2, U4, U5 and U6) to which over 100 proteins associate (Zhou, Licklider et al. 2002; Chen, Moore et al. 2007). Binding of the U1 small nuclear ribonucleoprotein particle (snRNP) to pre-mRNA forms the “early”, or E, complex (Wahl, Will et al. 2009). Subsequent incorporation of additional U snRNPs and associated protein factors lead to formation of complexes A, B and C, which represent discreet steps in the splicing process (Wahl, Will et al. 2009).
The U1 snRNA, along with three U1-specific proteins (U1A, U1-70K and U1C) and seven ‘Smith antigen’ (Sm) proteins (Sm B/B’, Sm C, Sm D1, Sm D2, Sm D3, Sm F, and Sm G), forms the U1 small nuclear ribonucleoprotein particle (snRNP), which binds via complementary base pairing to the 5′ exon/intron junction with the consensus sequence CAG|GUAAGU (Ast 2004). Interestingly, expressed snRNA variants have been described in a number of organisms, including Dictyostelium discoideum (Hinas, Larsson et al. 2006), Drosophila melanogaster (Chen, Lullo et al. 2005), the silk moth, Bombyx mori (Sierra-Montes, Freund et al. 2002; Sierra-Montes, Pereira-Simon et al. 2003; Sierra-Montes, Pereira-Simon et al. 2005; Smail, Ayesh et al. 2006) and humans (Kyriakopoulou, Larsson et al. 2006). In the case of human U1-like sequences, Kyriakopoulou et al. 2006 identified four expressed variants, U1A4, U1A5, U1A6, and U1A7, which were hypermethylated, bound Sm proteins in varying degrees and formed high molecular weight complexes.
In the present work, we characterized the ability of the human U1 variants (U1A4, U1A5, U1A6 and U1A7) to associate with proteins of the U1 snRNP. We found that all five variants appeared to assemble into high molecular weight complexes to varying degrees. These non-canonical U1-like snRNAs also bound in vitro to U1-70K and U1A proteins, as determined by native acrylamide gel electrophoresis and immunoprecipitations (IPs); however, competition assays indicated that the canonical U1 snRNA was able to effectively outcompete each of the variant snRNAs for binding to U1-70K. These results suggest that the sequence divergence among the expressed U1-like snRNAs may inhibit their ability to compete with the canonical U1 snRNA for binding to proteins of the U1 snRNP.
2. Materials and Methods
2.1. Identification of hU1 variants in an expressed sequence tag database
Sequences of the U1 snRNA variants from Kyriakopoulou et al. (2006) were input as queries in the National Center for Biotechnology Information nr/nt and Express Sequence Tags (EST) database using the nucleotide Basic Local Alignment Search Tool (BLASTn). ESTs with 100% and 95% identity to each of the variants are listed in Table 1. Variant sequences were aligned to the canonical hU1 using the optimal global pairwise alignment in BioEdit (v.7.0.9.0).
Table 1.
Identification of U1 snRNA variant sequences in the NCBI Expressed Sequence Tag (EST)database.
2.2. In vitro assembly of hU1-like RNA sequences into high molecular weight complexes
Templates for in vitro transcription of hU1-like sequences were generated by overlap PCRs in two steps. First, long primers containing approximately 80 nucleotides of each hU1-like snRNA and overlapping by approximately 20 nucleotides were used as templates to prime complementary strands with 30 cycles of the following thermocycling conditions: 95°C for 15 seconds, 68°C for 15 seconds and 72°C for 20 seconds. The T7 promoter sequence and 3′ end of each variant was added in a subsequent PCR using 1 μl of template from the first PCR with the following cycling parameters: 30 cycles of 95°C for 15 seconds, 68°C for 15 seconds and 72°C for 20 seconds. The primer sequences used to generate templates for in vitro transcription of the variants are included in Supplementary Table 1. All hU1 snRNAs were transcribed in vitro in the presence of 2 μl of 32P-UTP using Megascript T7 in vitro transcription kits (Invitrogen), separated on 1% agarose gels and visualized with ethidium bromide to verify the purity of the snRNAs. Importantly, In vitro transcribed U1 snRNAs have been previously shown to restore splicing to extracts depleted of endogenous U1 snRNA (Will, Rumpler et al. 1996). To perform in vitro RNP assembly reactions in a total volume of 25 μl, 1 μl containing, on average, 600 CPM of 32P-labeled variant was incubated at 30°C in the presence of 20 μM HEPES, 3.2 mM MgCl, 0.5 mM ATP/ 20 μM Creatine Phosphate, 0.6 units of RNasin and 15 μl of HeLa nuclear extract (a kind gift of Dr. Adrian Krainer; Cold Spring Harbor Laboratory). Reactions were stopped at 0′ and 60′ by the addition of heparin sulfate (40 ng/ml final concentration) and 5 min of incubation at 30°C. Subsequently, RNA-protein complexes were separated in 1.6% native Tris-glycine agarose gels. Gels were dried and visualized by autoradiography on Kodak Biomax films (Eastman Kodak Co.).
2.3. U1A and U1-70K protein binding assays
A plasmid encoding histidine-tagged U1A protein was kindly provided by Dr. Carol Lutz (Rutgers University). His-U1A plasmid was transformed and U1A protein was expressed in E. coli following the manufacturer’s recommendations for the pRSET family of vectors (Invitrogen). U1A protein was purified over a nickel-agarose column using the Ni-NTA kit (Invitrogen). A purified U1-70K fragment encoding amino acids 63-205 (the U1 snRNA binding domain) containing a maltose binding protein fusion on the N-terminus was a kind gift of Dr. Eric Greidinger (University of Miami). Protein-U1 binding assays were performed by adding 70-100 ng containing on average 600 CPM of 32P-labeled variant to 5 μg of purified, recombinant protein and incubated under splicing conditions described above for 60 minutes at 30°C. Protein-RNA complexes were resolved on 6% native TBE acrylamide gels and visualized as described above. The presence of bound U1-70K and U1A proteins were confirmed by western blotting. As a negative control, U1 snRNA was incubated in the absence of purified protein and included on each gel.
2.4. Variant snRNA competition assays with U1-70K
Competition assays between canonical (U1 snRNA) and non-canonical (U1A4, U1A5, U1A6 and U1A7) isoforms were performed as described for gel shift assays with U1 snRNAs and U1-70K, with the exception that competition assays contained 50 or 500 times the cold (unlabeled) non-canonical variant in the splicing reactions. Unlabeled, canonical hU1 snRNA at 50 or 500 times the molar ratio of the labeled variant was included in each competition assay. As a negative control for the gel shift, U1 snRNA was incubated in the absence of purified U1-70K and included on each gel.
2.5. IP of hU1 variants
Anti-U1A, anti-U1-70K, and anti-U1C antibodies were gifts of Dr. Douglas Black (University of California, Los Angeles). The anti-Sm (Y12) antibody was a gift of Dr. Joan Steitz (Yale University). A total of 50 μl of antibodies against U1 snRNP proteins were bound to 150 μl of protein A sepharose for two hours at room temperature on an end-over-end shaker, washed three times in phosphate buffered saline (PBS) containing 0.05% Tween-20 (PBS-T). Subsequently, 70-100 ng of in vitro transcribed and radiolabeled snRNA variant RNA containing on average 600 CPM of 32P as well as 50 μl of Hela nuclear extract were added to antibody-bound protein A sepharose and incubated at 4°C for two hours with end-over-end rotation. Protein A sepharose-antibody-RNA complexes were washed three times with 1 ml of PBS-T. Radiolabeled U1 snRNA variants were isolated by standard phenol-chloroform extraction, separated on 1.6% native Tris-glycine agarose gels, and visualized by autoradiography. Pre-immune rabbit serum was used as a negative control for the IPs.
3. Results
3.1. A subset of U1 variants are found as expressed sequence tags
Previous investigations have revealed the expression of human U1 variant sequences in human cell lines (HeLa cells) and tissues (Kyriakopoulou, Larsson et al. 2006). To further verify the expression of these U1-like RNAs, we performed a search using BLASTn. The variants (hU1A4, hU1A5, hU1A6 and hU1A7) were used as queries to search the human EST database. A limit of 95% minimum identity was used. Interestingly, we identified three hU1A4 ESTs with 100% identity to the U1A4 sequence and two ESTs with 100% identity to hU1A6 (Table 1). An additional 11 sequences are at least 95% identical to hU1A6. Given that there is only one genomic locus corresponding to hU1A6, it remains to be determined whether the ESTs matching hU1A6 with 95% identity represent additional U1 variants or if these sequences result from mutations within the tissues or cell lines from which the ESTs were derived. Two ESTs were also found that range in size from 50 nucleotides (nt) to 75 nt and match different portions of hU1A7 (nucleotide positions 25-99 and 106-165). Nucleotide positions 25-99 and 106-165 within hU1A7 contain a total of 12 mismatches with the canonical hU1A, indicating that these ESTs are much more closely related to hU1A7 than hU1A. We found no ESTs matching hU1A5. Overall, our data support the observations made by Kyriakopoulou et al. (2006) that hU1A4, hU1A6, and hU1A7 represent transcribed RNAs.
3.2. U1 variants form high molecular weight assemblies in vitro
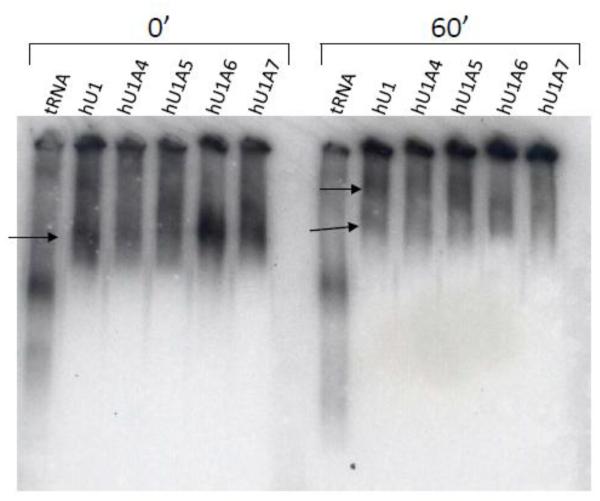
We used in vitro native gel assays to determine whether the U1-like sequences could assemble into ribonucleoprotein complexes. All five radiolabeled hU1 sequences were incubated under splicing conditions in the presence of HeLa nuclear extracts (a gift of Dr. Adrian Krainer; Cold Spring Harbor Laboratory). The canonical U1 snRNA was included to show the putative positions of snRNPs at 0 minutes and 60 minutes (Figure 1; arrows). In vitro transcribed, internally radiolabeled tRNA was used as a negative control. At 0 minutes, we observed mobility shifts to higher molecular weight than free U1 snRNA (data not shown). In addition, all variants at the 0 minute time point appeared to form complexes similar in mobility to that of the canonical U1 snRNA. Similarly, all variants shifted to higher molecular weight assemblies after 60 minutes, although it should be noted that the size of the RNPs containing hU1A6 and hU1A7 varied across experiments. In some cases, hU1A6 and hU1A7 formed complexes that matched the size of hU1 snRNA while in other experiments the two variants migrated with lower mobility than hU1 (Figure 1). The hU1A4 and hU1A5 snRNAs reproducibly assembled into complexes that matched the size of those generated by hU1 snRNA. HU1A4 also separated into the two discrete bands formed by hU1A. Conversely, hU1A5, hUA16, and hU1A7 separated into high molecular weight bands that did not match hU1 or hU1A4. Importantly, tRNAs underwent unique mobility shifts that were not observed with the snRNAs (Figure 1).
Figure 1. U1-like snRNAs differentially form high molecular weight complexes in vitro.
Human U1-like variants were incubated with nuclear extract under splicing conditions for 0 and 60 minutes, and high molecular weight assemblies were visualized by native gel electrophoresis. While all variants form complexes, the sizes of these assemblies vary, with hU1A6 and hU1A7 forming the fewest complexes at 60 minutes.
3.3. U1-like snRNAs differentially bind U1A protein
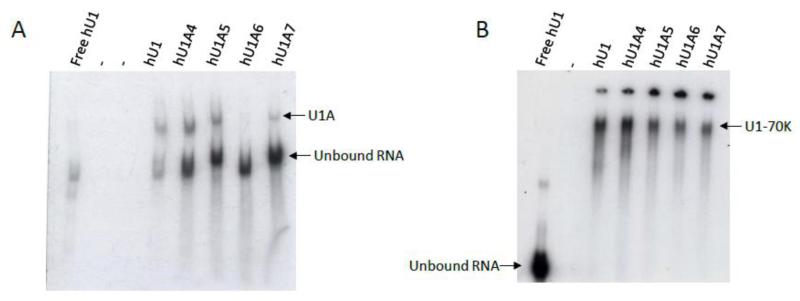
The ability of the U1 variants to associate with the U1A protein was assessed using gel shift assays. Recombinant U1A was incubated in the presence of each U1 RNA under splicing conditions and separated in native polyacrylamide-TBE gels. While hU1 bound efficiently to U1A protein, the U1 variants displayed a range of binding capacities (Figure 2A). Strikingly, hU1A6 appeared nearly incapable of associating with U1A protein, and hU1A7 bound only minimally (Figure 2A). Not surprisingly, these two snRNAs contain the highest number of mutations within the U1A binding site when compared to canonical hU1 (6 and 5 nucleotide differences for hU1A6 and hU1A7, respectively, compared to hU1). Despite the fact that hU1A5 possesses just one nucleotide change within the U1A protein binding loop, hU1A5 interacted with U1A protein at levels similar to hU1A4, with most snRNA remaining in the unbound form (Figure 2A).
Figure 2. U1-like variants bind U1A and U1-70K proteins with varying affinity. A.
Canonical hU1 snRNA is the most efficient at binding U1A protein (compare signal of bound to unbound RNA). Of the variants, all but hU1A6 bind U1A protein to some degree, although much of the free hU1 remains in the unbound form. B. Both hU1 and hU1A4 efficiently bind U1-70K while the other variants bind minimally to U1-70K. The origin of loading is visible at the top of the gel. Free hU1 in the absence of protein was included in each gel.
3.4. Binding capacity of hU1 variants for U1-70K protein
Gel shift assays were also used to determine the capability of the hU1 variants to associate with U1-70K. As expected, incubation of hU1 in the presence of U1-70K resulted in a mobility shift of radiolabeled hU1 to a higher molecular weight (Figure 2B). Interestingly, hU1A4, which has just a single nucleotide difference compared to hU1 within the U1-70K binding site, binds U1-70K with efficiency comparable to that of hU1. Conversely, the hU1A6 and hU1A7 variants, which contain multiple nucleotide changes within the U1-70K binding stem-loop, appeared to associate the least with U1-70K. Interestingly, although eight out of 13 consensus nucleotides on the U1-70K stem-loop sequence within hU1A5 are mutated (62% divergence), the hU1A5 snRNA was capable of binding U1-70K, albeit with low efficiency (Figure 2B).
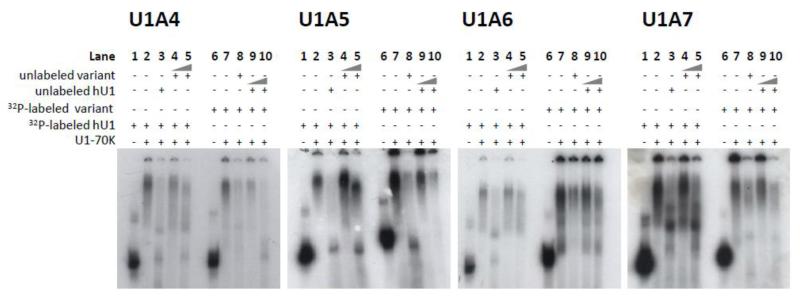
We further investigated the ability of the U1-like snRNAs to bind U1-70K using competition assays. Each hU1 sequence was radiolabeled in vitro and incubated in the presence of U1-70K alone (Figure 3; lanes 2 and 7), with a 50-fold molar excess of a non-labeled version of the same snRNA (Figure 3; lanes 3 and 8), or with 50- or 500-fold molar excess of unlabeled variant (Figure 3; lanes 4-5) or canonical hU1 snRNA (Figure 3; lanes 9-10). Incubation of radiolabeled variants in the presence of U1-70K resulted in a shift to a higher molecular weight (Figure 3; compare lanes 1 with 2 and lanes 6 with 7 in each panel). A total of 50-fold excess of unlabeled specific competitor greatly reduced the signal of labeled variant, suggesting that the equilibrium of U1-70K binding had been shifted toward unlabeled snRNAs (Figure 3; compare lanes 2 with 3 and lanes 7 with 8 in each panel). While hU1 efficiently outcompeted hU1 variants even at 50-fold molar excess, the hU1A4 variant was the most efficient competitor of hU1 (Figure 3; lanes 4 and 5). Although the gel shift experiments indicated that hU1A5 and hU1A6 were capable of binding U1-70K with low efficiency, hU1A5 and hU1A6 were unable to strongly outcompete hU1, even at 500-fold molar excess (Figure 3; lanes 4 and 5). Similarly, hU1 successfully competed with hU1A5 and hU1A6 for U1-70K binding at 50-fold and 500-fold excess (Figure 3; compare lanes 7 and 9). Surprisingly, hU1A7 was more effective at supplanting hU1 than hU1A5 or hU1A6 (Figure 3). Similarly, hU1 competed out hU1A7 less efficiently than hU1A5 and hU1A6 (Figure 3).
Figure 3. Variant U1 snRNAs compete inefficiently with the canonical hU1 in binding U1-70K.
Each gel contains (from left to right) 1. unbound radiolabeled hU1 snRNA, 2. radiolabeled hU1 snRNA incubated in the presence of U1-70K, 3. radiolabeled hU1 incubated in the presence of U1-70K and a 50 fold molar excess of unlabeled hU1, 4. radiolabeled hU1 incubated in the presence of U1-70K and a 50 fold molar excess of unlabeled variant snRNA, 5. radiolabeled hU1 incubated in the presence of U1-70K and a 500 fold molar excess of unlabeled variant snRNA, 6. unbound radiolabeled variant snRNA, 7. radiolabeled variant snRNA incubated in the presence of U1-70K, 8. radiolabeled variant incubated in the presence of U1-70K and a 50 fold molar excess of unlabeled variant, 9. radiolabeled variant incubated in the presence of U1-70K and a 50 fold molar excess of unlabeled hU1, and 10. radiolabeled variant incubated in the presence of U1-70K and a 500 fold molar excess of unlabeled hU1.
3.5. HU1 variants differentially associate with U1 snRNP proteins in vitro
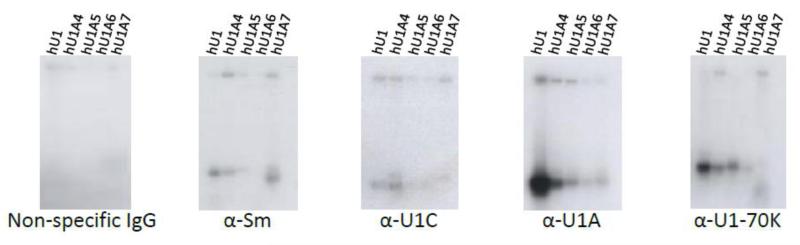
We also used IP with antibodies targeting components of the U1 snRNP, including anti-SmB, anti-U1C, anti-U1A and anti-U1-70K antibodies, to determine the relative binding ability of the variants for these proteins. Non-specific IgG was included as a negative control. Overall, U1A and U1-70K antibodies were most effective at IP of hU1 snRNAs (Figure 4). This is not surprising considering that Sm proteins form a heptameric ring structure during binding to hU1, and U1C does not associate directly with U1 (Sillekens, Beijer et al. 1988; Nelissen, Heinrichs et al. 1991). As expected, hU1 was recovered with the highest efficiency in all pulldowns (Figure 4). HU1A4 was recovered to the greatest extent in the anti-Sm, anti-U1Cand anti-U1A IPs (Figure 4). HU1A4 and hU1A5 were pulled down with similar efficiency by anti-U1-70K. Very low signal was observed in HU1A6 IPs with anti-U1A and anti-U1-70K, but no hU1A6 was detected upon IP with anti-Sm and anti-U1C (Figure 4). Similarly, hU1A7 was observed in anti-Sm and anti-U1A pulldowns (Figure 4), but not in anti-U1C or anti-U1-70K IPs (Figure 4). These results further corroborate our gel shift assays and suggest that the capacity of the hU1 variants to bind snRNP proteins is dramatically reduced in comparison to the canonical hU1 snRNA.
Figure 4. Immunoprecipitation of hU1-like sequences with antibodies to U1 snRNP components.
Antibodies used for each immunoprecipitation (IP) are indicated below each gel. The canonical hU1 snRNA is IPed the most efficiently, followed by hU1A4. The other variants are IPed to varying degrees, but with a much lower capability than hU1 or hU1A4.
4. Discussion
The presence of expressed snRNA sequence variants appears to be a common feature of multiple, divergent taxa (Sierra-Montes, Freund et al. 2002; Sierra-Montes, Pereira-Simon et al. 2003; Chen, Lullo et al. 2005; Sierra-Montes, Pereira-Simon et al. 2005; Hinas, Larsson et al. 2006; Kyriakopoulou, Larsson et al. 2006; Smail, Ayesh et al. 2006); however, information about the potential binding capacity or functional ability of these snRNA-like molecules is limited. Investigations in Drosophila melanogaster indicated that U5 variants may be incorporated into spliceosomes and showed distinct developmental expression profiles (Chen, Lullo et al. 2005). Similar studies in the silk moth, Bombyx mori, indicated that specific U2 variants were differentially associated with spliceosomes (Somarelli, Mesa et al. 2009) and that U1- and U2-like snRNAs exhibited variable spatio-temporal expression (Sierra-Montes, Pereira-Simon et al. 2005). In addition, many of these insect snRNA variants differ from the canonical sequence by just a few nucleotides, with little disruption to the secondary structures (Chen, Lullo et al. 2005; Smail, Ayesh et al. 2006). Taken together, these results suggest that the snRNA variants incorporate into functional spliceosomes. Pereira-Simon et al. (2004) hypothesize that Bombyx mori snRNA variants may act to regulate tissue-specific or developmental splicing. Similarly, Chen et al. (2005) postulate that snRNA variants may function in splicing of development or tissue-specific pre-mRNAs or may be involved in alternative splicing. In the case of human snRNAs, Kyriakopoulou et al. (2006) found that hU1-like sequences could form high molecular weight complexes and were IPed with antibodies against trimethyl guanosine caps and Sm proteins. Overall, these data suggest some functional relevance for snRNA variants. Yet, elegant experiments using hU1A7 and survival of motor neuron (SMN) exon 7 minigenes showed that the canonical hU1, rather than hU1A7, bound SMN 5′ splice sites (Roca and Krainer 2009).Our data support the hypothesis that the hU1 variants analyzed here are unable to efficiently bind U1 snRNP proteins when compared to the canonical hU1 snRNA. Unlike the insect snRNA variants that have very few mutations, the human variants may represent processed pseudogenes that are unable to effectively form snRNPs.
Not surprisingly, hU1A4, which has the highest identity to hU1A, was the most capable of associating with U1 snRNP proteins and forming spliceosomal complexes. Along these lines, a BLASTn search of the National Center for Biotechnology Information EST database using the canonical hU1A as a query revealed the presence of over one hundred U1-like sequences with at least 95% coverage and 95% identity to the canonical hU1A. Given this seemingly widespread variability in expressed U1 sequences, we cannot rule out that hU1A4 and/or any of the U1-like snRNAs with high identity to hU1 may play a role in constitutive and/or alternative splicing. Additional functional assays are needed to address the importance of variant snRNAs in splicing.
Highlights.
Human U1 small nuclear RNA variants differentially form ribonucleoprotein particles
The U1 variants bind the U1A and U1-70K proteins in vitro with limited capacity
The canonical U1 efficiently outcompetes the variants for binding to U1-70K
Acknowledgements
This study was supported by NIH/NIGMS R25 GM061347 (to AM) and NIH S06 GM08205 (to RJH). JAS acknowledges American Cancer Society post-doctoral fellowship PF-11-036-01-DDC. SS acknowledges NIH R21 CA170786.
Abbreviations
- RNA
ribonucleic acid
- snRNA
small nuclear RNA
- snRNP
small nuclear ribonucleoprotein particle
- Sm
Smith antigen
- EST
expressed sequence tag
- BLAST
basic local alignment search tool
- tRNA
transfer RNA
- IP
immunoprecipitation
- IgG
immunoglobulin G
- NCBI
National Center for Biotechnology Information
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5(10):773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- Chen L, Lullo DJ, et al. Identification and analysis of U5 snRNA variants in Drosophila. RNA. 2005;11(10):1473–1477. doi: 10.1261/rna.2141505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YI, Moore RE, et al. Proteomic analysis of in vivo-assembled pre-mRNA splicing complexes expands the catalog of participating factors. Nucleic Acids Res. 2007;35(12):3928–3944. doi: 10.1093/nar/gkm347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinas A, Larsson P, et al. Identification of the major spliceosomal RNAs in Dictyostelium discoideum reveals developmentally regulated U2 variants and polyadenylated snRNAs. Eukaryot Cell. 2006;5(6):924–934. doi: 10.1128/EC.00065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, Schor IE, et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol. 2013;14(3):153–165. doi: 10.1038/nrm3525. [DOI] [PubMed] [Google Scholar]
- Kyriakopoulou C, Larsson P, et al. U1-like snRNAs lacking complementarity to canonical 5′ splice sites. RNA. 2006;12(9):1603–1611. doi: 10.1261/rna.26506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen RL, Heinrichs V, et al. Zinc finger-like structure in U1-specific protein C is essential for specific binding to U1 snRNP. Nucleic Acids Res. 1991;19(3):449–454. doi: 10.1093/nar/19.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25(17):1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca X, Krainer AR. Recognition of atypical 5′ splice sites by shifted base-pairing to U1 snRNA. Nat Struct Mol Biol. 2009;16(2):176–182. doi: 10.1038/nsmb.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A, Carninci P, et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat Rev Genet. 2007;8(6):424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Sierra-Montes JM, Freund AV, et al. Multiple forms of U2 snRNA coexist in the silk moth Bombyx mori. Insect Mol Biol. 2002;11(1):105–114. doi: 10.1046/j.0962-1075.2001.00313.x. [DOI] [PubMed] [Google Scholar]
- Sierra-Montes JM, Pereira-Simon S, et al. A diversity of U1 small nuclear RNAs in the silk moth Bombyx mori. Insect Biochem Mol Biol. 2003;33(1):29–39. doi: 10.1016/s0965-1748(02)00164-9. [DOI] [PubMed] [Google Scholar]
- Sierra-Montes JM, Pereira-Simon S, et al. The silk moth Bombyx mori U1 and U2 snRNA variants are differentially expressed. Gene. 2005;352:127–136. doi: 10.1016/j.gene.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Sillekens PT, Beijer RP, et al. Human U1 snRNP-specific C protein: complete cDNA and protein sequence and identification of a multigene family in mammals. Nucleic Acids Res. 1988;16(17):8307–8321. doi: 10.1093/nar/16.17.8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smail SS, Ayesh K, et al. U6 snRNA variants isolated from the posterior silk gland of the silk moth Bombyx mori. Insect Biochem Mol Biol. 2006;36(6):454–465. doi: 10.1016/j.ibmb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Somarelli JA, Mesa A, et al. U2 snRNA variants are differentially incorporated into spliceosomes. Entomological Research. 2009;39(5):135–145. [Google Scholar]
- Wahl MC, Will CL, et al. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Rumpler S, et al. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 1996;24(23):4614–4623. doi: 10.1093/nar/24.23.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, et al. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419(6903):182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]