Abstract
Background. Detailed knowledge on protein repertoire of a pathogen during host infection is needed for both developing a better understanding of the pathogenesis and defining potential therapeutic targets. Such data, however, have been missing for Staphylococcus aureus, a major human pathogen.
Methods. We determined the surface proteome of methicillin-resistant S. aureus (MRSA) clone USA300 derived directly from murine systemic infection.
Results. The majority of the in vivo-expressed surface-associated proteins were lipoproteins involved in nutrient acquisition, especially uptake of metal ions. Enzyme-linked immunosorbent assay (ELISA) of convalescent human serum samples revealed that proteins that were highly produced during murine experimental infection were also produced during natural human infection. We found that among the 7 highly abundant lipoproteins only MntC, which is the manganese-binding protein of the MntABC system, was essential for MRSA virulence during murine systemic infection. Moreover, we show that MntA and MntB are equally important for MRSA virulence.
Conclusions. Besides providing experimental evidence that MntABC might be a potential therapeutic target for the development of antibiotics, our in vivo proteomics data will serve as a valuable basis for defining potential antigen combinations for multicomponent vaccines.
Keywords: Staphylococcus aureus, MRSA, proteomics, bacterial pathogenesis, vaccine and antibiotic development
Staphylococcus aureus is a leading cause of hospital- and community-acquired infections [1]. It is a versatile pathogen capable of causing a wide spectrum of diseases ranging from minor skin infections to life-threatening diseases, such as endocarditis, osteomyelitis, and sepsis [2]. This versatility is enabled by a large arsenal of virulence factors encoded on the genome [3]. Surface-associated proteins are important virulence factors as they are involved in multiple processes of bacterial pathogenesis, such as mediating adhesion to host cells or sequestering nutrients in nutrient-limiting host environments [4]. Moreover, because of their extracellular localization, surface-associated proteins are attractive targets for both small-molecule antibiotics and large-molecule antibody-based therapeutics. The major classes of surface-associated proteins of S. aureus are sortase-anchored proteins [5, 6] and lipoproteins [7, 8]. To date, the contribution of sortase-anchored proteins to the S. aureus pathogenesis has been extensively studied [9, 10]. In contrast, except for their role as ligands for the mammalian Toll-like receptor 2 [11], the significance of individual lipoproteins for infection has not been well characterized.
Several transcriptional analyses have provided insights into how S. aureus regulates the expression of virulence factors under different environmental conditions [12, 13]. However, these data cannot be extrapolated to a complete understanding of the proteome during host infection because protein production can be regulated post-translationally. Detailed knowledge on the protein repertoire of a pathogen during infection is critical not only for elucidating the pathogenesis but also for defining therapeutic targets. Such data, however, are available only for a few human pathogens and comprehensive proteomics study of S. aureus during host infection has not been performed to date [14, 15]. A major challenge in studying the proteome of S. aureus within an intact host has been the recovery of a sufficient number of bacterial cells from infected organs [16, 17]. To circumvent this, alternative approaches have been employed. They include analysing the antibody repertoire of infected subjects to identify antigens produced by S. aureus during host infection [18, 19], or conducting proteomic analysis under in vitro conditions that were thought to mimic the in vivo environment such as low iron concentration [20]. Each approach, however, has limitations. The first approach is only able to detect a subset of proteins that are highly immunogenic during infection, whereas the second approach does not recapitulate the complexity of an in vivo environment.
Here we report the first characterization of the S. aureus proteome derived directly from infection of an intact animal. We characterized the surface proteome of the methicillin-resistant s. aureus (MRSA) USA300 strain, the most prevalent MRSA strain causing community-acquired infections in the United States [21, 22], using a murine systemic infection model. Our data contribute to a better understanding of the pathogenesis of MRSA and help in defining potential therapeutic targets.
MATERIALS AND METHODS
Ethics Statement
All procedures involving mice were compliant with the ILAR guidelines and were approved by the IACUC at Genentech, Inc.
Proteomic Analysis of MRSA in Vitro and in Vivo
The USA300_0114 (NRS384) strain (obtained from NARSA) was grown in 4 mL of tryptic soy broth (TSB) media (Sigma-Aldrich) overnight, then diluted 1:50 into 50 mL of TSB media in a 250 mL nonbaffled Erlenmeyer flask. Bacteria were grown at 37°C with shaking (170–230 rpm) and harvested at an OD600 of 0.4. Cells were washed and resuspended in sterile phosphate-buffered saline (PBS) to a concentration of 5 × 108 colony-forming units (CFU)/mL. In total, 5 × 107 CFU of bacteria were inoculated into 12 week-old female C57BL/6 mice (The Jackson Laboratory) via tail-vein injection. Six days postinfection, mice were killed and kidneys were removed and homogenized in ice-cold PBS containing 0.05% Triton-X. After differential centrifugation at 4°C, pellets containing bacterial cells were washed and resuspended in ice-cold PBS containing 0.5% BSA and 200 µg/mL of human immunoglobulin G (IgG; Jackson ImmunoResearch). In total, 100 µg/mL of biotinylated anti-S. aureus antibody (Abcam) was added to the reaction mixture and incubated on ice for 10 minutes. After washing, streptavidin microbeads (Miltenyl Biotech) were added (75 µL/mL reaction mixture) and incubated on ice for 20 minutes. Bacteria were separated from kidney debris using an autoMACS Pro Separator (Miltenyl Biotech) using ice-cold buffer and cooling tube racks in order to maintain sample integrity. For in vitro proteomics study, bacteria were grown either in nutrient-restricted Roswell Park Memorial Institute (RPMI) media (RPMI 1640, Sigma-Aldrich) or nutrient-rich TSB media to an OD600 of 0.4 or 4.0, washed and resuspended in PBS. Samples enriched for S. aureus surface-associated proteins were prepared by incubating bacteria in PBS containing 30% sucrose and 10 µg/mL lysostaphin for 30 minutes at 37°C. After centrifugation, supernatant was collected and proteins were concentrated by methanol/chloroform precipitation. Details on sample processing, LC-MS/MS, database search, and protein identification are provided in the Supplementary Materials and Methods.
Human Serum ELISA Reactivity Assay
Human serum samples from patients recovering from S. aureus infections were obtained from Dr Henry F. Chambers from the University of California San Francisco. Recombinant proteins were produced and purified from Escherichia coli (Genentech). All lipoproteins were produced as nonlipidated form in the cytoplasm as the endogenous signal sequence including the conserved cysteine residue was removed from the protein expression constructs. Purified proteins were diluted to 2 μg/mL in PBS and dispensed into 96-well Nunc Maxisorp plates (Nunc, Rochester, NY). The plates were coated overnight at 4°C, rinsed in PBS, and then blocked for 1-hour at room temperature with PBS containing 1% bovine serum albumin (BSA, Sigma-Aldrich). Next, each plate received 100 µL of serially diluted human serum. The initial dilution was 1/100 followed by 3-fold serial dilutions. All dilutions were performed in PBS containing 1.0% BSA and 0.05% Polysorbate 20 (Sigma-Aldrich). After a 1-hour incubation the plates were washed and then incubated with 100 μL of a 1:5000 dilution of horseradish peroxidase-conjugated Fc specific anti-human IgG (Jackson ImmunoResearch Laboratories) for 30 minutes at room temperature. Following the incubation the plates were washed and developed with TMB substrate (Kirkegaard and Perry Laboratories, Inc). Plates were read on a SpectraMax plate reader (Molecular Devices, Sunnyvale, CA) at OD450. Serum titers were determined by calculating the sample dilution at OD450 = 1, which is within the linear range of full serum dilution curves.
Construction of Bacterial Mutants
In-frame gene deletion, gene inactivation, and gene repair were performed on SF8300, a clinical strain representative of the epidemic clone USA300-0114 [23], using oligonucleotides listed in Supplementary Table 1 and the pKOR1 system as described elsewhere [24, 25]. Summary of constructed strains is provided in Table 1.
Table 1.
List of Bacterial Strains Described in This Study
| Strain | Relevant Characters | Source/ References |
|---|---|---|
| SF8300 wild-type | Clinical strain of community-associated MRSA genotype USA300-0114 | [21] |
| SF8300 ΔprsA | In-frame deletion of SAUSA300_1790 gene | This study |
| SF8300 Δ2136 | In-frame deletion of SAUSA300_2136 gene | This study |
| SF8300 Δopp-1A | In-frame deletion of SAUSA300_2411 gene | This study |
| SF8300 ΔsaeS | In-frame deletion of SAUSA300_0693 gene | This study |
| SF8300 Δ0798 | In-frame deletion of SAUSA300_0798 gene | This study |
| SF8300 ΔsirA | In-frame deletion of SAUSA300_0117 gene | This study |
| SF8300 ΔmntC | In-frame deletion of SAUSA300_0618 gene | This study |
| SF8300 mntCstop | SAUSA300_0618 gene: 8th codon TTA changed to TAA and 12th codon from TTA changed to TAA | This study |
| SF8300 mntBstop | SAUSA300_0619 gene: 34th codon TTA changed to TAA and 37th codon TTA changed to TAA | This study |
| SF8300 mntAstop | SAUSA300_0620 gene: 43rd codon CCT changed to TAA | This study |
| SF8300 mntCstop_repaired | SF8300 mntCstop mutations repaired by allelic replacement mutagenesis with mntC wild-type allele containing silent mutations at 143th codon from GAT to GAC and 145th codon from GAC to GAT | This study |
Virulence Analysis of Bacterial Mutants
Bacterial mutants were grown in TSB media to an OD600 of 0.4, washed, and resuspended in sterile PBS. 2 × 106 CFU of bacteria were inoculated into 7-week-old, female A/J mice (The Jackson Laboratory) via tail-vein injection. In contrast to major laboratory mouse strains such as C57BL/6 and BALB/c, A/J mice have a functional Nramp1 locus [26]. Seven days postinfection, mice were killed, and kidneys were removed and homogenized in PBS to enumerate bacterial load. For lethal challenge, 5 × 108 CFU of bacteria were inoculated into 7-week-old, female A/J mice (The Jackson Laboratory) via tail-vein injection, and animals were monitored for 14 days to determine the survival rate.
RESULTS AND DISCUSSION
Proteomic Analysis of MRSA Derived Directly From a Murine Systemic Infection
We have established a protocol to rapidly and efficiently define surface proteome of S. aureus recovered directly from infected tissues. First, we chose an experimental animal infection model that results in high organ bacterial load. The murine systemic infection model is well established for studying S. aureus pathogenesis where robust bacterial proliferation occurs in kidneys [27]. In the first 48 hours of infection a steady increase in kidney bacterial load occurs before reaching a maximum of about 108 CFU/pair of kidneys, which is maintained for at least 1 week postinfection (data not shown). We harvested kidneys 6 days postinfection in order to study the surface proteome of MRSA in an established infection. Second, we employed immunomagnetic cell sorting using S. aureus-specific antibodies in order to achieve sufficient enrichment of MRSA cells over a vast excess of kidney debris. On average, we recovered 5 × 107 CFU of bacteria from kidneys of 5 mice (data not shown). Samples enriched in surface-associated proteins were prepared by treating whole bacteria with lysostaphin [28]. Mass spectrometry analysis identified 342 S. aureus proteins, among which 57 were predicted to contain signal sequences (Supplementary Table 2). Presence of high number of cytoplasmic proteins in cell wall preparation has also been observed in several other proteomics studies on cell surface proteins of S. aureus that were recovered from liquid cultures in vitro [20, 29, 30]. Although potential mechanisms for translocating proteins without signal sequence to the cell surface might exist, the presence of cytoplasmic proteins on the cell surface might be a simple consequence of autolysis or cell lysis during sample preparation.
MRSA Surface Proteome During Infection Is Dominated by Proteins Involved in Nutrient Uptake
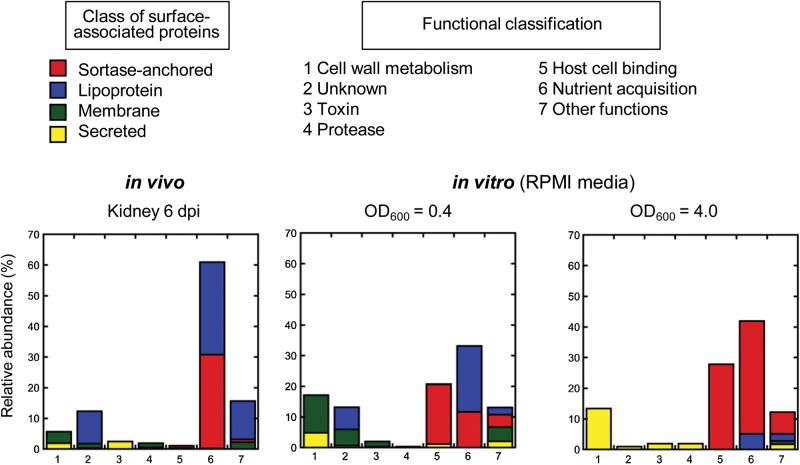
Among the proteins with predicted signal sequence, lipoproteins were the most abundant class of proteins followed by sortase-anchored proteins (45.5% and 32.3% of total number of identified peptides, respectively; Figure 1, Supplementary Table 2). The remaining surface proteome was composed of secreted proteins, many of which are cytolytic toxins like hemolysin or leukotoxin, and integral membrane proteins such as MecA that confers resistance to β-lactam antibiotics in MRSA strains (Figure 1, Supplementary Table 2).
Figure 1.
Surface proteome of MRSA USA300 derived from in vivo infection and nutrient-limiting liquid culture in vitro. Relative abundance of surface-associated proteins is shown based on percentile of number of peptides detected for each class of protein relative to the total number of detected peptides. Proteins are grouped on predicted cellular location (red, sortase-anchored protein; blue, lipoprotein; green, integral membrane protein; yellow, secreted protein) and function (1, cell wall metabolism; 2, unknown function; 3, toxin; 4, protease; 5, host cell binding; 6, nutrient acquisition; 7, other functions). Abbreviations: dpi, days postinfection; MRSA, methicillin-resistant Staphylococcus aureus; RPMI, Roswell Park Memorial Institute media.
One of the key factors that distinguishes host environments from the standard laboratory growth conditions is nutrient limitation. For example, the concentration of free iron in tissue fluids and blood is in the range of 10−18 M, whereas bacteria are reported to require iron at 0.4 to 4.0 mM for their survival [31, 32]. Previous transcriptome analysis of MRSA USA300 revealed that genes encoding proteins associated in iron transport are highly upregulated during incubation in human blood and serum [13]. When we grouped the 57 S. aureus surface-associated proteins that were produced in murine kidneys 6 days postinfection based on their functions, we found that the surface proteome of MRSA USA300 during host infection is indeed dominated by proteins involved in nutrient acquisition (Figure 1). Specifically, 8 of the 10 most highly abundant surface-associated proteins, based on the number of total peptides identified [33, 34], were annotated to be involved in nutrient uptake: Five are annotated to be involved in iron uptake (IsdA [35], IsdB [35], IsdC [35], SAUSA300_2136, SirA [36]), one for manganese/zinc (MntC) [37], and one for oligopeptides (Opp-1A) [38] (Figure 1, Supplementary Table 1). SAUSA300_0798 is annotated to be substrate-binding protein of ATP-binding cassette (ABC) transporter and therefore likely to be involved in nutrient acquisition as well; however, its substrate specificity is unknown. In addition to the aforementioned 8 proteins, we identified five lipoproteins (SAUSA300_2235, SAUSA300_1978, ModA, SAUSA300_2351, SAUSA300_0721) that were annotated to be involved in nutrient acquisition (Supplementary Table 2). Previous quantitative proteomic study revealed that 8 proteins were highly induced by the MRSA COL strain under iron-limited conditions in vitro [20]. Notably, 7 of these proteins (IsdA, IsdB, IsdC, SirA, SACOL2277 corresponding to SAUSA300_2235, SACOL2167 corresponding to SAUSA300_2136, SACOL0799 corresponding to SAUSA300_0721), were identified in our proteomics study (Supplementary Table 2).
To analyze the degree of overlap between growth in mouse kidneys and growth in vitro under nutrient limitations, we also determined the surface proteome of the MRSA USA300 strain harvested after exponential and stationary growth in RPMI, a nutrient-limiting growth media [39]. From a total of 686 proteins that were identified from bacteria harvested after stationary growth, 133 were predicted to have signal sequence (Supplementary Table 3). Twenty-five proteins are annotated to be involved in nutrient uptake, which include all the 13 surface-associated proteins that were produced in vivo (Supplementary Table 2 and Supplementary Table 3). However, a stark contrast to in vivo was the high abundance of sortase-anchored proteins involved in host cell adhesion, especially Protein A (Spa; immunoglobulin G binding protein A; Figure 1, Supplementary Table 3). Based on the number of identified peptides, proteins involved in host cell adhesion made up 20%–30% of the surface proteome of S. aureus grown in nutrient-limiting media in vitro, whereas they made up only about 1% of the surface proteome of bacteria recovered from infected kidneys (Figure 1). By contrast, no peptide fragments of Protein A were detected in vivo (Supplementary Table 2). Although we cannot completely rule out the presence of Protein A in bacteria derived in vivo, as the amount of produced protein could be under the detection limit of our study, our results clearly suggest that production of Protein A can be significantly regulated during host infection.
Proteins Identified Using the Murine Infection Model Are Likely to Be Produced Under Different Host Environments
To verify whether the surface-associated proteins of S. aureus that were produced in murine kidneys are produced under different host environments, we determined the surface proteome of the USA300 MRSA strain derived from liver, where the bacterial load reached a CFU of about 108 at day 6 postinfection, which was comparable to that of kidneys (data not shown). We were unable to determine the surface proteome of bacteria present in the other organs and blood because the number of bacteria recovered were several orders of magnitude smaller than that recovered from the kidney or liver (data not shown). Among the 254 proteins identified from liver-derived bacteria, 24 were predicted to contain signal sequence (Table 2). All these 24 surface-associated proteins were identified in kidney-derived bacteria as well, indicating that the repertoire of highly abundant S. aureus surface-associated proteins produced in the host is strikingly similar between different organs.
Table 2.
List of MRSA USA300 Proteins With Predicted Signal Sequence Identified in Murine Liver 6 Days Postinfection
| Accession No. | Name | Annotation | MW (kDa) | Peptidesa | Surface Display |
|---|---|---|---|---|---|
| SAUSA300_1029 | isdA | Iron transport associated domain-containing protein | 38.75 | 49 | Sortase-anchored |
| SAUSA300_0618 | mntC | ABC transporter substrate-binding protein | 34.74 | 38 | Lipoprotein |
| SAUSA300_2411 | opp-1A | Oligopeptide permease peptide-binding protein | 60.02 | 38 | Lipoprotein |
| SAUSA300_1028 | isdB | Iron transport associated domain-containing protein | 72.19 | 33 | Sortase-anchored |
| SAUSA300_0693 | saeP | Hypothetical protein | 16.05 | 19 | Lipoprotein |
| SAUSA300_2136 | … | Iron compound ABC transporter iron compound-binding protein | 36.59 | 16 | Lipoprotein |
| SAUSA300_0117 | sirA | Iron compound ABC transporter iron compound-binding protein | 36.74 | 10 | Lipoprotein |
| SAUSA300_2403 | … | Putative lipoprotein | 17.31 | 8 | Lipoprotein |
| SAUSA300_2328 | … | Hypothetical protein | 13.34 | 7 | Lipoprotein |
| SAUSA300_0916 | … | Hypothetical protein | 19.33 | 6 | Lipoprotein |
| SAUSA300_0372 | … | Hypothetical protein | 21.31 | 5 | Lipoprotein |
| SAUSA300_2235 | … | Iron compound ABC transporter iron compound-binding protein | 34.01 | 4 | Lipoprotein |
| SAUSA300_1677 | sasI | Cell wall surface anchor family protein | 100.93 | 4 | Sortase-anchored |
| SAUSA300_2230 | modA | Molybdenum ABC transporter molybdenum-binding protein | 29.05 | 3 | Lipoprotein |
| SAUSA300_0769 | … | Hypothetical protein | 28.42 | 3 | Lipoprotein |
| SAUSA300_1790 | prsA | Foldase protein PrsA precursor | 35.64 | 2 | Lipoprotein |
| SAUSA300_0721 | … | Transferrin receptor | 37.85 | 2 | Lipoprotein |
| SAUSA300_1478 | … | Putative lipoprotein | 14.1 | 2 | Lipoprotein |
| SAUSA300_0692 | … | Hypothetical protein | 17.66 | 1 | Membrane |
| SAUSA300_1030 | isdC | Iron transport associated domain-containing protein | 24.85 | 1 | Sortase-anchored |
| SAUSA300_2315 | … | Hypothetical protein | 23.36 | 1 | Lipoprotein |
| SAUSA300_0798 | … | ABC transporter substrate-binding protein | 30.35 | 1 | Lipoprotein |
| SAUSA300_2297 | … | Hypothetical protein | 24.26 | 1 | Lipoprotein |
| SAUSA300_0032 | mecA | Penicillin-binding protein 2′ | 76.1 | 1 | Membrane |
| SAUSA300_0079 | … | Putative lipoprotein | 20.14 | 1 | Lipoprotein |
Abbreviations: ABC, ATP-binding cassette; MRSA, methicillin-resistant Staphylococcus aureus.
a No. of identified peptides. Peptide identifications were accepted if they could be established at >90% probability as specified by the Peptide Prophet algorithm as described in Supplementary Materials and Methods.
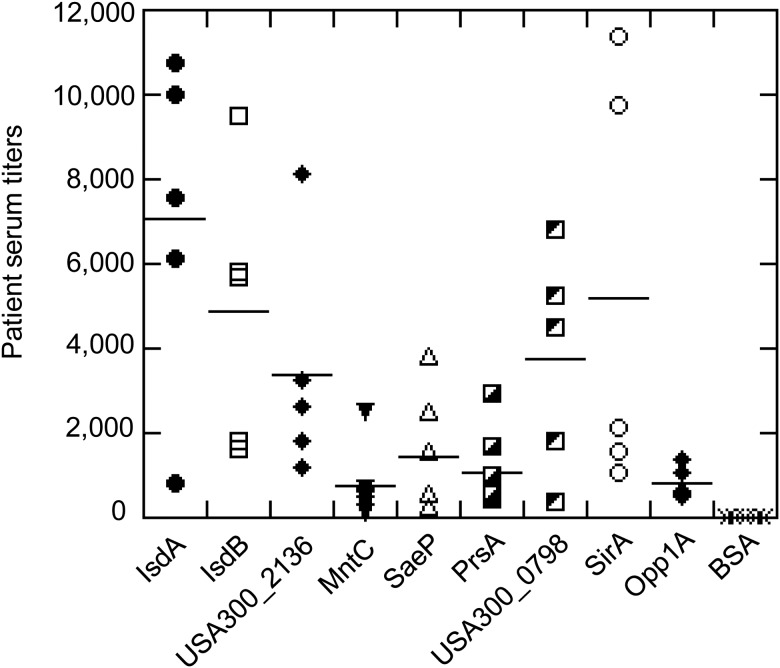
To determine if surface-associated proteins that were highly abundant during experimental murine infection are also expressed during natural human infection, we analyzed serum samples obtained from patients recovering from systemic S. aureus infections against recombinant IsdA, IsdB, and the 7 different lipoproteins we identified by ELISA (Figure 2). IgGs could be detected for all tested proteins indicating that surface proteins produced in the murine systemic infection model are also produced during human systemic infections (Figure 2).
Figure 2.
ELISA analysis of human convalescent sera against surface-associated proteins highly produced during murine systemic infection. Serum samples from individual convalescent patients (n = 5) were tested against 9 recombinantly produced proteins. Proteins are indicated either by their names or by their accession numbers based on USA300_FPR3757 genome. BSA protein was used as a negative control. Medians of patient serum titers at A450 = 1 are shown in horizontal bars. Abbreviations: BSA, bovine serum albumin; ELISA, enzyme-linked immunosorbent assay.
Contribution of Highly Abundant Lipoproteins to MRSA Virulence During Murine Systemic Infection
The contribution of sortase-anchored proteins such as IsdA, IsdB, and IsdC proteins to S. aureus virulence is well established. Inactivation of genes encoding IsdA, IsdB, and IsdC proteins resulted in a substantial reduction of S. aureus load in mouse kidneys after intravenous inoculation [9, 40]. By contrast, although lipoproteins represent a major class of surface-associated proteins produced during infection (Figure 1, Supplementary Table 2), the significance of individual lipoproteins for infection remains to be systematically evaluated.
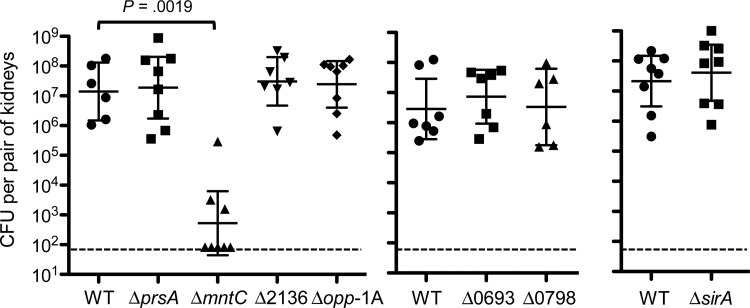
Therefore, we investigated the contribution of 7 lipoproteins that were most highly produced during murine systemic infection to MRSA pathogenesis. MntC, Opp-1A, SAUSA300_2136, SirA, and SAUSA300_0798 are substrate-binding proteins of ABC transporters involved in nutrient acquisition. The function of SaeP is unknown, but its gene is found in the sae locus that encodes the 2-component system SaeRS, which regulates the expression of a wide variety of virulence factors [41]. PrsA with a parvulin-type peptidyl-prolyl cis-trans isomerase domain is an essential protein in B. subtilis and serves as a folding catalyst for secreted proteins [42]. We inactivated each gene by in-frame deletion and assessed the virulence of individual mutants in a murine systemic infection model. Only deletion of the mntC gene resulted in significant attenuation of virulence as evident by more than 4-log decrease in kidney bacterial load at 7 days postinfection (Figure 3). In contrast, deletion of the 6 other lipoprotein genes did not have an effect on MRSA virulence in this infection model (Figure 3).
Figure 3.
Inactivation of the gene encoding the MntC lipoprotein results in a severe attenuation of MRSA USA300 virulence in a murine systemic infection model. Kidney bacterial load of USA300 MRSA lipoprotein deletion mutants (deleted genes are shown in their names or in their locus numbers based on USA300_FPR3757 genome) 7 days postintravenous infection of 2 × 106 CFU bacteria into A/J mice (n = 7). Data show the geometrical mean with 95% confidence intervals. Statistical analyses were performed by unpaired Mann–Whitney t tests. Dashed lines show the detection limit (83 CFU). Abbreviations: CFU, colony-forming units; MRSA, methicillin-resistant Staphylococcus aureus; WT, wild type.
MntABC Is an Important Virulence Factor of MRSA in a Murine Intravenous Infection Model
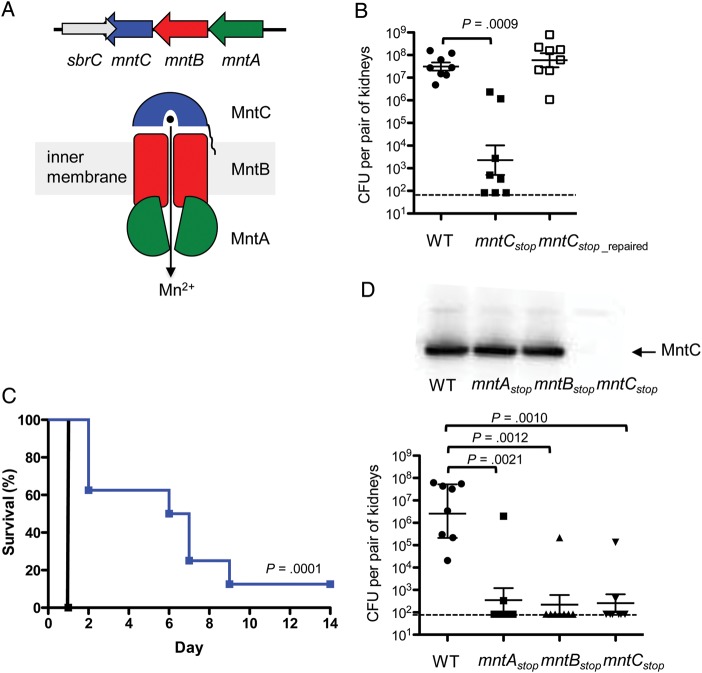
The mntC gene is part of the mntABC operon that encodes an ABC transporter that shares high homology with ABC transporters involved in the uptake of manganese and/or zinc from a variety of other bacterial species [37, 43]. Indeed, a previous study showed that a S. aureus mutant with a disrupted mntABC operon had lowered intracellular manganese concentration [37]. The MntABC complex is composed of an ATP-binding protein (MntA), an integral membrane transporter (MntB), and a metal binding lipoprotein (MntC; Figure 4A). Within the 3′ end of the mntC open reading frame lies the predicted terminator of sbrC, which encodes a small noncoding RNA (Figure 4A) [44]. To verify that the strong attenuation of the ΔmntC strain was a direct consequence mntC gene inactivation and not due to inactivation of sbrC, we constructed a second mntC mutant (hereafter denoted as mntCstop strain), in which the mntC gene was inactivated by changing its 8th and 12th codon to a stop codon thereby leaving the sbrC gene intact (Table 1). Again, the mntCstop strain was severely attenuated similar to the original ΔmntC strain after intravenous inoculation into mice (Figure 4B). The importance of the MntC protein in the virulence of MRSA during murine systemic infection was also evident in the prolonged survival of mice infected with the mntCstop strain, which showed a median survival time of 6.5 days (Figure 4C). In contrast, all mice infected with the wild-type strain succumbed to infection within 24 hours (Figure 4C). To confirm that the strong attenuation of the mntCstop strain was a direct consequence of inactivating the mntC gene and not due to a secondary mutation, we repaired the stop-codon insertion by inserting a wild-type allele of mntC by allelic replacement mutagenesis (Table 1). As expected, the repair of stop-codon insertion resulted in full restoration of virulence (Figure 4B).
Figure 4.
MntABC system is an essential virulence factor of MRSA during murine systemic infection. A, Operon organization of mntABC (top); schematic of the MntABC transport system (bottom). B, Kidney bacterial load of USA300 MRSA mntCstop mutant and the mntCstop_repaired strain 7 days post intravenous infection of 2 × 106 CFU bacteria into A/J mice (n = 7). Data show the geometrical mean with 95% confidence intervals. Statistical analyses were performed by unpaired Mann–Whitney t tests. Dashed lines show the detection limit (83 CFU). C, Survival of A/J mice after intravenous infection of 5 × 108 CFU USA300 MRSA wild-type (black line) and mntCstop mutant (blue line) strains (n = 8). Statistical analysis was performed by the Mantel-Cox test. D, Kidney bacterial load of USA300 MRSA stop-codon insertion mutants in the mntABC operon 7 days post intravenous infection of 2 × 106 CFU bacteria into A/J mice (n = 7). Data show the geometrical mean with 95% confidence intervals. Statistical analyses were performed by unpaired Mann–Whitney t tests. Dashed lines show the detection limit (83 CFU). Western blot analysis with anti-MntC antibodies shows that inactivation of mntA and mntB does not affect the expression of mntC (top panel). Abbreviations: CFU, colony-forming units; MRSA, methicillin-resistant Staphylococcus aureus; WT, wild type.
To confirm that the strong attenuation of the mntCstop strain was indeed caused by the lack of a functional MntABC system, we inactivated mntA and mntB genes separately by in-frame stop codon insertion near the beginning of each gene (Table 1). Both mntAstop and mntBstop strains showed strong attenuation similar to the mntCstop strain after intravenous inoculation in mice as evidenced by nearly complete clearance of these mutants in the kidney (Figure 4D). Western-blot analysis using polyclonal antibodies against MntC confirmed that the MntC protein is produced at wild-type level in these 2 bacterial mutants (Figure 4D). These results indicate that the MntABC system is a critical virulence factor of MRSA during murine systemic infection.
CONCLUDING REMARKS
Obtaining sufficient amount of nutrients is critical for the survival and replication of pathogens within the host, which restricts the availability of essential elements by a process termed nutritional immunity [45]. S. aureus counteracts host nutritional immunity by expressing multiple high affinity transport systems. Our proteomics analysis revealed that the major surface proteome of MRSA USA300 during host infection is indeed composed of proteins that are involved in nutrient acquisition. For instance, 5 of at least 10 distinct iron uptake systems encoded on the MRSA USA300 genome were produced during murine systemic infection based on the presence of lipoproteins that comprise part of ABC-type transport systems. Thus, the lack of attenuation of individual lipoprotein mutants with a single iron acquisition system inactivated could be due to functional redundancy of the transport systems. Functional redundancy among iron transport system makes targeting iron acquisition a challenging antimicrobial strategy because multiple transport systems will have to be inhibited simultaneously to achieve significant reduction in S. aureus growth in vivo.
In contrast to iron acquisition, manganese acquisition of S. aureus during murine intravenous infection seems to be dominated by a single pathway, MntABC, because its inactivation resulted in severe defects of S. aureus in establishing infection and killing infected mice. S. aureus encodes a second manganese uptake system, called MntH, which belongs to the Nramp family of membrane metal transporters [46]. Previous works demonstrated that S. aureus strains lacking both MntABC and MntH were attenuated in a murine skin abscess model [37] and in a murine systemic infection model [47]. Contribution of individual manganese uptake systems to S. aureus virulence has remained elusive, because mutant strains lacking MntABC or MntH individually were not attenuated in a murine skin abscess model [37]. One potential reason for the lack of attenuation of these individual mutants in the previous work is that the experiment was performed in a mouse strain with a defective Nramp1 locus, which was shown to be important to study manganese-dependent processes of pathogens in the host [48, 49].
MntC has been defined as a potential vaccine target based on the observation that the protein is stably produced throughout infection in mice according to an immunofluorescence microscopy of infected tissues [50]. It has also been demonstrated that immunization with MntC effectively reduced the bacterial load associated with S. aureus infection in an acute murine bacteremia model [50]. Besides MntC being a vaccine target, MntABC might be a potential target for antibiotic development due to the strong attenuation of MRSA USA300 with inactivated MntABC in a murine systemic infection model as demonstrated here. Further studies are warranted to better understand the role MntABC plays in MRSA pathogenesis to determine whether MntABC is a viable target for the treatment of MRSA infections.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. All work except constructing bacterial mutants was supported by internal funds at Genentech, Inc. Construction of bacterial mutants was supported by a National Institutes of Health grant (NIH/NCRR UCSF-CTSI grant UL1 RR024131) and NIH R01 AI087674 to B. A. D. The authors declare that the article's content is solely their responsibility and do not necessarily represent the official views of NIH.
Potential conflicts of interest. Q. P., S. D., D. A., C. B., M. X., G. N., D. L. S., M. K. A., M. W. T., E. J. B., and M. N. are employees of Genentech, Inc. and as such may be shareholders of company stock. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Klein E, Smith DL, Laxminarayan R. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999–2005. Emerg Infect Dis. 2007;13:1840–6. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–32. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Feng Y, Chen CJ, Su LH, et al. Evolution and pathogenesis of Staphylococcus aureus: lessons learned from genotyping and comparative genomics. FEMS Microbiol Rev. 2008;32:23–37. doi: 10.1111/j.1574-6976.2007.00086.x. [DOI] [PubMed] [Google Scholar]
- 4.Smeltzer MS, Lee CY, Harik N, Hart ME. Molecular basis of pathogenicity. In: Staphylococci in Human Diseases. In: Crossley KB, Jefferson GL, Archer GL, Fowler VG, editors. Oxford: Willey-Blackwell; 2009. pp. 65–108. [Google Scholar]
- 5.Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–3. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 6.Mazmanian SK, Ton-That H, Su K, Schneewind O. An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc Natl Acad Sci U S A. 2002;99:2293–8. doi: 10.1073/pnas.032523999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sutcliffe IC, Harrington DJ. Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology. 2002;148:2065–77. doi: 10.1099/00221287-148-7-2065. [DOI] [PubMed] [Google Scholar]
- 8.Sutcliffe IC, Russell RRB. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–8. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng AG, Kim HK, Burts ML, et al. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corrigan RM, Mialovic H, Foster TJ. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbiol. 2009;9:22. doi: 10.1186/1471-2180-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto M, Tawaratsumida K, Kariya H, et al. Lipoprotein is a predominant Toll-like receptor 2 ligand in Staphylococcus aureus cell wall components. Int Immunol. 2005;18:355–62. doi: 10.1093/intimm/dxh374. [DOI] [PubMed] [Google Scholar]
- 12.Garzoni C, Francois P, Huyghe A, et al. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics. 2007;8:171. doi: 10.1186/1471-2164-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malachowa N, Whitney AR, Kobayashi SD, et al. Global changes in Staphylococcus aureus gene expression in human blood. PloS One. 2011;6:e18617. doi: 10.1371/journal.pone.0018617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker D, Selbach M, Rollenhagen C, et al. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature. 2006;440:303–7. doi: 10.1038/nature04616. [DOI] [PubMed] [Google Scholar]
- 15.Kruh NA, Troudt J, Izzo A, Prenni J, Dobos KM. Portrait of a pathogen: the Mycobacterium tuberculosis proteome in vivo. PloS One. 2010;5:e13938. doi: 10.1371/journal.pone.0013938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Windle HJ, Brown PA, Kelleher DP. Proteomics of bacterial pathogenicity: Therapeutic implications. Proteomics Clin Appl. 2010;4:215–27. doi: 10.1002/prca.200900145. [DOI] [PubMed] [Google Scholar]
- 17.Bumann D. Pathogen proteomes during infection: A basis for infection research and novel control strategies. J Proteomics. 2010;73:2267–76. doi: 10.1016/j.jprot.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Etz H, Minh DB, Henics T, et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc Natl Acad Sci U S A. 2002;99:6573–8. doi: 10.1073/pnas.092569199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brady AB, Leid JG, Camper AK, Costerton JW, Shirtliff ME. Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect Immun. 2006;74:3415–26. doi: 10.1128/IAI.00392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hempel K, Herbst FA, Moche M, Hecker M, Becher D. Quantitative proteomic view on secreted, cell surface-associated, and cytoplasmic proteins of the methicillin-resistant human pathogen Staphylococcus aureus under iron-limited conditions. J Proteome Res. 2010;10:1657–66. doi: 10.1021/pr1009838. [DOI] [PubMed] [Google Scholar]
- 21.Diep BA, Gill SR, Chang RF, et al. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–9. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- 22.O'Hara FP, Amrine-Madsen H, Mera RM, et al. Molecular characterization of Staphylococcus aureus in the United States 2004–2008 reveals the rapid expansion of USA300 among inpatients and outpatients. Microbiol Drug Resistance. 2012;18:555–61. doi: 10.1089/mdr.2012.0056. [DOI] [PubMed] [Google Scholar]
- 23.Diep BA, Palazzolo-Balance AM, Tattevin P, et al. Contribution of Panton-Valentine leukocidin in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. PloS One. 2006;3:e3198. doi: 10.1371/journal.pone.0003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diep BA, Chan L, Tattevin P, et al. Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton-Valentine leukocidin-induced lung inflammation and injury. Proc Natl Acad Sci U S A. 2010;107:5587–92. doi: 10.1073/pnas.0912403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Malo D, Vogan K, Vidal S, et al. Halotype mapping and sequence analysis of the mouse Nramp gene predict susceptibility to infection with intracellular parasites. Genomics. 1994;23:51–61. doi: 10.1006/geno.1994.1458. [DOI] [PubMed] [Google Scholar]
- 27.Gorrill RH, Klyhn KM, McNell EM. The initiation of infection in the mouse kidney after intravenous injection of bacteria. J Pathol Bacteriol. 1961;91:157–72. doi: 10.1002/path.1700910120. [DOI] [PubMed] [Google Scholar]
- 28.Schindler CA, Schulhardt VT. Lysostaphin: a new bacteriolytic agent for the Staphylococcus. Proc Natl Acad Sci U S A. 1964;51:414–21. doi: 10.1073/pnas.51.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gatlin CL, Pieper R, Huang ST, et al. Proteomic profiling of cell envelope-associated proteins from Staphylococcus aureus. Proteomics. 2006;6:1530–49. doi: 10.1002/pmic.200500253. [DOI] [PubMed] [Google Scholar]
- 30.Ventura CL, Malachowa N, Hammer CH, et al. Identification of novel Staphylococcus aureus two-component leukotoxin using cell surface proteomics. PloS One. 2010;5:e11364. doi: 10.1371/journal.pone.0011634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 2004;6:390–7. doi: 10.1016/j.micinf.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–37. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 33.Bondarenko PV, Chelius D, Shaler TA. Identification and relative quantitation of protein mixtures by enzymatic digestion followed by capillary reversed-phase liquid chromatography-tandem mass spectrometry. Anal Chem. 2002;74:4741–9. doi: 10.1021/ac0256991. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 35.Mazmanian SK, Skaar EP, Gaspar AH, et al. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–9. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 36.Heinrichs JH, Gatlin LE, Kunsch C, Choi GH, Hanson MS. Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J Bacteriol. 1999;181:1436–43. doi: 10.1128/jb.181.5.1436-1443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horsburgh MJ, Wharton SJ, Cox AG, et al. MntR modulates expression of the PerR regulon and superoxide resistance in Staphulococcus aureus through control of manganese uptake. Mol Microbiol. 2002;44:1269–86. doi: 10.1046/j.1365-2958.2002.02944.x. [DOI] [PubMed] [Google Scholar]
- 38.Coulter SN, Schwan WR, Ng EY, et al. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 39.Stoll H, Dengjel J, Nerz C, Götz F. Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect Immun. 2005;73:2411–23. doi: 10.1128/IAI.73.4.2411-2423.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HK, DeDent A, Cheng AG, et al. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine. 2010;28:6382–92. doi: 10.1016/j.vaccine.2010.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Adhikari RP, Novick RP. Regulatory organization of staphylococcal sae locus. Microbiology. 2007;154:949–59. doi: 10.1099/mic.0.2007/012245-0. [DOI] [PubMed] [Google Scholar]
- 42.Vitikainen M, Lappalainen I, Seppala R, et al. Structure-function analysis of PrsA reveals roles for the parvulin-like and flanking N- and C-terminal domains in protein folding and secretion in Bacillus subtilis. J Biol Chem. 2004;279:19302–14. doi: 10.1074/jbc.M400861200. [DOI] [PubMed] [Google Scholar]
- 43.Claverys JP. A new family of high-affinity ABC manganese and zinc permeases. Res Microbiol. 2001;152:231–43. doi: 10.1016/s0923-2508(01)01195-0. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen JS, Christiansen MH, Bonde M, et al. Searching for small σB-regulated genes in Staphylococcus aureus. Arch Microbiol. 2011;193:23–34. doi: 10.1007/s00203-010-0641-1. [DOI] [PubMed] [Google Scholar]
- 45.Cassat JE, Skaar EP. Metal ion acquisition in Staphylococcus aureus: overcoming nutritional immunity. Semin Immunopathol. 2012;34:215–35. doi: 10.1007/s00281-011-0294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Forbes JR, Gros P. Divalent-metal transport by Nramp proteins at the interface of host-pathogen interactions. Trends Microbiol. 2001;9:397–403. doi: 10.1016/s0966-842x(01)02098-4. [DOI] [PubMed] [Google Scholar]
- 47.Kehl-Fie TE, Zhang Y, Moore JL, et al. MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect Immun. 2013;81:3395–405. doi: 10.1128/IAI.00420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaharik ML, Cullen VL, Fung AM, et al. The Salmonella enterica serovar typhimurium divalent cation transport systems MntH and Sit ABCD are essential for virulence in an Nramp1G169 murine typhoid model. Infect Immun. 2004;72:5522–5. doi: 10.1128/IAI.72.9.5522-5525.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papp-Wallace KM, Maguire ME. Manganese transport and the role of manganese in virulence. Annu Rev Microbiol. 2006;60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- 50.Anderson AS, Scully IL, Timofeyeva Y, et al. Staphylococcus aureus manganese transport protein C is a highly conserved cell surface protein that elicits protective immunity against S. aureus and Staphylococcus epidermidis. J Infect Dis. 2012;205:1688–96. doi: 10.1093/infdis/jis272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.