Abstract
Invasive group A streptococcal (GAS) strains often have genetic differences compared to GAS strains from nonsterile sites. Invasive, “hypervirulent” GAS strains can arise from a noninvasive progenitor following subcutaneous inoculation in mice, but such emergence has been rarely characterized in humans. We used whole genome analyses of multiple GAS isolates from the same patient to document the molecular basis for emergence of a GAS strain with an invasive phenotype during human infection. In contrast to previous theories, we found that elimination of production of the cysteine protease SpeB was not necessary for emergence of GAS with an invasive, “hypervirulent” phenotype.
Keywords: group A Streptococcus, bacterial pathogenesis, virulence, whole genome sequencing, gene expression
Group A Streptococcus (GAS) is a major human pathogen that causes a wide variety of diseases ranging from benign colonization, to mild cases of pharyngitis or cellulitis, to highly lethal syndromes such as necrotizing fasciitis and the streptococcal toxic shock syndrome. Over the past decade it has been recognized that invasive GAS strains often have subtle genetic differences compared to GAS strains isolated from nonsterile sites such as the throat and skin [1, 2]. For example, analysis of serotype M3 GAS strains found that 37% of all invasive strains, but only 4.6% of pharyngitis strains, contained polymorphisms in genes encoding the 2-component gene regulatory system composed of the control of virulence regulator (CovR) and sensor kinase (CovS; also known as CsrRS) [1]. Similarly, in a study of a broad array of GAS serotypes, 46% of GAS isolates causing streptococcal toxic shock syndrome had covRS mutations compared to 2% of noninvasive GAS strains [2]. The CovRS system mainly serves to repress GAS virulence factor production such that GAS strains with CovRS mutations are more lethal in mice following intraperitoneal infection and proliferate to a higher degree in human blood compared to GAS strains with an intact CovRS system [3, 4]. Thus, GAS strains with CovRS mutations are considered hypervirulent and are termed to have an invasive phenotype [3, 5–7].
Studies in mice have shown that GAS strains with an invasive phenotype can emerge from CovRS wild-type strains following subcutaneous inoculation, mainly through complete inactivation of the CovS protein [3, 7]. Mice, however, are not naturally infected by GAS, and numerous GAS virulence factors, such as streptokinase, which binds to and activates plasminogen in humans, are not active in mice [8]. To date, there has only been a single documentation of a GAS strain with an invasive phenotype arising from its CovRS wild-type progenitor during human infection, and this occurred in the setting of a pharyngeal rather than a skin/soft-tissue infection [9]. Thus, it is not known whether the subcutaneous mouse model of invasive GAS infection accurately reproduces what occurs in humans infected with GAS. We took advantage of the availability of paired noninvasive and invasive GAS isolates in a patient with a skin/soft tissue infection to investigate the molecular mechanisms underlying the emergence of the hypervirulent, invasive GAS phenotype in the human host.
CASE REPORT
A 66 year-old man with colorectal carcinoma and chronic renal insufficiency presented to the MD Anderson Cancer Center with a 3-day history of ulceration and erythema in his right lower extremity. Upon admission, he was febrile to 38.9°C, blood pressure was 95/52, heart rate was 105/min, and respiratory rate was 28/min. He was noted to have 2 ulcers with frank purulence in the right lower extremity. White blood cell count was 17 000/µL with 93% neutrophils. Creatinine was 4.04 mg/dL compared with a previous baseline of 2.02 mg/dL. Cultures were taken from each ulcer and from the blood. He was diagnosed with severe sepsis, admitted to the intensive care unit, started on broad-spectrum antibiotics, and given fluid resuscitation. All cultures were positive for group A Streptococcus (GAS), and the patient was treated with 14 days of ceftriaxone. No surgical intervention was performed, and the patient recovered after an extended hospital stay.
RESULTS
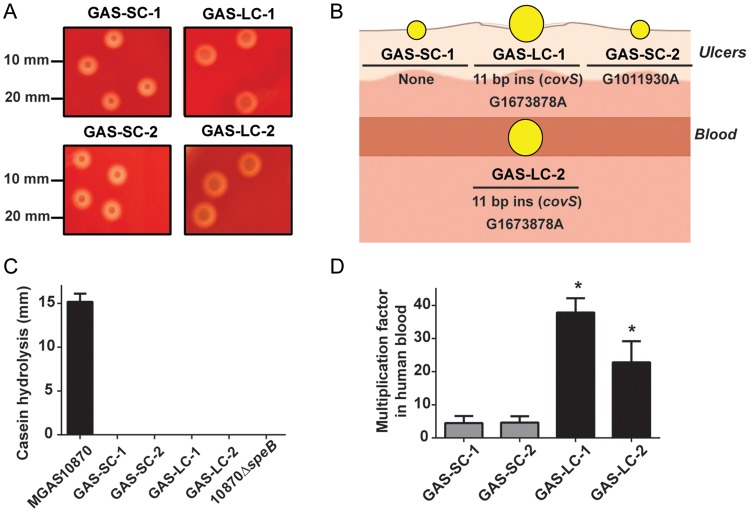
GAS organisms from 1 ulcer were noted to have a homogenous small-colony phenotype (isolate GAS-SC-1), whereas organisms from the other skin lesion were a mixture of small- and large-colony phenotypes (GAS-SC-2 and GAS-LC-1, respectively; Figure 1A). GAS organisms from the blood had a uniform large-colony phenotype (GAS-LC-2; Figure 1A). Genomic DNA was isolated using the DNeasy kit (Qiagen) from overnight cultures taken from a single colony, and emm typing was performed using the Centers for Disease Control and Prevention protocol. All 4 isolates had an identical emm sequence and matched emm type 1.0 (serotype M1). Whole genome sequencing was performed on the MiSeq platform (Illumina) using 251 bp paired-end sequencing. The raw reads in FASTQ format were aligned to the serotype M1 strain MGAS5005 (GenBank accession number NC_007297) using CLC Genomics Workbench (CLC Bio) [10]. Minimum depth of coverage was 120-fold indicating an excellent ability to determine polymorphisms (sequencing details are available in Supplementary Table 1). All 4 isolates contained the 3 prophages found in MGAS5005, including the prophage encoding the DNA-degrading Sda1 protein (also called SdaD2) [10].
Figure 1.
Characterization of GAS isolates. A, Comparison of colony phenotypes for SC and LC isolates. B, Schematic of GAS isolates from this study. Location of isolation (either ulcers or blood) is shown as are genetic polymorphisms relative to the other 3 isolates identified by whole genome analyses and confirmed by Sanger sequencing. C, SpeB activity measured by casein hydrolysis assay. Strains MGAS10870 (SpeB producing isolate) and its isogenic SpeB-inactivated derivative are included as positive and negative controls, respectively. Strains were grown in triplicate on 3 separate occasions, and data graphed are mean ± SD. D, Comparison of SC and LC isolates during growth in human blood. Data graphed are mean ± SD of isolates grown in triplicate in blood from 3 separate donors (ie, 9 data points per strain). *Indicates that the multiplication factors for each LC isolate was significantly higher (P < .05) compared to the SC isolates as determined by ANOVA using Tukey post hoc test to account for multiple comparisons (calculations performed in Prism GraphPad version 6). Abbreviations: ANOVA, analysis of variance; GAS, group A Streptococcus; LC, large colony; SC, small colony; SD, standard deviation.
We identified 2 consistent genetic variations between isolates of the small- and large-colony phenotypes (Figure 1B). First, the large-colony isolates contains an 11 bp duplication in the covS gene beginning at bp 98 (ins290204 based on the MGAS5005 genome, Supplementary Table 2) that results in a premature stop codon at amino acid 39 as compared to the usual CovS protein, which is 500 amino acids in length. Inasmuch as CovS inactivation is well described to produce a large-colony phenotype in serotype M1 GAS due to increased production of the hyaluronic capsule, it is highly likely the observed phenotypic differences between the large- and small-colony isolates were due to this 11-bp insertion [3]. The other change was G1673878A, which is located in the region between genes encoding a putative transposase (M5005_spy1712) and a putative hypothetical protein (M5005_spy1713). Additionally, the GAS-SC-2 isolate had a unique G1011930A change (resulting in an Ala to Val amino acid substitution) in the ssb-2 gene, which encodes a single stranded DNA binding protein. All identified single nucleotide polymorphisms and insertions were confirmed using Sanger sequencing.
Next, we sought to determine whether there were genetic changes present in all the isolates compared to the reference strain MGAS5005. A total of 138 genetic changes were universally present in the 4 isolates (Supplementary Table 1). Importantly, all 4 isolates contained an insertion of a single T nucleotide at position 697 in the regulator of proteinase B (ropB) gene, which encodes a transcriptional regulator that is absolutely required for expression of the broad-spectrum cysteine protease SpeB [11]. This insertion creates a premature stop codon in the ropB gene, which would be expected to result in an isolate unable to produce SpeB [11]. Indeed, we used a casein hydrolysis assay, which is a marker of SpeB production [11], and found that all 4 isolates produced no detectable SpeB (Figure 1C).
It has been postulated that invasive, hypervirulent GAS emerges from a SpeB-positive progenitor strain by CovS inactivation, which in turn results in decreased SpeB production thereby facilitating GAS dissemination [12]. Thus, we next sought to determine whether CovS inactivation in a SpeB-negative progenitor also results in an invasive phenotype. Because increased resistance to phagocytic killing is a hallmark of the invasive GAS phenotype [4], we compared the ability of the large-colony and small-colony GAS isolates to proliferate in human blood. Growth of the isolates in human blood was measured in 3 healthy human volunteers as described elsewhere [13] using a human subjects protocol approved by the Houston Methodist Hospital Institutional Review Board. The large-colony isolates multiplied an average of 6-fold greater in human blood vs the small-colony isolates (P < .01), consistent with an invasive phenotype (Figure 1D).
DISCUSSION
As the limitations of animal models of bacterial infection are increasingly recognized, data from human infections are becoming progressively more important in understanding bacterial pathogenesis [1]. One key conclusion of this work was that complete inactivation of the CovS protein, without significant additional genetic changes, was responsible for emergence of GAS with an invasive phenotype during human infection. This finding corroborates those of Sumby et al [3] who used whole genome sequencing to demonstrate that, in mice, complete CovS inactivation is sufficient to drive emergence of GAS strains with an invasive phenotype following subcutaneous inoculation of a serotype M1 strain with an intact CovRS system. Also, based on mouse infection models, Walker et al [5] postulated that GAS strains with an invasive phenotype are selected for at the infection site rather than following entry into the bloodstream. Our finding of a large-colony, CovS-inactivated strain in one of the patient's ulcers supports the idea that GAS strains with an invasive phenotype begin to emerge from their CovRS wild-type progenitors prior to entry of the organism into the bloodstream (Figure 1B).
Although our human data confirm 2 central findings of animal models of GAS pathogenesis, the whole genome sequencing approach also calls into question a previous theory regarding the molecular mechanisms underlying the invasive GAS phenotype. It has been proposed that SpeB is critical for GAS infection at the local site but that elimination of SpeB activity promotes GAS dissemination by allowing the organism to accumulate host plasmin on its cell surface [6]. Thus, the paradigm has been that invasive, SpeB-negative GAS strains emerge from a SpeB-positive progenitor following establishment of infection [5]. However, our data show that a GAS strain with an invasive phenotype can emerge from a SpeB-negative background. In addition to eliminating SpeB activity, CovS inactivation also increases production of a large variety of GAS virulence factors, such as the hyaluronic acid capsule [3]. Thus, upregulation of CovS-repressed virulence factors, rather than elimination of SpeB activity, may be the driving force for emergence of CovS-inactivated, hypervirulent GAS during human infection.
Our finding of an invasive GAS strain with mutations in both RopB and CovS is not unique. In a study of 164 invasive GAS strains from Japan, 10 had mutations in both proteins, whereas 11 strains with mutations in CovS and RopB were identified in a study of invasive serotype M3 strains from Ontario [1, 2, 14]. Because we did not recover a GAS isolate from a site of carriage (such as the throat or uninfected skin), we cannot rule out the possibility that the initial colonizing strain may have had functional RopB and CovS proteins and that mutations in both proteins were subsequently selected for during infection. However, the genetic content of the GAS variants studied herein clearly shows that the RopB inactivation preceded CovS inactivation such that the covS mutation arose from a SpeB-negative background. It has been shown that CovS inactivation decreases GAS adherence to epithelial cells, reduces GAS biofilm formation, and inhibits the ability of GAS to colonize mouse skin [15]. Similarly, having an intact RopB protein has been shown to increase GAS virulence in a necrotizing fasciitis mouse infection model [11]. Thus, the range of genetic content at the CovS and RopB loci can be considered reflective of the distinct selective pressures faced by GAS at the diverse locations that GAS colonizes and infects in humans. Future studies may be able to better understand the factors driving emergence of GAS with diverse genetic content by studying a large numbers of isolates from the same patient.
In summary, we used whole genome analyses to characterize the emergence of GAS with an invasive phenotype during human infection. Our findings emphasize the importance of close collaboration between infectious diseases consultants and the clinical microbiology laboratory in increasing understanding of the molecular basis of bacterial disease in humans.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. The authors thank the clinical microbiology laboratory at MD Anderson for their efforts in identifying the GAS isolates.
Financial support. This work was supported the National Institutes of Allergy and Infectious Diseases at the National Institutes of Health by [5R01AI089891 to S. A. S.]. A. R. F. is a scholar of the Harold Amos Medical Faculty Development Program.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Shea PR, Beres SB, Flores AR, et al. Distinct signatures of diversifying selection revealed by genome analysis of respiratory tract and invasive bacterial populations. Proc Natl Acad Sci U S A. 2011;108:5039–44. doi: 10.1073/pnas.1016282108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikebe T, Ato M, Matsumura T, et al. Highly frequent mutations in negative regulators of multiple virulence genes in group A streptococcal toxic shock syndrome isolates. PLoS Pathog. 2010;6:e1000832. doi: 10.1371/journal.ppat.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran-Winkler HJ, Love JF, Gryllos I, Wessels MR. Signal transduction through CsrRS confers an invasive phenotype in group A Streptococcus. PLoS Pathog. 2011;7:e1002361. doi: 10.1371/journal.ppat.1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker MJ, Hollands A, Sanderson-Smith ML, et al. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 2007;13:981–5. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
- 6.Cole JN, McArthur JD, McKay FC, et al. Trigger for group A streptococcal M1T1 invasive disease. Faseb J. 2006;20:1745–7. doi: 10.1096/fj.06-5804fje. [DOI] [PubMed] [Google Scholar]
- 7.Kansal RG, Datta V, Aziz RK, Abdeltawab NF, Rowe S, Kotb M. Dissection of the molecular basis for hypervirulence of an in vivo-selected phenotype of the widely disseminated M1T1 strain of group A Streptococcus bacteria. J Infect Dis. 2010;201:855–65. doi: 10.1086/651019. [DOI] [PubMed] [Google Scholar]
- 8.Sun H, Ringdahl U, Homeister JW, et al. Plasminogen is a critical host pathogenicity factor for group A streptococcal infection. Science. 2004;305:1283–6. doi: 10.1126/science.1101245. [DOI] [PubMed] [Google Scholar]
- 9.Garcia AF, Abe LM, Erdem G, Cortez CL, Kurahara D, Yamaga K. An insert in the covS gene distinguishes a pharyngeal and a blood isolate of Streptococcus pyogenes found in the same individual. Microbiology. 2010;156:3085–95. doi: 10.1099/mic.0.042614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sumby P, Porcella SF, Madrigal AG, et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–82. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 11.Olsen RJ, Laucirica DR, Watkins ME, et al. Polymorphisms in regulator of protease B (RopB) alter disease phenotype and strain virulence of serotype M3 group A Streptococcus. J Infect Dis. 2012;205:1719–29. doi: 10.1093/infdis/jir825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cole JN, Barnett TC, Nizet V, Walker MJ. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol. 2011;9:724–36. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 13.Flores AR, Jewell BE, Fittipaldi N, Beres SB, Musser JM. Human disease isolates of serotype M4 and M22 group A Streptococcus lack genes required for hyaluronic acid capsule biosynthesis. MBio. 2012;3:e00413-12. doi: 10.1128/mBio.00413-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beres SB, Carroll RK, Shea PR, et al. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci U S A. 2010;107:4371–6. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollands A, Pence MA, Timmer AM, et al. Genetic switch to hypervirulence reduces colonization phenotypes of the globally disseminated group A Streptococcus M1T1 clone. J Infect Dis. 2010;202:11–9. doi: 10.1086/653124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.