Abstract
Previous studies reported associations between neuropathogenesis and human immunodeficiency virus (HIV) compartmentalization in cerebrospinal fluid (CSF) and between sexual transmission and human immunodeficiency virus type 1 (HIV) compartmentalization in semen. It remains unclear, however, how compartmentalization dynamics change over time. To address this, we used statistical methods and Bayesian phylogenetic approaches to reconstruct temporal dynamics of HIV migration between blood and CSF and between blood and the male genital tract.
We investigated 11 HIV-infected individuals with paired semen and blood samples and 4 individuals with paired CSF and blood samples. Aligned partial HIV env sequences were analyzed by (1) phylogenetic reconstruction, using a Bayesian Markov-chain Monte Carlo approach; (2) evaluation of viral compartmentalization, using tree-based and distance-based methods; and (3) analysis of migration events, using a discrete Bayesian asymmetric phylogeographic approach of diffusion with Markov jump counts estimation. Finally, we evaluated potential correlates of viral gene flow across anatomical compartments.
We observed bidirectional replenishment of viral compartments and asynchronous peaks of viral migration from and to blood over time, suggesting that disruption of viral compartment is transient and directionally selected. These findings imply that viral subpopulations in anatomical sites are an active part of the whole viral population and that compartmental reservoirs could have implications in future eradication studies.
Keywords: Human Immunodeficiency Virus, Compartmentalization, Migration; Cerebrospinal Fluid, Semen, Coalescence
Human immunodeficiency virus (HIV) compartmentalization is defined by a restriction of viral migration between distinct anatomic compartments [1]. Such compartmentalization can affect HIV-associated pathogenesis such as neurocognitive disease [2, 3], development of drug resistance, and sexual transmission [4–12]. Furthermore, understanding viral dynamics may be important for vaccine design and efforts for a sterilizing cure. Phylogenetic analyses have permitted, with growing precision [13], the reconstruction of HIV type 1 (HIV) evolutionary history within a host. Such analyses have found a relationship between HIV compartmentalization and disease progression, as well as characterized the response to antiretroviral therapy (ART) within various compartments [14–16]. It remains unclear, however, how compartmentalization dynamics change over time.
The introduction of relaxed molecular clock phylogenetic inference has permitted an improvement in the inference of the temporal dynamics of viral evolutionary processes [17]. These time-scaled methods have been successfully used at the interhost level to investigate the epidemiological and geographical dispersal dynamics of influenza A virus [18] and dengue virus [19]. Longitudinally obtained HIV sequences analyzed with such techniques can also be used to investigate the direction and timing of a specific transmission event [20, 21] and to describe the dynamics of the HIV subpopulations within the host [22–27]. In the current study, we use discrete Bayesian phylogeographic methods to analyze longitudinally isolated HIV sequences generated from blood specimens paired with specimens from either the genital or the nervous compartment to characterize intrahost dynamics of HIV compartmentalization, especially viral migration and repopulation of sampled compartments.
METHODS
Sequence Data
We selected published studies that had well-characterized partial env sequences generated via clonal sequencing methods [3, 9, 28, 29] or single-genome amplification [5] from HIV RNA in paired blood and seminal or cerebrospinal fluid (CSF) specimens collected at ≥3 time points. There was no condition for the time between sample collections. Available partial HIV env sequences from longitudinal paired blood and seminal or CSF specimens were downloaded from the Los Alamos sequence database [3, 5, 9, 28, 29]. All C2-V3 env sequences are publicly available at GenBank, with the following accession numbers: FJ159856 to FJ160261 [3]; AF098718 to AF098734, AF256230 to AF256465, AF373037 to AF373043, AF535219 to AF535859, AY005164 to AY005179, U00821 to U00843, U13381 to U13388, and U96502 to U96608 [4]; JN393317 to JN393413, JN393423 to JN393504, JN886090 to JN886221, JN886222 to JN886300, JN886323 to JN886407, JN886408 to JN886476, and JN886499 to JN886563 [29]; and AF535485 to AF535859, AF535376 to AF535428, AF535272 to AF535375, and AF535429 to AF535484 [28]. Fifteen individuals were included with serial samples from at least 3 time points (Table 1). A total of 543 partial env sequences derived from seminal plasma specimens and 682 partial env sequences from blood plasma specimens from 11 subjects [5, 9, 28, 29] were analyzed to evaluate compartmentalization between blood plasma and male genital tract HIV populations. Additionally, 184 CSF and 222 blood plasma viral clones from 4 additional individuals were included in the study to evaluate viral compartmentalization between the blood plasma and the CSF [3].
Table 1.
Data Summary
| Subject | Sequences |
Time Points, No. | Sequences per Time Point, No., Mean (Range) | Follow-Up Duration, d | HIV RNA Load, Log10 Copies/mL, Mean (Range) |
CD4+ T-Cell Count, Cells/mm3, Mean (Range) | Sequence Length, Nucleotides, No., Mean | GenBank Accession Nos. | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | BP | SP/CSF | BP | SP/CSF | ||||||||
| G1 | 48 | 27 | 21 | 3 | 16 (11–21) | 48 | NA | NA | NA | 609 | AF256230–AF256286 | [20] |
| G8 | 55 | 30 | 25 | 3 | 18 (17–20) | 63 | NA | NA | NA | 627 | AF256405–AF256465 | |
| WA | 243 | 152 | 91 | 11 | 22 (17–30) | 198 | NA | NA | NA | 426 | AF535485–AF535859 | [30] |
| WB | 52 | 35 | 17 | 3 | 17 (11–25) | 334 | NA | NA | NA | 648 | AF535376–AF535428 | |
| WE | 64 | 37 | 27 | 3 | 21 (19–24) | 2071 | NA | NA | NA | 627 | AF535272–AF535375 | |
| WF | 52 | 29 | 23 | 5 | 10 (8–13) | 1096 | NA | NA | NA | 639 | AF535429–AF535484 | |
| L3 | 151 | 79 | 72 | 4 | 38 (19–45) | 195 | 4.6 (4.1–4.9) | 3.7 (3.0–4.0) | 895 (847–958) | 414 | JN886090–JN886221 | [31] |
| L7 | 164 | 79 | 85 | 4 | 41 (36–50) | 98 | 5.0 (4.9–5.2) | 2.5 (1.9–3.0) | 428 (400–484) | 413 | JN886222–JN886300 | |
| P9 | 179 | 97 | 82 | 4 | 45 (40–50) | 1155 | 4.3 (4.0–4.6) | 3.3 (1.2–5.8) | 638 (384–952) | 413 | JN393317– JN393413 | |
| X3 | 134 | 69 | 65 | 3 | 45 (43–46) | 70 | 4.8 (4.5–5.2) | 3.6 (3.3–3.8) | 1123 (890–1186) | 413 | JN886408–JN886476 | |
| M8 | 83 | 48 | 35 | 3 | 28 (8–48) | 1052 | 5.4 (5–5.8) | 4.0 (3.8–4.2) | 365 (352–377) | 333 | GU597090–GU597321 | [14] |
| P409 | 117 | 59 | 58 | 3 | 39 (38–40) | 45 | 5.1 (5.0–5.4) | 4.1 (1.7–5.4) | 325 (270–394) | 420 | FJ159856–FJ160261 | [7] |
| P486 | 119 | 60 | 59 | 3 | 40 (39–40) | 11 | 5.2 (5.0–5.5) | 5.4 (5.1–5.6) | 461 (NA) | 426 | FJ159856–FJ160261 | |
| P686 | 55 | 44 | 11 | 3 | 18 (10–23) | 9 | 4.6 (4.4–4.8) | 2.5 (2.3–2.6) | 200 (NA) | 426 | FJ159856–FJ160261 | |
| P839 | 115 | 59 | 56 | 3 | 38 (37–40) | 51 | 3.7 (3.5–4.0) | 3.1 (2.8–3.6) | 436 (293–579) | 420 | FJ159856–FJ160261 | |
Abbreviations: BP, blood plasma; CSF, cerebrospinal fluid; HIV, human immunodeficiency virus type 1; NA, not available; SP, seminal plasma.
Sequencing Analyses
Multiple sequence alignments of the partial HIV env region were reconstructed with MAFFT [32] and then manually edited using BioEdit [33]. Sequences that could not be unambiguously aligned were removed.
Phylogenetic/Evolutionary Reconstruction
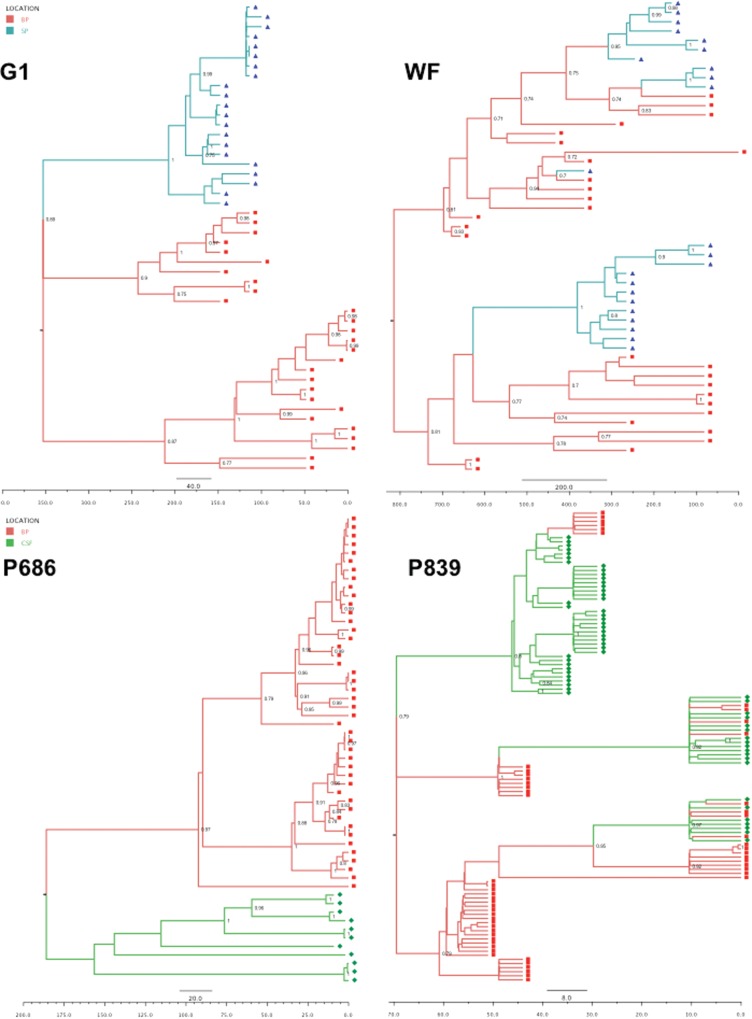
A Bayesian Markov chain Monte Carlo (BMCMC) approach was used to reconstruct phylogenies. Sampling dates were used to calibrate the time scale of the trees in days. To incorporate site of sampling, we used the discrete Bayesian asymmetric diffusion approach [34] implemented in the BEAST software package [35]. Briefly, BMCMC chains of 50 million generations were performed for each analysis with a GTR + Γ4 substitution model under an extended Bayesian skyline plot coalescent model. All analyses were performed using an uncorrelated lognormal relaxed molecular clock [35]. Tracer, version 1.5 (available at: http://beast.bio.ed.ac.uk/Tracer), was used to check for convergence. Trees were visualized in FigTree, version 1.4.0, after selecting the location state set for the nodes and their posterior probability (Figure 1).
Figure 1.
Time-scaled Bayesian phylogenetic trees. Branches are colored according to the most probable location state of their descendant nodes, in red (blood plasma), blue (seminal plasma), and green (cerebrospinal fluid [CSF]). Squares, diamonds, and triangles represent blood-, semen-, and CSF-derived variants. Posterior probabilities of >.70 are indicated at the root of each branch. Time scale (in days before last time points) are also indicated above each tree.
Location State Transition
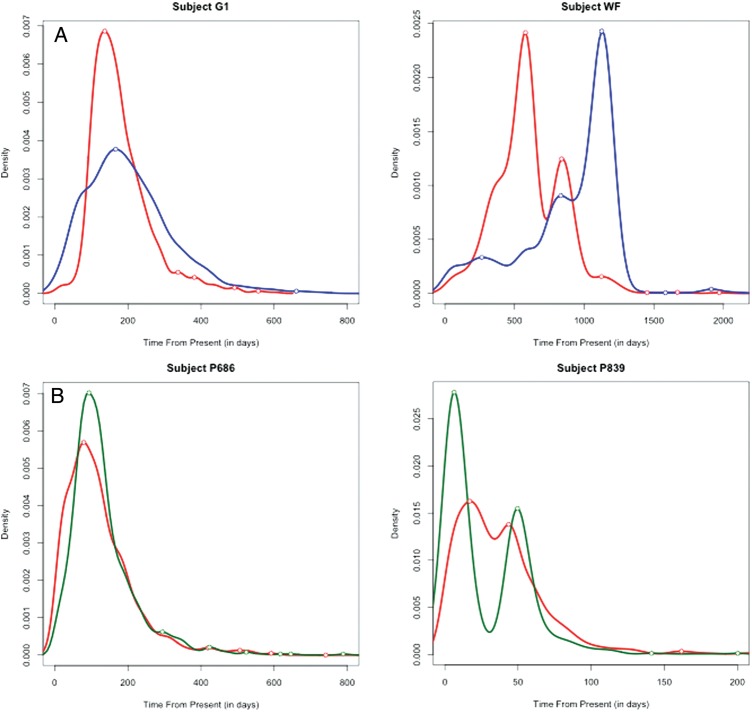
We annotated sequences with the compartment of isolation as a trait in the diffusion model and then estimated Markov jump counts [36, 37] along the branches of the posterior tree distribution [38], using a continuous-time Markov chains model. This method provided the expectations for the location state transitions between anatomical compartments, the Markov jump counts, and 95% highest posterior densities between the anatomic compartments (Table 3) and the Markov jump densities through time from these compartments (Figure 2).
Table 3.
Longitudinal Analyses of Compartmentalization of Human Immunodeficiency Virus env Sequences From Cerebrospinal Fluid (CSF) or Semen and Blood Plasma
| Subject | SM Test | Migration Events, Total No. |
Migration Events From Plasma to CSF, No. (%)c | Migration Events From CSF to Plasma, No. (%)c | Gene Flow, Migration Events per d | |
|---|---|---|---|---|---|---|
| SM Testa | Markov Jump Count (95% HPD)b | |||||
| P409 | 0 | 19 | 31 (16–57) | 16 (52) | 15 (48) | 0.69 |
| P487 | 0 | 22 | 25 (11–40) | 12 (48) | 13 (52) | 2.27 |
| P686 | 0 | 3 | 1 (1–4) | 0 | 1 (100) | 0.11 |
| P839 | 0 | 13 | 17 (9–26) | 4 (24) | 13 (76) | 0.33 |
| Migration Events From Plasma to Semen, No. (%)c | Migration Events From Semen to Plasma, No. (%)c | |||||
| G1 | 0 | 3 | 1 (1–3) | 0 | 1 (100) | 0.02 |
| G8 | 0 | 11 | 19 (8–24) | 11 (52) | 10 (48) | 0.30 |
| WA | 0 | 50 | 74 (50–102) | 34 (42) | 43 (58) | 0.37 |
| WB | 0.633 | 12 | 27 (17–52) | 15 (56) | 12 (44) | 0.08 |
| WE | 0.056 | 18 | 53 (34–65) | 29 (55) | 24 (45) | 0.03 |
| WF | 0 | 5 | 5 (4–11) | 3 (60) | 2 (40) | 0.01 |
| L3 | 0 | 20 | 24 (9–48) | 12 (50) | 12 (50) | 0.12 |
| L7 | 0 | 26 | 34 (16–46) | 22 (65) | 12 (35) | 0.35 |
| M8 | 0.016 | 9 | 37 (13–69) | 16 (43) | 21 (57) | 0.04 |
| P9 | 0 | 20 | 17 (11–29) | 9 (53) | 8 (47) | 0.01 |
| X3 | 0.307 | 35 | 29 (14–62) | 17 (59) | 12 (41) | 0.41 |
a Total number of migration events was calculated by the Slatkin-Maddison (SM) test as implemented in HyPhy (57).
b Location state transitions estimated by Markov jump counts, with 95% high posterior densities (HPDs).
c Migration direction according to Bayesian discrete phylogeographic analysis. Gene flow was defined as the number of location state transitions per day.
Figure 2.
Markov jump count density plot of viral migration between compartments. A, Subjects with paired blood and semen samples. B, Subjects with paired blood and cerebrospinal fluid (CSF) samples. Migration from blood (BP), CSF, and seminal plasma (SP) are colored in red, green, and blue, respectively.
Compartmentalization Analysis
We evaluated a battery of distance-based and tree-based methods to test each individual data set for the presence or absence of viral compartmentalization [39], using the HyPhy software package [40].
Pairwise Distance Methods
We calculated the Wright measure of population subdivision (FST) [41–45], which compares the mean pairwise distance between sequences from within 1 compartment with the pairwise distance between sequences from different compartments [41]. Statistical significance was derived via a population-structure randomization test. We also estimated the nearest-neighbor statistic (Snn) for each subset of sequences [43]. This is a measure of how frequent neighboring sequences in the phylogenetic tree were isolated from the same compartment. For our analyses, the distance matrices were calculated using the Tamura-Nei93 genetic distance [44].
Tree-Based Method
The Slatkin-Maddison test [45] was used to evaluate compartmentalization, using a tree-based method. This method determines the minimum number of migration events between compartments that will result in the phylogenetic tree under evaluation. Statistical support was determined by comparing this number of migration events to the number of events that would be expected in a randomly structured population [39, 45]. These results were compared with the number of migration events for 1000 randomizations of compartment labels on the same tree. Evidence of restricted gene flow (ie, compartmentalization) was documented (at a P value of .05) when <5% of the replicates required as many or fewer migration events as in the observed tree.
Statistics
Statistical analyses were performed using the Prism 6.0c software package for Mac (GraphPad Software, San Diego, CA) and R (R Development Core Team, 2010). Comparisons between distance-based and tree-based compartmentalization tests were evaluated with a Wilcoxon nonparametric pairwise t test. Correlation between location state transitions (ie, Markov jump count) and inferred migration events obtained from the Slatkin-Maddison test within each individual were performed with a nonparametric Spearman correlation test.
RESULTS
Study Participants and Collected Data Summary
Sequence data for paired blood and seminal plasma specimens were collected from 11 ART-naive men [5, 9, 28, 46], and sequence data for paired blood plasma and CSF specimens were collected from 4 ART-naive men [3]. A total of 904 partial HIV env sequences were collected from blood plasma specimens (range, 27–152 sequences for each individual), 543 from seminal plasma specimens (range, 17–91 sequences for each individual), and 184 from CSF specimens (range, 11–59 sequences for each individual). All included partial env sequences were generated using clonal sequencing methods [3, 28, 46], except for blood plasma sequences from Butler et al, which were generated by single-genome amplification [5]. The mean env sequence length was 484 nucleotides (range, 333–648 nucleotides), and sequences were sampled longitudinally with a mean of 4 time points per subject (range, 3–11 time points) over a mean follow-up of 433 days (range, 9–2071 days; Table 1). Briefly, blood plasma HIV RNA levels ranged between 2.5 and 5.4 log10 copies/mL (mean, 4.6 log10 copies/mL), and mean seminal plasma and CSF HIV RNA were 3.4 log10 copies/mL (range, 2.5–4.0 log10 copies/mL) and 3.8 (range, 2.5–5.4 log10 copies/mL), respectively. The mean CD4+ T-cell count was 541 cells/mL (range, 200–1123 cells/mL).
Markov Jump Counts Correlate With Inferred Migration Events, According to the Slatkin-Maddison Tree-Based Test of Compartmentalization
Using previously developed distance-based and tree-based tests of compartmentalization (Table 2), we found evidence for segregation of viral populations between CSF and blood at 10 of 12 sampled time points (83%). Maintenance of compartmentalization throughout all time points was observed for 3 individuals (P487, P686, and P839). In the 11 individuals with sequence data from blood and semen specimens, 20 (43%) of 46 time points demonstrated compartmentalization. None of these subjects maintained compartmentalization throughout all of the analyzed time points.
Table 2.
Analysis of Compartmentalization of Human Immunodeficiency Virus Type 1 env Between Cerebrospinal Fluid and Blood Plasma
| Subject | Days of Sampling | Test of Compartmentalization |
Compa | ||||
|---|---|---|---|---|---|---|---|
| FST |
Snn | SM | |||||
| Hudson-Slatkin- Maddison | Slatkin | H Boos K n | |||||
| CSF and blood plasma | |||||||
| P409 | 0 | 0 | 0 | 0 | 0 | 0 | Yes |
| 15 | 0.424 | 0.424 | 0.424 | 0.692 | 0.683 | No | |
| 45 | 0 | 0 | 0 | 0 | 0 | Yes | |
| P487 | 0 | 0 | 0 | 0 | 0 | 0 | Yes |
| 4 | 0.001 | 0.001 | 0.001 | 0.106 | 0.009 | No | |
| 11 | 0 | 0 | 0 | 0 | 0 | Yes | |
| P686 | 0 | 0 | 0 | 0 | 0 | 0.011 | Yes |
| 7 | 0 | 0 | 0 | 0 | 0 | Yes | |
| 9 | 0 | 0 | 0 | 0 | 0 | Yes | |
| P839 | 0 | 0 | 0 | 0 | 0 | 0 | Yes |
| 7 | 0 | 0 | 0 | 0 | 0 | Yes | |
| 51 | 0 | 0 | 0 | 0 | 0.008 | Yes | |
| Seminal and blood plasma | |||||||
| G1 | 0 | 0 | 0 | 0 | 0 | 0 | Yes |
| 34 | 0 | 0 | 0 | 0 | 0 | Yes | |
| 48 | 0 | 0 | 0 | 0 | 0.075 | No | |
| G8 | 0 | 0.002 | 0.002 | 0.002 | 0 | 0 | Yes |
| 34 | 0.243 | 0.243 | 0.243 | 0.041 | 0.092 | No | |
| 63 | 0.151 | 0.151 | 0.151 | 0.012 | 0.302 | No | |
| WA | 0 | 0 | 0 | 0 | 0.003 | 0 | Yes |
| 61 | 0.717 | 0.717 | 0.717 | 0.006 | 0.018 | No | |
| 84 | 0 | 0 | 0 | 0 | 0 | Yes | |
| 98 | 0.864 | 0.864 | 0.864 | 0.037 | 0.212 | No | |
| 105 | 0.2 | 0.2 | 0.2 | 0.003 | 0.138 | No | |
| 113 | 0.41 | 0.41 | 0.41 | 0.012 | 0.279 | No | |
| 119 | 0.818 | 0.818 | 0.818 | 0.047 | 0.376 | No | |
| 128 | 0.169 | 0.169 | 0.169 | 0.014 | 0.215 | No | |
| 133 | 0.013 | 0.013 | 0.013 | 0.009 | 0.096 | No | |
| 170 | 0.001 | 0.001 | 0.001 | 0.007 | 0.164 | No | |
| 198 | 0 | 0 | 0 | 0 | 0.002 | Yes | |
| WB | 0 | 0.138 | 0.083 | 0.138 | 0.138 | 0.012 | No |
| 122 | 0.35 | 0.778 | 0.35 | 0.35 | 1 | No | |
| 334 | 0.077 | 0.18 | 0.077 | 0.077 | 1 | No | |
| WE | 0 | 0.582 | 0.582 | 0.582 | 0.817 | 0.834 | No |
| 1341 | 0.025 | 0.025 | 0.025 | 0.001 | 0.003 | Yes | |
| 2071 | 0.46 | 0.46 | 0.46 | 0.923 | 1 | No | |
| WF | 0 | 0 | 0 | 0 | 0 | 1 | No |
| 366 | 0.006 | 0.006 | 0.006 | 0.086 | 0.012 | No | |
| 547 | 0 | 0 | 0 | 0 | 1 | No | |
| 912 | 0 | 0 | 0 | 0.038 | 0.002 | Yes | |
| 1096 | 0 | 0 | 0 | 0 | 0.013 | Yes | |
| L3 | 0 | 0.172 | 0.172 | 0.172 | 0.172 | 0.009 | No |
| 28 | 0.001 | 0.001 | 0.001 | 0.001 | 0 | Yes | |
| 169 | 0.006 | 0.006 | 0.006 | 0.01 | 0 | Yes | |
| 195 | 0 | 0 | 0 | 0 | 0 | Yes | |
| L7 | 0 | 0 | 0 | 0 | 0 | 0 | Yes |
| 35 | 0.035 | 0.035 | 0.035 | 0.024 | 0.005 | Yes | |
| 50 | 0.001 | 0.001 | 0.001 | 0 | 0 | Yes | |
| 98 | 0.028 | 0.028 | 0.028 | 0.078 | 0.052 | No | |
| M8 | 0 | 0 | 0 | 0 | 0 | 0.033 | Yes |
| 320 | 0.35 | 0.35 | 0.35 | 0.154 | 0 | No | |
| 1093 | 0.004 | 0.004 | 0.004 | 0 | 0.001 | Yes | |
| P9 | 0 | 0 | 0 | 0 | 0 | 0 | Yes |
| 9 | 1 | 1 | 1 | 0.142 | 0 | No | |
| 737 | 0 | 0 | 0 | 0 | 0 | Yes | |
| 1155 | 0 | 0 | 0 | 0 | 0 | Yes | |
| X3 | 0 | 0.604 | 0.1 | 0.604 | 0.604 | 0.036 | No |
| 21 | 0.176 | 0.215 | 0.176 | 0.176 | 0.004 | No | |
| 70 | 0.509 | 0.012 | 0.509 | 0.509 | 0.274 | No | |
Abbreviations: FST, Wright measure of population subdivision; SM, Slatkin-Maddison; Snn, nearest-neighbor statistic.
a Sample is defined as compartmentalized (Comp) if all tests show a P value of < .05.
To investigate the spatiotemporal dynamics of viral gene flow between compartments, we applied a discrete Bayesian diffusion model to the data, which allowed us to estimate the ancestral spatial location of HIV variants during the course of infection. In other words, we were able to define compartment of origin of sampled subpopulations and the migration of these subpopulations over time. To validate our analytical method, expected location state transitions between blood and CSF or between blood and semen were first compared with the number of inferred migration events according to the Slatkin-Maddison test of compartmentalization, and we observed a significant correlation between those 2 methods (Spearman R, 0.61; P = .002; Supplementary Figure 1). This correlation was consistent among sequence data sets for paired CSF and blood specimens and paired semen and blood specimens.
Application of a Discrete Bayesian Diffusion Approach to Reconstruct the Gene Flow Between Compartments
By estimating the ancestral spatial locations of HIV variants, we inferred the direction of viral gene flow from blood to anatomic compartment fluid (CSF or seminal plasma) or from anatomic compartment fluid to blood. Direction was defined as a location state transition from the ancestral location, similar to a “migration” of viral genetics. As examples, we showed that blood plasma appeared to be the main location from which seminal plasma variants are seeded throughout the migration history of individual WF (Figure 1B). On the contrary, for individuals G1 (Figure 1A) and P686 (Figure 1C), we found distinct compartment-specific clusters of variants in the blood and the semen (G1) or CSF (P686). These tree topologies are consistent with compartmentalized and independently evolving subpopulations without replenishment from one location to another.
In most cases, there was a reciprocal replenishment between blood and anatomical compartments, with no difference in the number of migration events from blood to anatomic compartment or from compartments to blood (mean, 13 migration events from blood to semen or CSF [range, 0–34 events] and 13.3 migrations from CSF or genital tract to blood [range, 1–43 events]; P = not significant, by the Wilcoxon pairwise t test). With regard to CSF, we observed 2-directional migration between blood and CSF compartments for all but 1 individual (P686). Similarly, bidirectional gene flow was observed between semen and blood for all but 1 individual (G1; Table 3). Interestingly, both P686 and G1, who had unidirectional gene flow observed (from blood only), had very few overall inferred potential migration events (1–3 events; Table 3).
Patterns of HIV Migration and Compartment Disruption During the Course of Infection
To investigate the dynamics of viral migration between compartments during the course of infection, we summarized the Markov jump density across compartments over time (Figure 2). This approach allowed us to define HIV dynamics in a natural time scale that complements previously described interpretation of Bayesian phylogenetic tree topologies and taxon origins (Figure 1). We identified different peaks of Markov jump density, which reflect distinct waves of viral migration between compartments during the course of infection (Figure 2). For example, for subject WF, we observed 2 main peaks of viral migration from blood to the male genital tract (579 and 838 days before the last sampling), as well as 1 major peak from genital tract to blood (1125 days before the last sampling date; Figure 2). For subject P839, we also observed distinct peaks from blood to CSF (17 and 44 days before the last sampling) and from CSF to blood (6 and 50 days before the last sampling) over time (Figure 2). These results demonstrate temporality of the bidirectional viral migration event between blood and other anatomical compartments, as previously inferred by the location state transitions (see the discussion above). Therefore, these results suggest that disruption of viral compartmentalization is a dynamic process, occurring intermittently among HIV-infected individuals.
Potential Correlates of Viral Gene Flow Across Anatomical Compartments
Next, we determined the potential correlates of disruption of compartmentalization and factors that could influence viral gene flow across compartments. For this purpose, we considered the viral gene flow per unit of time (mean, 0.34 migration events/day [0.01–2.27]; Table 3) and available clinical and virological data. Specifically, we estimated the potential impact of HIV RNA levels and CD4+ T-cell counts for both CSF and male genital tract compartment analysis. We also evaluated the association of CSF pleocytosis with blood-CSF compartment disruption. Although the sample size was small, we observed trends toward an association between disruption of compartmentalization and CSF pleocytosis (P = .08).
DISCUSSION
A comprehensive characterization of how HIV colonizes, adapts to, and migrates between various tissues in infected individuals can help elucidate how it is transmitted sexually and how the virus causes certain diseases, such as neurocognitive dysfunction. By definition, viral compartments are characterized by a restriction of viral gene flow between different anatomic compartments [1]. Understanding how HIV moves between and repopulates compartments could also be useful for understanding reservoirs in compartments that must be considered when designing eradication studies. Here, we applied a discrete diffusion model to a BMCMC phylogenetic reconstruction of all available compartment-specific HIV sequence data. A Markov jump count analysis allowed us to summarize not only the number of migration events but also to determine the temporal characteristics of HIV gene flow between blood plasma and male genital tract and CSF compartments. This method, which has been used and validated at a population scale to investigate epidemiological and spatiotemporal patterns of rapidly evolving viruses such as dengue virus or influenza virus [18, 19, 47], had not been previously applied to intrahost HIV spatiotemporal dynamics. Here, we present the methods and demonstrate the utility of this approach in the evaluation of within host HIV viral compartment dynamics.
To quantify the dissemination process of the virus across the body, we estimated the Markov jump count of location state transitions along tree branches, which identified multiple migration events occurring in both directions during the course of infection. These reciprocal migrations across central nervous system and male genital tract barriers illustrate the complex intermixing of HIV subpopulations within the host, suggesting that the replenishment of anatomic compartments is a complex and dynamic bidirectional process. Next, using a discrete Bayesian diffusion approach, we identified several unique tree topologies, reflecting different evolutionary and viral gene flow patterns during the course of infection. The finding of independently evolving subpopulations (eg, for subject G1 and P686; Figure 1) argues for well-segregated and independently evolving variants in the different compartments, whereas the presence of multiple location state transitions (eg, for subjects WF and P839; Figure 1) suggests a different process, in which viral populations often mix between compartments. These results remind us that the viral subpopulation residing within a particular cell type, tissue, or organ is not always a self-limited independently evolving subpopulation but is an integrated part of the whole HIV effective population.
The Bayesian phylogeographic inference framework used in this study incorporates not only spatial but also temporal dynamics of viral gene flow. This approach allowed us to further investigate the temporal dynamics of viral dispersal between compartments during the course of HIV infection (Figure 2). Again, we observed a bidirectional replenishment of viral compartments: viral gene flow moved relatively equally from blood to the anatomic compartments and vice versa. These results may contrast with the patterns of spread expected from previous studies that argued for the blood compartment as the predominant reservoir of viral replenishment [3]. Specifically, we were able to distinguish asynchronous peaks of viral migration from and to blood over time, suggesting that viral disruption could be transient and directionally selected. Together, these findings imply that viral gene flow across compartments is far from a steady or consistent process and can vary considerably during the course of infection. It also argues for the need to investigate the numerous factors that could drive viral migration and related compartment disruption over the course of infection, especially for planned eradication strategies.
Previous studies have provided insight into factors that may influence HIV migration (or not) between compartments, such as pleocytosis in CSF [3], which were reconfirmed in this analysis. Other factors were also considered—the HIV load in blood plasma, genital tract, and CSF, as well as the CD4+ T-cell count. None of these factors were correlated with viral migration events, but this lack of association could be a consequence of the limited size of the data set. Future studies involving careful clinical characterization of the subjects will be necessary to identify factors associated with HIV migration within the host, especially if all 3 compartments (ie, blood, CSF, and semen) could be sampled simultaneously. Another possible limitation of the study was the use of clonal and single genome sequencing, which provide only a limited sampling of the viral population in blood and compartments, introducing possible sampling bias. As with all polymerase chain reaction (PCR)–based standard sequencing methods, we also need to consider potential PCR recombination in the data that were obtained from population-based (or “bulk”) sequencing of cloned samples [30] and its potential lack of sensitivity [31]. In addition to a limit of sensitivity, it is also critical to account for biases that may be introduced by founder effect of the bulk PCR procedure [48]. Reliable detection of minor variants with next-generation sequencing method will help to more finely explore HIV migration and compartmentalization.
The limited depth of sampling may also affect conclusions about the directionality of viral migration events and about compartmentalization itself, as determination of the population structure dynamics are highly dependent upon adequate sampling. Future studies should use next-generation sequencing techniques to extensively sample the HIV populations in compartment fluids.
In summary, we used newly developed Bayesian phylogeographic methods to evaluate the dynamics and determinants of HIV compartmentalization. Our results suggest the need to consider that anatomical sites not only can restrict viral gene flow, but that viral subpopulations can migrate back and forth between anatomic compartments i.e., not always from blood. This could have profound implications in eradication studies that do not consider anatomic compartments in their design.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Financial support. This work was supported by the Department of Veterans Affairs, the National Institutes of Health (grants P30-AI027763 [to the University of California, San Francisco Gladstone Institute of Virology & Immunology Center for AIDS Research], AI100665, DA034978, AI36214, AI7462, AI69432, AI47745, MH097520, AI027763, and MH62512 and award 7UM1AI068636-07), the James B. Pendleton Charitable Trust, amfAR (grant 108537, with support from FAIR), and the Interdisciplinary Research Fellowship in NeuroAIDS (award R25-MH081482).
Potential conflicts of interest. D. D. R. has served as a consultant for Biota, Chimerix, BMS, Gilead, Gen-Probe, Monogram, Sirenas, and Prism. D. M. S. has received grant support from ViiV Pharmaceuticals and consultant fees from Gen-Probe and Testing Talent Services. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Nickle DC, Shriner D, Mittler JE, Frenkel LM, Mullins JI. Importance and detection of virus reservoirs and compartments of HIV infection. Curr Opin Microbiol. 2003;6:410–6. doi: 10.1016/s1369-5274(03)00096-1. [DOI] [PubMed] [Google Scholar]
- 2.Cowley D, Gray LR, Wesselingh SL, Gorry PR, Churchill MJ. Genetic and functional heterogeneity of CNS-derived tat alleles from patients with HIV-associated dementia. J Neurovirol. 2011;17:70–81. doi: 10.1007/s13365-010-0002-5. [DOI] [PubMed] [Google Scholar]
- 3.Smith DM, Zárate S, Shao H, et al. Pleocytosis is associated with disruption of HIV compartmentalization between blood and cerebral spinal fluid viral populations. Virology. 2009;385:204–8. doi: 10.1016/j.virol.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pillai SK, Good B, Pond SK, et al. Semen-specific genetic characteristics of human immunodeficiency virus type 1 env. J Virol. 2005;79:1734–42. doi: 10.1128/JVI.79.3.1734-1742.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler DM, Delport W, Kosakovsky Pond SL, et al. The origins of sexually transmitted HIV among men who have sex with men. Sci Transl Med. 2010;2:18re1. doi: 10.1126/scitranslmed.3000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Philpott S, Burger H, Tsoukas C, et al. Human immunodeficiency virus type 1 genomic RNA sequences in the female genital tract and blood: compartmentalization and intrapatient recombination. J Virol. 2005;79:353–63. doi: 10.1128/JVI.79.1.353-363.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delwart EL, Mullins JI, Gupta P, et al. Human immunodeficiency virus type 1 populations in blood and semen. J Virol. 1998;72:617–23. doi: 10.1128/jvi.72.1.617-623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson JA, Ping L-H, Dibben O, et al. HIV-1 Populations in Semen Arise through Multiple Mechanisms. PLoS Pathog. 2010;6:e1001053. doi: 10.1371/journal.ppat.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta P, Leroux C, Patterson BK, et al. Human immunodeficiency virus type 1 shedding pattern in semen correlates with the compartmentalization of viral Quasi species between blood and semen. J Infect Dis. 2000;182:79–87. doi: 10.1086/315644. [DOI] [PubMed] [Google Scholar]
- 10.Boeras DI, Hraber PT, Hurlston M, et al. Role of donor genital tract HIV-1 diversity in the transmission bottleneck. Proc Natl Acad Sci U S A. 2011;108:E1156–63. doi: 10.1073/pnas.1103764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bull M, Learn G, Genowati I, et al. Compartmentalization of HIV-1 within the female genital tract is due to monotypic and low-diversity variants not distinct viral populations. PLoS One. 2009;4:e7122. doi: 10.1371/journal.pone.0007122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overbaugh J, Anderson RJ, Ndinya-Achola JO, Kreiss JK. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res Hum Retroviruses. 1996;12:107–15. doi: 10.1089/aid.1996.12.107. [DOI] [PubMed] [Google Scholar]
- 13.Rambaut A, Posada D, Crandall KA, Holmes EC. The causes and consequences of HIV evolution. Nat Rev Genet. 2004;5:52–61. doi: 10.1038/nrg1246. [DOI] [PubMed] [Google Scholar]
- 14.Castro-Nallar E, Pérez-Losada M, Burton GF, Crandall KA. The evolution of HIV: inferences using phylogenetics. Mol Phylogenet Evol. 2012;62:777–92. doi: 10.1016/j.ympev.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nickle DC, Jensen MA, Shriner D, et al. Evolutionary indicators of human immunodeficiency virus type 1 reservoirs and compartments. J Virol. 2003;77:5540–6. doi: 10.1128/JVI.77.9.5540-5546.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shankarappa R, Margolick JB, Gange SJ, et al. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–9. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nunes MRT, Faria NR, Vasconcelos HB, et al. Phylogeography of dengue virus serotype 4, Brazil, 2010–2011. Emerg Infect Dis. 2012;18:1858–64. doi: 10.3201/eid1811.120217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.English S, Katzourakis A, Bonsall D, et al. Phylogenetic analysis consistent with a clinical history of sexual transmission of HIV-1 from a single donor reveals transmission of highly distinct variants. Retrovirology. 2011;8:54. doi: 10.1186/1742-4690-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rachinger A, Groeneveld PHP, van Assen S, Lemey P, Schuitemaker H. Time-measured phylogenies of gag, pol and env sequence data reveal the direction and time interval of HIV-1 transmission. AIDS Lond Engl. 2011;25:1035–9. doi: 10.1097/QAD.0b013e3283467020. [DOI] [PubMed] [Google Scholar]
- 22.Worobey M, Gemmel M, Teuwen DE, et al. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455:661–4. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pybus OG, Rambaut A, Harvey PH. An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics. 2000;155:1429–37. doi: 10.1093/genetics/155.3.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drummond A, Pybus OG, Rambaut A. Inference of viral evolutionary rates from molecular sequences. Adv Parasitol. 2003;54:331–58. doi: 10.1016/s0065-308x(03)54008-8. [DOI] [PubMed] [Google Scholar]
- 25.Hudson RR. How often are polymorphic restriction sites due to a single mutation? Theor Popul Biol. 1989;36:23–33. doi: 10.1016/0040-5809(89)90021-x. [DOI] [PubMed] [Google Scholar]
- 26.Grenfell BT, Pybus OG, Gog JR, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–32. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- 27.Rosenberg NA, Nordborg M. Genealogical trees, coalescent theory and the analysis of genetic polymorphisms. Nat Rev Genet. 2002;3:380–90. doi: 10.1038/nrg795. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Frey S, Gupta P, et al. Compartmentalization and migration between HIV-1 viral populations in blood and semen. 2002 [Google Scholar]
- 29.Gianella S, Mehta SR, Strain MC, et al. Impact of seminal cytomegalovirus replication on HIV-1 dynamics between blood and semen. J Med Virol. 2012;84:1703–9. doi: 10.1002/jmv.23398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lahr DJG, Katz LA. Reducing the impact of PCR-mediated recombination in molecular evolution and environmental studies using a new-generation high-fidelity DNA polymerase. BioTechniques. 2009;47:857–66. doi: 10.2144/000113219. [DOI] [PubMed] [Google Scholar]
- 31.Hughes JP, Totten P. Estimating the accuracy of polymerase chain reaction-based tests using endpoint dilution. Biometrics. 2003;59:505–11. doi: 10.1111/1541-0420.00060. [DOI] [PubMed] [Google Scholar]
- 32.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol Clifton NJ. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 33.Hall TA. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8. [Google Scholar]
- 34.Edwards CJ, Suchard MA, Lemey P, et al. Ancient hybridization and an Irish origin for the modern polar bear matriline. Curr Biol CB. 2011;21:1251–8. doi: 10.1016/j.cub.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minin VN, Suchard MA. Counting labeled transitions in continuous-time Markov models of evolution. J Math Biol. 2008;56:391–412. doi: 10.1007/s00285-007-0120-8. [DOI] [PubMed] [Google Scholar]
- 37.Talbi C, Lemey P, Suchard MA, et al. Phylodynamics and human-mediated dispersal of a zoonotic virus. PLoS Pathog. 2010;6:e1001166. doi: 10.1371/journal.ppat.1001166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Brien JD, Minin VN, Suchard MA. Learning to count: robust estimates for labeled distances between molecular sequences. Mol Biol Evol. 2009;26:801–14. doi: 10.1093/molbev/msp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zárate S, Pond SLK, Shapshak P, Frost SDW. Comparative study of methods for detecting sequence compartmentalization in human immunodeficiency virus type 1. J Virol. 2007;81:6643–51. doi: 10.1128/JVI.02268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinforma Oxf Engl. 2005;21:676–9. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 41.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–9. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9:138–51. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 43.Hudson RR. A new statistic for detecting genetic differentiation. Genetics. 2000;155:2011–4. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–26. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 45.Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–13. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gianella S, Strain MC, Rought SE, et al. Associations between virologic and immunologic dynamics in blood and in the male genital tract. J Virol. 2012;86:1307–15. doi: 10.1128/JVI.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holmes EC. Evolutionary history and phylogeography of human viruses. Annu Rev Microbiol. 2008;62:307–28. doi: 10.1146/annurev.micro.62.081307.162912. [DOI] [PubMed] [Google Scholar]
- 48.Vandenbroucke I, Van Marck H, Mostmans W, et al. HIV-1 V3 envelope deep sequencing for clinical plasma specimens failing in phenotypic tropism assays. AIDS Res Ther. 2010;7:4. doi: 10.1186/1742-6405-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.