Abstract
Objective. We evaluated Toll-like receptors (TLRs) single nucleotide polymorphisms (SNPs) for associations with HIV-1 acquisition, set-point and disease progression in African couples.
Methods. Seven candidate and 116 haplotype-tagging SNPs (tagSNPs) were genotyped in 504 HIV-1 infected cases, and 343 seronegative controls.
Results. TLR9 1635A/G was associated with reduced HIV-1 acquisition among HIV-seronegative controls with high but not low HIV-1 exposure (odds ratio [OR] = 0.7; P = .03 and OR = 0.9, P = .5, respectively). TLR7 rs179012 and TLR2 597C/T reduced set-point; the latter modified by time since HIV-1 acquisition. TLR8 1A/G reduced disease progression.
Conclusions. TLR SNPs impact HIV-1 outcomes with epidemiologic factors modifying these relationships.
Keywords: Acute Infection, Genetics, Heterosexual HIV-1 transmission, Progression, Risk Factors, Viral load, Toll-like Receptors, HIV-1 set-point, HIV-1 acquisition
Disparities in susceptibility to HIV-1 acquisition and control of viremia and disease progression following infection are partially explained by human genetic variation (ie, CCR5-Δ32 and HLA alleles). Toll-like receptor (TLR) genes may alter HIV-1 outcomes by influencing recognition of pathogen-associated molecular patterns and initiating innate immune responses that vary longitudinally and may contribute to functional impairment and immune exhaustion. TLR single nucleotide polymorphisms (SNPs) have been associated with clinical HIV-1 outcomes, however, discrepant results underscore the need for validation [1–7]. Furthermore, these studies have generally not included populations from Africa, where HIV-1 is most prevalent and where different TLR variant frequencies may modify HIV-1 associations. To better understand TLR-mediated responses against HIV-1, we evaluated TLR SNP associations with heterosexual HIV-1 acquisition, set-point and disease progression among African HIV-1 serodiscordant couples.
METHODS
Participant Selection
Participants came from prospective HIV-1 serodiscordant heterosexual couple cohorts from sub-Saharan Africa: the Partners in Prevention HSV-2/HIV-1 and Couples Observational Studies. Both studies assessed HIV-1 transmission quarterly over 1–2 years of follow-up and collected longitudinal behavioral and laboratory data from both partners as previously described [8, 9]. Human subjects research review and approval was obtained at the University of Washington, study sites and affiliated institutions. All participants provided written informed consent.
We used a nested case-control study to analyze TLR SNP associations with HIV-1 acquisition. We matched 129 HIV-1 transmitting couples to 246 non-transmitting couples based on HIV-1 exposure scores, which increased with HIV-1 levels of infected partners, frequency of unprotected sex, decreasing age, female gender, and lack of male circumcision [9, 10]. Other factors such as treatable sexually transmitted infections were not modeled due to low prevalence among uninfected participants. To increase representation of HIV-1 seronegative individuals, we included additional seronegative participants having the highest persistent HIV-1 exposure levels in these cohorts. For this analysis, HIV-1 seroprevalent cases included participants with HIV-1 at study enrollment (unknown infection date) whereas seroconverter cases acquired HIV-1 during follow-up.
HIV-1 Set-point and Disease Progression
Plasma HIV-1 RNA levels were determined using the COBAS AmpliPrep/COBAS TaqMan HIV-1 RNA assay, version 1.0 (Roche Diagnostics, Indianapolis, IN), with a quantification limit of 240 copies/mL. For seroconverters, HIV-1 set-point was defined as the average log10 plasma HIV-1 RNA measurement taken 4–18 months after infection (median = 6 measurements per person). For seroprevalent cases, set-point was based on consecutive measurements within 1 log10 copies/mL determined before antiretroviral therapy (ART) initiation or CD4 count < 200 cells/mm3 (median = 4 measurements per person). HIV-1 disease progression among seroprevalent cases was defined as time to (1) ART initiation, (2) CD4 count < 200 cells/mm3, or (3) death from medical causes.
Genotyping
DNA was isolated from archived blood using Puregene DNA purification (Qiagen, Valencia, CA), with genotyping using an Illumina Custom Oligo Pooled Assay for 124 SNPs (9 in TLR2, 13 TLR3, 22 TLR4, 40 TLR7, 25 TLR8, 3 TLR9, 4 MYD88, and 8 TIRAP). These included seven candidate SNPs previously associated with HIV-1 infection and 116 haplotype-tagging SNPs (tagSNPs) chosen to represent common variation across TLR genes (Supplementary Table 1). Haplotypes were inferred from the Yoruba HapMap population, and tagSNPs with minor allele frequency >5% were selected. Overall, six SNPs were excluded for >10% missing results (n = 3), for being monomorphic (n = 1), or failing Hardy-Weinberg Equilibrium (n = 1) or X-linked autosomal tests (n = 1). Of 847 genotyped samples, 15 were excluded for X-chromosome sex discrepancies and 5 for >10% genotypic missingness.
Statistical Analysis
HIV-1 acquisition analyses compared TLR genotypes in all seropositive cases to seronegative controls using logistic regression. Secondarily, an ‘extreme phenotype design’ was restricted to highly exposed seronegative controls to improve sensitivity [10]. Set-point analyses were performed using linear regression among all seropositive individuals, with interaction analyses by seroconverter or seroprevalent HIV-1 status to assess differential effects of TLR polymorphisms by time since infection. Finally, we determined associations of TLR variants with disease progression using Cox regression and Kaplan-Meier curves.
Regression models were conducted in the R GenABEL package (http://www.genabel.org/) and were adjusted for sex, age, acyclovir use and population stratification using principal components from a separate study [9]. X-chromosome SNP associations were evaluated combining all participants and separately for males and females. Evaluation of candidate SNPs represents confirmatory analyses of factors previously implicated in HIV-1 infection, therefore, we report uncorrected P-values for those SNPs. For exploratory analyses of 111 TLR tagSNPs we calculated Bonferroni-corrected P-values with a significance cutoff of Pcorrected < .00045 (α = .05). Since different inheritance models lack independence, we did not adjust P-values for testing multiple models.
RESULTS
Study Participants
Among genotyped individuals, 343 were HIV-1 seronegative controls (243 with high and 100 with lower exposure) and 504 were seropositive cases (129 seroconverter and 375 seroprevalent) (Supplementary Table 2). Approximately 75% of cases and controls were from East Africa. Overall, cases and controls had similar sex distributions with 160 (47%) controls and 265 (52%) infected cases being female (P = .2). HIV-1 exposure scores ranged from 0 to 7 and were similar among seroconverters and highly exposed controls (median = 5 [IQR: 3–5] vs 5 [IQR: 5–6]) but were lower among low exposure controls (2 [IQR: 1–3], P < .001). Highly exposed controls were similar to seroconverters in age (31 vs 30 years, P = .4), but had lower prevalence of male circumcision (33% vs 43% P = .2) and increased frequency of any unprotected sex (50% vs 44%, P = .3). Compared to low exposure controls, highly exposed controls were more likely to report unprotected sex (50% vs 22%, P < .001) and to have HIV-1 infected partners with higher HIV-1 levels (5.0 vs 3.9 log10 copies/mL, P < .001).
Follow-up among HIV-1 seronegative controls was similar among participants with high and low exposure (21 [IQR: 15–24] vs 21 months [IQR: 18–24], respectively). Among, HIV-1 seroconverters, median time before the first seropositive HIV-1 test was 9 months (IQR: 3–15) and median post-seroconversion follow-up was 12 months (IQR: 12–12). Median follow-up for HIV-1 seroprevalent cases was 22 months (IQR: 17–24).
TLR Associations With HIV-1 Acquisition
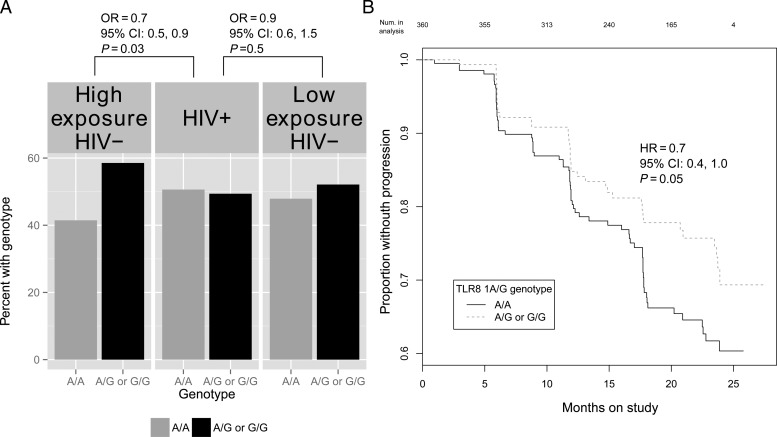
Comparing HIV-1 infected cases to all seronegative controls, the candidate TLR9 1635G allele was associated with reduced HIV-1 acquisition risk (OR = 0.8; 95% CI, .6–.9; P = .04). The association was stronger when cases were compared to highly exposed controls (OR = 0.7, 95% CI, .5–.9; P = .03) but was lost when compared to low exposure controls (OR = 0.9, 95% CI, .6–1.5; P = .5) (Figure 1A).
Figure 1.
A, Frequency of the TLR9 1635A/G (rs352140) single nucleotide polymorphism by HIV-1 infection status and level of HIV-1 exposure. HIV-1 exposure scores were used to represent an individual's risk of infection at enrollment and were based on a Cox proportional hazards regression model that included HIV-1 plasma RNA levels of the infected partner, unprotected sex, age, sex, and male circumcision. B, Kaplan-Meier plot of HIV-1 progression by TLR8 1A/G (rs3764880) genotype. HIV-1 progression is defined as CD4 < 200 cells/mL3, ART initiation, or death among HIV-1 seroprevalent cases with enrollment CD4+ T cell counts greater than 200.
TLR Associations With HIV-1 Set-point
Median plasma HIV-1 set-point among all cases was 4.7 log10 copies/mL (IQR: 4.1–5.2) and was similar among seroprevalent (4.8; IQR: 4.4–5.3) and seroconverter cases (4.5; IQR: 3.6–5.0). The TLR7 rs179012 G allele, a haplotype-tagging intronic SNP, was associated with 0.4 log10 copies/mL lower set-points (95% CI, .2–.6; Pcorrected = .03), regardless of duration of infection (Table 1). This association was stronger among females (β = −.5; 95% CI, −.7,−.2; P = .0002) but not males (β = −.3; 95% CI, −.6, −.01; P = .05). Homozygosity of the candidate TLR2 597 T allele was associated with lower set-point among seroprevalent cases (β = −.4; 95% CI, −.1,−.7; P = .005), but not seroconverters (β = .3; 95% CI, −.4,1.1; P = .4)(interaction P-value = .02).
Table 1.
Associations Between TLR Polymorphisms and Plasma HIV-1 RNA Set-point Among All HIV-1 Seropositive Participants and Among HIV-1 Seroprevalent and Seroconverters Cases Separately
| Gene Variant | Gender | Cases | Na | MAFb |
Median HIV-1 RNA Set-point by Genotype (IQR) |
Mean Difference in HIV-1 Set-point (95% CI)c,d | |||
|---|---|---|---|---|---|---|---|---|---|
| T | CC | CT | TT | ||||||
| TLR2 rs3804099 (candidate SNP: 597 C/T, synonymous change in exon 3) Chromosome: 4 | All | 359 | 0.3 | 4.6 (4.3–5.2) | 4.6 (4.1–6.2) | 4.4 (3.9–4.9) | −0.20 (−.5, .09) | P = .2 | |
| All | Prevalent | 249 | 0.3 | 4.8 (4.5–6.2) | 4.9 (4.4–5.4) | 4.4 (3.7–4.7) | -0.42 (−.72, -.13) | P = .005 | |
| Incident | 110 | 0.29 | 4.3 (3.7–4.9) | 4.4 (3.7–5.0) | 5.0 (4.1–5.3) | 0.34 (−.42, 1.10) | P = .4 | ||
| G | AA/A− | AG | GG/G− | ||||||
| All | 359 | 0.25 | 4.8 (4.4–5.3) | 4.5 (3.9–5.0) | 4.5 (3.9–4.9) | −0.37 (−.56, −.19) | P = .00008e | ||
| All | Prevalent | 249 | 0.24 | 4.9 (4.5–5.4) | 4.6 (4.3–5.0) | 4.6 (4.0–4.9) | −0.34 (−.54, −.15) | P = .0006 | |
| Incident | 110 | 0.28 | 4.7 (3.9–5.1) | 4.1 (2.6–5.2) | 4.4 (3.7–4.8) | −0.38 (−.79, .03) | P = .07 | ||
| G | AA | AG | GG | ||||||
| TLR7 rs179012 (Intronic tagSNP) Chromosome: X | All | 193 | 0.23 | 4.9 (4.4–5.4) | 4.5 (3.9–5.0) | 4.6 (3.7–4.7) | −0.49 (−.74, −.23) | P = .0002 | |
| Female | Prevalent | 148 | 0.23 | 5.0 (4.6–5.0) | 4.6 (4.3–5.0) | 4.6 (3.9–4.7) | -0.40 (−.64, −.15) | P = .0001 | |
| Incident | 45 | 0.26 | 4.8 (4.0–5.1) | 4.1 (2.6–5.2) | 3.6 (3.6–3.6) | −0.64 (−1.36, .09) | P = .08 | ||
| G | A− | G− | |||||||
| All | 166 | 0.26 | 4.8 (4.3–5.2) | 4.5 (4.0–4.9) | −0.29 (−.58, −.01) | P = .05 | |||
| Male | Prevalent | 101 | 0.25 | 4.8 (4.5–5.3) | 4.6 (4.0–5.0) | −0.26 (−.59, .08) | P = .14 | ||
| Incident | 65 | 0.29 | 4.6 (4.0–5.1) | 4.4 (3.9–4.8) | −0.22 (−.73, .29) | P = .41 | |||
a Includes HIV-1 infected participants for whom HIV-1 set-point met the definition of consecutive plasma HIV-1 RNA measurements within one log10 copies/mL and who had genotype data.
b Minor allele frequency.
c Plasma HIV-1 RNA set-point log10 copies/mL.
d Mean differences and 95% confidence intervals from ordinary least squares regression models adjusted for sex, age, and population stratification. For TLR7 rs179012, we report results under a dominant model of inheritance and a recessive model of inheritance for TLR2 rs3804099.
e Bonferroni adjusted P-value = .03.
TLR Associations With HIV-1 Progression
At enrollment, HIV-1 seroprevalent cases with the candidate non-synonymous TLR8 1A/G polymorphism had higher median CD4+ T cells/mm3 (442; IQR: 334–636) than those with the AA genotype (405; IQR: 325–535) and this association was significant under a dominant model of inheritance (β = 49.3; 95% CI, 2.6–96.1; P = .04). When evaluating associations with HIV-1 disease progression, TLR8 1A/G was protective among all participants (hazard ratio [HR] = 0.7; 95% CI, .4–1.0; P = .05) and was stronger among females (HR = 0.5; 95% CI, .3–.9; P = .03) (Figure 1B). This association remained after participants with ART initiation or death as their disease progression end-point were removed.
DISCUSSION
Innate immune responses through TLR signaling may be important in HIV-1 infection. We found that polymorphisms in TLR2 and TLR7 were associated with HIV-1 set-point, TLR8 with disease progression, and TLR9 with HIV-1 acquisition. These relationships may be modified by HIV-1 exposure levels and duration of HIV-1 infection, providing a potential context for understanding previous study results.
In our study, TLR9 1635A/G was 1.4-times more prevalent among HIV-1 exposed seronegative controls than infected cases. This is the first report of an association between this SNP and reduced HIV-1 acquisition, but is consistent with previous protective associations with HIV-1 progression and set-point [2, 6]. We did not find an association between this SNP and disease progression or set point. This may be due to differences in ethnicity (African vs Caucasian) or background characteristics (heterosexual, MSM or IDU) of these study populations. Perinatal HIV-1 transmission studies found this locus to associate with increased acquisition risk, possibly reflecting differences in heterosexual and vertical transmission [7]. One possible reason for protective effects of this SNP among adults is that it may overcome HIV-1 gp120 suppression of IFN-α responses to TLR9 agonists in pDCs, thereby maintaining production of antiviral and inflammatory responses against HIV-1 [11]. This association was more pronounced when comparing HIV-1 infected cases to controls with high, but not low, HIV-1 exposure, demonstrating that detection of host risk factors for sexual HIV-1 acquisition may be influenced by exposure levels among seronegative participants [10].
We found TLR2 and TLR7 SNP associations with HIV-1 set-point. The TLR2 597C/T SNP was associated with 0.4 log10 copies/mL lower viral load among HIV-1 seroprevalent cases but not among recent seroconverters, suggesting that effects of this SNP may vary over the course of infection. Although TLR2 recognizes bacteria, fungi, and parasites, it could influence responses to HIV-1 by enhancing chronic immune activation through translocation of gut-associated microbial factors [12].
TLR7 rs179012 was associated with 0.4 log10 copies/mL lower set-point among all HIV-infected participants. One hypothesis for similar TLR7 associations during early and chronic infection is that trafficking of HIV-1 to early endosomes instead of lysozomes elicits chronic immune activation through persistent release of IFN-α in the absence of pro-inflammatory cytokines [13], resulting in more targets for HIV-1 throughout acute and chronic infection. A recent study of TLR signaling pathways suggests that responses to a TLR2 agonist varied by duration of infection; in contrast, TLR7 agonist responses were detectable during acute and chronic infection [14]. Further studies may elucidate how TLR signaling through these variant receptors impact plasma HIV-1 levels throughout infection.
Finally, consistent with previous studies, TLR8 1A/G was associated with slower HIV-1 progression due to CD4+ T cell decline [4]. This SNP is associated with elevated TNF-α, reduced IL-10 production, and elevated prostaglandin E2 and leukotriene B4, which are associated with reduced HIV-1 replication and disease progression [4].
An advantage of our study was having longitudinal data from both partners in heterosexual African HIV-1 serodiscordant couples, including chronically infected individuals, HIV-1 seroconverters, and HIV-1 exposed seronegative individuals with varying exposure levels. This allowed us to evaluate if TLR SNP associations differed by time since HIV-1 infection and to evaluate resistance to infection among highly exposed HIV-1 seronegative participants, which may help avoid spurious associations [10]. Several cohort limitations could have obfuscated associations. First, most HIV-1 seroprevalent participants had HSV-2, possibly confounding TLR-HIV-1 associations through synergistic effects of HSV-2/HIV-1. However, sensitivity analyses suggest that HSV-2 was not important in this analysis. Second, selecting tagSNPs from Yorubans could limit representation of common variants among East and southern Africans.
Two previous GWA studies, including one among participants from our study, did not identify TLR SNP associations with HIV-1 acquisition or set-point [9, 15], but did not consider a priori information regarding TLR associations with HIV-1 outcomes and applied rigorous multiple testing corrections for all SNPs. In our GWAS, we had 80% power to detect associations with relative risks >3.2 for variants with minor allele frequency = 5% and, thus, may have missed modest associations. In the current candidate gene study, we sought to confirm associations for candidate SNPs and were powered to detect modest associations. Additionally, both GWA studies used standard GWAS chips designed for European populations and less than half of the variants considered in the current analysis of Africans were analyzed. Lower linkage disequilibrium in Africans could accentuate the impact of missed variants.
In summary, our results further suggest that TLR polymorphisms alter the course of HIV-1 acquisition and infection. However, validation in other study populations or through functional assays is needed. Research designed to evaluate functional mechanisms might test how these polymorphisms influence pro-inflammatory cytokine levels and IFN-α following stimulation with TLR agonists. An integrated evaluation of innate immune functional pathways mediating these phenotypes may identify new targets for HIV-1 preventative and therapeutic interventions.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. All authors participated in critical review of the report. R. M., A. B., M. B., and J. L. designed the study. C. C., G. d. B, G. J. S., A. R., N. M., and J. L. participated in patient recruitment. R. M., A. B., C. C., G. d. B, K. B. S., G. J. S., A. R., N. M., K. B., J. M, M. J. M., and J. L. were involved in data collection and analysis, and laboratory processing. R. M., A. B., and J. L. wrote the report. R. M. and A. B. did the statistical analyses. All authors reviewed and commented on the report at all stages and approved the final version.
The authors thank the women and men who participated in the study, and the clinical, recruitment, retention, data, and laboratory teams involved in the study in Africa and Seattle.
Partners in Prevention HSV-2/HIV-1 Transmission Study Team (for genomic studies): University of Washington Coordinating Center and Central Laboratories, Seattle, USA: Connie Celum (principal investigator), Anna Wald (protocol co-chair), Jairam R. Lingappa (medical director), Jared M. Baeten, Mary S. Campbell, Lawrence Corey, Robert W. Coombs, James P. Hughes, Amalia Magaret, M. Juliana McElrath, Rhoda Morrow, James I. Mullins.
Study site principal investigators and study coordinators relevant to this analysis: Cape Town, South Africa (University of Cape Town): David Coetzee; Eldoret, Kenya (Moi University, Indiana University): Kenneth Fife, Edwin Were; Gaborone, Botswana (Botswana Harvard Partnership): Max Essex, Joseph Makhema; Kampala, Uganda (Infectious Disease Institute, Makerere University): Elly Katabira, Allan Ronald; Kisumu, Kenya (Kenya Medical Research Institute, University of California San Francisco): Elizabeth Bukusi, Craig Cohen; Moshi, Tanzania (Kilimanjaro Christian Medical College, Harvard University): Saidi Kapiga, Rachel Manongi; Nairobi, Kenya (University of Nairobi, University of Washington): Carey Farquhar, Grace John-Stewart, James Kiarie; Orange Farm, South Africa (Reproductive Health Research Unit, University of the Witwatersrand): Sinead Delany-Moretlwe, Helen Rees; Soweto, South Africa (Perinatal HIV Research Unit, University of the Witwatersrand): Guy de Bruyn, Glenda Gray, James McIntyre; Thika, Kenya (University of Nairobi, University of Washington): Nelly Rwamba Mugo.
Financial support. This work was supported by the Bill and Melinda Gates Foundation grants 26469 and 41185, NIH/NIAID grants AI073115 and AI27757, and by the University of Washington Center for AIDS Research (CFAR), an NIH funded program (P30 AI027757) which is supported by the following NIH Institutes and Centers (NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, NIA). RDM was supported by the University of Washington STD/AIDS Research Training Program (T32AI007140) from the National Institutes of Health, U.S. Public Health Service. AWB was supported by a training fellowship from the NIH/National Human Genome Research Institute (T32 HG00035). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Bochud P-Y, Hersberger M, Taffé P, et al. Polymorphisms in Toll-like receptor 9 influence the clinical course of HIV-1 infection. AIDS. 2007;21:441–6. doi: 10.1097/QAD.0b013e328012b8ac. [DOI] [PubMed] [Google Scholar]
- 2.Pine SO, McElrath MJ, Bochud P-Y. Polymorphisms in toll-like receptor 4 and toll-like receptor 9 influence viral load in a seroincident cohort of HIV-1-infected individuals. AIDS. 2009;23:2387–95. doi: 10.1097/QAD.0b013e328330b489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oh D-Y, Baumann K, Hamouda O, et al. A frequent functional toll-like receptor 7 polymorphism is associated with accelerated HIV-1 disease progression. AIDS. 2009;23:297–307. doi: 10.1097/QAD.0b013e32831fb540. [DOI] [PubMed] [Google Scholar]
- 4.Oh D-Y, Taube S, Hamouda O, et al. A functional toll-like receptor 8 variant is associated with HIV disease restriction. J Infect Dis. 2008;198:701–9. doi: 10.1086/590431. [DOI] [PubMed] [Google Scholar]
- 5.Ricci E, Malacrida S, Zanchetta M, et al. Toll-like receptor 9 polymorphisms influence mother-to-child transmission of human immunodeficiency virus type 1. J Transl Med. 2010;8:49. doi: 10.1186/1479-5876-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soriano-Sarabia N, Vallejo A, Ramírez-Lorca R, et al. Influence of the Toll-like receptor 9 1635A/G polymorphism on the CD4 count, HIV viral load, and clinical progression. J Acquir Immune Defic Syndr. 2008;49:128–35. doi: 10.1097/QAI.0b013e318184fb41. [DOI] [PubMed] [Google Scholar]
- 7.Beima-Sofie KM, Bigham AW, Lingappa JR, et al. Toll-like Receptor (TLR) variants are associated with infant HIV-1 acquisition and peak plasma HIV-1 RNA level. AIDS. 2013;27:2431–9. doi: 10.1097/QAD.0b013e3283629117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Celum C, Wald A, Lingappa JR, et al. Acyclovir and Transmission of HIV-1 from Persons Infected with HIV-1 and HSV-2. N Engl J Med. 2010;362:427–39. doi: 10.1056/NEJMoa0904849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lingappa JR, Petrovski S, Kahle E, et al. Genomewide Association Study for Determinants of HIV-1 Acquisition and Viral Set Point in HIV-1 Serodiscordant Couples with Quantified Virus Exposure. PLoS One. 2011;6:e28632. doi: 10.1371/journal.pone.0028632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackelprang RD, Baeten JM, Donnell D, et al. Quantifying ongoing HIV-1 exposure in HIV-1 serodiscordant couples to identify individuals with potential host resistance to HIV-1. J Infect Dis. 2012;206:1299–308. doi: 10.1093/infdis/jis480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinelli E, Cicala C, Van Ryk D, et al. HIV-1 gp120 inhibits TLR9-mediated activation and IFN-α secretion in plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2007;104:3396–401. doi: 10.1073/pnas.0611353104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sandler NG, Douek DC. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat Rev Microbiol. 2012;10:655–66. doi: 10.1038/nrmicro2848. Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 13.O'Brien M, Manches O, Sabado RL, et al. Spatiotemporal trafficking of HIV in human plasmacytoid dendritic cells defines a persistently IFN-α – producing and partially matured phenotype. J Clin Invest. 2011;121:1088–1101. doi: 10.1172/JCI44960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chang JJ, Lacas A, Lindsay RJ, et al. Differential regulation of toll-like receptor pathways in acute and chronic HIV-1 infection. AIDS. 2012;26:533–41. doi: 10.1097/QAD.0b013e32834f3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petrovski S, Fellay J, Shianna KV, et al. Common human genetic variants and HIV-1 susceptibility: a genome-wide survey in a homogeneous African population. AIDS. 2011;25:513. doi: 10.1097/QAD.0b013e328343817b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.