Abstract
The ubiquitin-proteasome system (UPS) is impaired in Huntington’s disease, a devastating neurodegenerative disorder. Sulforaphane, a naturally-occurring compound, has been shown to stimulate UPS activity in cell cultures. To test whether sulforaphane enhances UPS function in vivo, we treated UPS function reporter mice ubiquitously expressing the green fluorescence protein (GFP) fused to a constitutive degradation signal that promotes its rapid degradation in the conditions of a healthy UPS. The modified GFP is termed GFPu. We found that both GFPu and ubiquitinated protein levels were significantly reduced and the three peptidase activities of the proteasome were increased in the brain and peripheral tissues of the mice. Interestingly, sulforaphane treatment also enhanced autophagy activity in the brain and the liver. To further examine whether SFN promotes mutant huntingtin (mHtt) degradation, we treated HD cells with sulforaphane and found that sulforaphane not only enhanced mHtt degradation but also reduced mHtt cytotoxicity. Sulforaphane-mediated mHtt degradation was mainly through the UPS pathway since the presence of a proteasome inhibitor abolished this effect. Taken together, these data indicate that sulforaphane activates protein degradation machineries in both the brain and peripheral tissues and may be a therapeutic reagent for HD and other intractable disorders.
Keywords: mutant huntingtin, sulforaphane, ubiquitin proteasome system, autophagy, protein degradation, neurodegeneration
Introduction
Huntington’s disease (HD) is a fatal autosomal dominant inherited neurodegenerative disorder caused by an unstable expansion of CAG trinucleotide repeats coding for polyglutamine (polyQ) in the huntingtin (Htt) gene (Zoghbi & Orr 2000, Bates 2002). HD is one of the most deleterious diseases, causing progressive physical and mental decline and placing a significant financial burden on both families and societies. Neuropathologically, HD is characterized by progressive neuronal death and the misfolding of Htt into soluble oligomers and insoluble aggregates that accumulate in neurons (Li & Li 2011). Unfortunately, there are currently no treatments available to prevent the onset of HD or halt or reverse its progression. Therefore, there is an urgent need for effective and feasible therapeutic agents for HD.
Compromised oxidative stress defense systems and functional insufficiency of protein degradation machinery have been implicated in HD pathogenesis (Jin et al. 2013, Li et al. 2011). The reactive oxygen species that are generated by aerobic metabolism and environmental stressors can chemically modify proteins and alter their biological functions. Cells possess protein repair pathways to rescue oxidized proteins and restore their functions. However, if these repair processes fail, oxidized proteins may become cytotoxic and should be degraded.
Two major protein degradation pathways exist in eukaryotic cells: the ubiquitin-proteasome system (UPS) and autophagy (Atg) pathways (Li et al. 2008, Rubinsztein 2006). The UPS is mainly responsible for the degradation of short-lived proteins. Most unwanted proteins subjected to proteasomal degradation are recognized by the presence of a polyubiquitin chain, which allows them to be transported to the proteasome for degradation. Atg generally mediates bulk degradation and requires the formation of double-membrane-bound structures (autophagosomes), which fuse with lysosomes. The contents are then degraded by the lysosomal hydrolases. Current data support that both pathways are impaired in a status of insufficiency in HD (Seo et al. 2004, Ravikumar et al. 2004). First, generalized inhibition of the proteasome and Atg is found not only in the brain cells but also in the skin fibroblasts derived from HD patients (Seo et al. 2004, Ravikumar et al. 2004). Second, mutant Htt (mHtt) cannot be efficiently degraded and forms aggregates both in vivo and in vitro (Chang et al. 2006). Third, overexpression of the proteasome activator, PA28γ, increases HD neuron viability (Seo et al. 2007). Finally, enhancement of Atg activity by compounds such as rapamycin reduces mHtt toxicity and improves cell viability in HD animal models (Ravikumar et al. 2004). These observations highlight the significance of protein degradation in HD pathogenesis, indicating that enhancement of the protein degradation activities is a feasible therapeutic strategy. To date, however, there is a paucity of compounds that can not only enhance the clearance of mHtt but also penetrate the blood brain barrier and work in brain cells.
Sulforaphane (SFN), an isothiocyanate, is a natural product originally isolated from broccoli or other cruciferous vegetables and has been shown to be an effective anti-cancer agent in a variety of cancer cells and animal models (Zhang et al. 1992). The major mechanism by which SFN protects cells is believed to function through its direct antioxidant effects or indirect induction of Phase-II metabolizing enzymes through Nrf2/ARE transcription factor pathway (Fahey & Talalay 1999, Kensler et al. 2007). These Phase-II enzymes inactivate many carcinogens and reactive oxygen species, thereby protecting cells against DNA damage and subsequent malignant transformation. Oxidative stress is implicated in a number of neurodegenerative disorders and several lines of research have investigated whether SFN can be used to treat these diseases. Data from studies of Parkinson’s disease and Alzheimer’s disease have indicated that SFN has a beneficial effect on these diseases in related animal models when administered peripherally (Morroni et al. 2013, Kim et al. 2013). However, recent data have shown that SFN upregulates expression of the 26S proteasome subunit, PSMB5, and enhances proteasome activity in cell cultures (Kwak et al. 2007, Gan et al. 2010). Therefore, SFN may have a dual effect on cells: suppression of excessive production of free radicals and maintaining protein homeostasis. As oxidative stress and accumulation of misfolded mHtt have been causally linked to HD (Goswami et al. 2006), it is possible that SFN will be a neuroprotective reagent for HD. Here we demonstrated that SFN enhances both proteasomal and autophagic activities in vitro and in a transgenic mouse model and reduces mHtt-caused neurotoxicity and mHtt accumulation in cell models of HD, suggesting that SFN is a potential therapeutic reagent for treating the disorder.
Materials and Methods
Cell culture and transfection
HeLa and HEK293 cells were grown in DMEM supplemented with 10% FBS and antibiotics. The UPS function reporter construct encoding the green fluorescent protein (GFP) fused with a degradation signal (degron) CL1 has previously been described (Kumarapeli et al. 2005, Liu et al. 2008). The modified GFP is referred to as GFPu for the UPS reporter. GFPu was transfected in HeLa cells using a Safectine RU50 DNA transfection kit according to the manufacturer’s protocol (Syd Labs, USA).
The methods to culture and differentiate the inducible neuronal progenitor cell lines stably expressing GFP-tagged Htt-exon1 containing 74Q have been previously described (Dong et al. 2011, Dong et al. 2012c).
GFPu transgenic mice
The GFPu transgenic mouse has been previously described (Kumarapeli et al. 2005). All animal maintenance and experimental procedures were conducted in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals and was approved by the Institutional Animal Care and Use Committee (IACUC) of the University of South Dakota. Only male mice were used in the experiments. Mice were maintained in a temperature- and humidity-controlled environment with a 12 h light:12 h dark cycle and with ad libitum access to food and water.
Western blot analysis
The cells prepared for western blot analysis were washed twice with ice-cold PBS and were then lysed with a cell lysis buffer (50 mM Tris-HCl, pH6.8, 150 mM NaCl, 20 mM EDTA, 1 mM EGTA, 0.5% SDS, 0.5% NP-40, 0.5% Sarkosyl) supplemented with fresh protease inhibitor cocktail (Sigma) for 20 min on ice with intermittent agitation. Protein extracts were resolved on 10 or 12% SDS-PAGE gels and transferred onto nitrocellulose membrane (Whatman GmbH) under standard transfer conditions as described previously (Dong et al. 2012b, Dong et al. 2012c). The antibodies used were anti-GFP (1:2000, Santa Cruz Biotechnology), actin (1:100, Santa Cruz Biotechnology), ubiquitin (1:1000, Cell Signaling), LC3 (1:1000, Cell Signaling), GAPDH (1:1000, Cell Signaling), and anti-FLAG antibodies (1:1000, Sigma). Detections were performed using HRP-conjugated anti-rabbit, -mouse, or -goat antibody (Santa Cruz Biotechnology) and Immobilon Western Chemiluminescent HRP substrate (Millipore).
The proteasome activity assays
The proteasome activity assays were performed according to previously described methods (Kisselev & Goldberg 2005). Briefly, tissues were lysed in proteasome activity assay buffer (50mM Tris-HCl, pH 7.5, 250mM sucrose,5mM MgCl2, 0.5mM EDTA, 2mM ATP, and 1mM dithiothreitol) by passing ten times through a 27-gauge needle attached to a 1-ml syringe. After centrifugation at 10,000g for 10 min at 4°C, the pellets were discarded. Protein concentrations of the supernatants were determined using the BCA protein assay kit (Pierce). Approximately 20 µg of the total protein of cell lysates were transferred to a 96-well plate. The following fluorogenic substrates were added to the cell lysates: a final concentration of 40 µM of Suc-Leu-Leu-Val-Tyr-AMC for measuring chymotrypsin-like activity of the proteasome; Z-Leu-Leu-Glu-AMC (40 µM) to measure the caspase-like activity of the proteasome; and Ac-Arg-Leu-Arg-AMC for the proteasome trypsin-like activity (10 µM). Fluorescence (380-nm excitation, 460-nm emission) was monitored on a microplate fluorometer after 1 h incubation at 37°C.
Cycloheximide (CHX) chase
The methods for generating and culturing HEK293 cells stably expressing mHtt containing 94Q have been previously described (Dong et al. 2012a). The cells were incubated with 10 µg/ml CHX and 10 µM SFN for different times before the cells were lysed for western blot analysis of the degradation of the mHtt protein.
Cell viability and caspase activity assays
Cell viability analysis was performed as described previously (Dong et al. 2012b, Dong et al. 2012c) with a TACS™ MTT assay kit (Trevigen) according to the manufacturers’ protocols. Caspase-activity was assessed according to a recently described method (Dong et al. 2012b).
Statistical analysis
Statistical comparisons of data between two groups were evaluated using two-tailed student’s t test. P < 0.05 was regarded as statistically significant. Two-way analysis of variance (ANOVA) was conducted on the experimental data using two types of cells and time as grouping factors.
Results
SFN reduces the level of GFPu, a surrogate substrate for the UPS, in vitro and in vivo
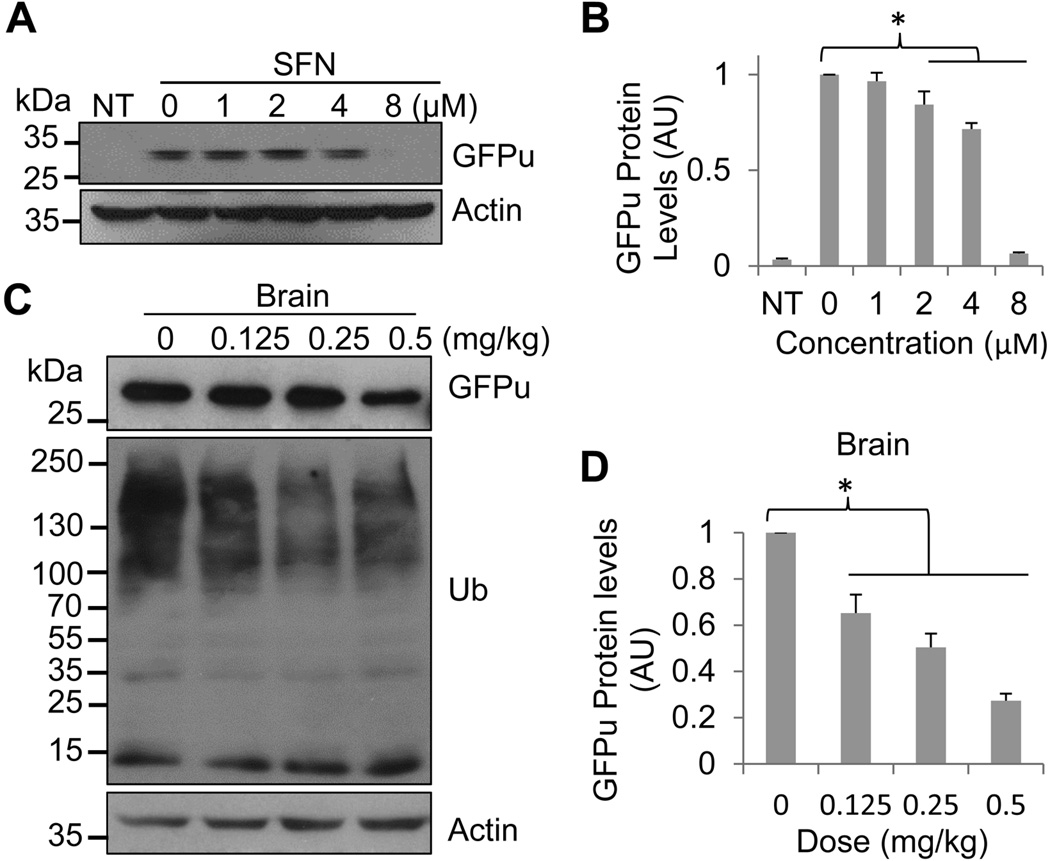
To verify whether SFN enhances proteasomal function, we utilized a UPS function reporter, a green fluorescent protein (GFP) fused with a degradation signal (degron) CL1 at its C-terminus that targets it for ubiquitination and degradation by the UPS. The modified GFP is referred to as GFPu. Since GFPdgn serves as a surrogate substrate for the UPS, its level inversely reflects UPS function. To examine whether SFN reduces GFPu protein level in cell culture, HeLa cells were transfected with a GFPu plasmid construct (Bence et al. 2001) and then treated with increasing concentrations of SFN 24 h after the transfection. Western blot analysis showed that treatment of HeLa cells with high concentrations of SFN resulted in a pronounced dose-dependent reduction of GFPu proteins (Figs. 1A & 1B), suggesting that SFN enhances GFPu degradation in cell culture.
Fig. 1.
SFN reduces the GFPu protein level in vitro and in vivo.
A. Western blot analysis of GFPu protein levels from cells transfected with a GFPu plasmid and treated with SFN. HeLa cells were transiently transfected with either an empty vector (non-transfected, NT) or a GFPu construct, and 24 h after the transfection cells were treated with indicated concentrations of SFN for 8 h before cell lysates were collected for western blot analysis of the GFPu protein using a GFP-specific antibody.
B. Quantitation of GFPu protein levels in A. GFPu protein bands were measured and normalized against actin protein levels (in arbitrary unit, AU). Data are shown as mean ± SD; n = 3. *p < 0.05 between 0 and 2, 4, 8 µM treatments.
C, E, and G. Western blot analysis of GFPu protein levels from the brains of GFPu transgenic mice treated with the indicated doses of SFN. Mice were injected daily either with vehicle as a control or with the indicated dose of SFN. Three days after injections, the mice were sacrificed and the GFPu protein levels in the cerebral cortex (C), heart (E), and liver (G) were analyzed by western blot.
D, F and H are quantitations of GFPu protein levels in (C), (E), and (G), respectively. GFPu protein bands were measured and then normalized against actin levels. Data are shown as mean ± SD; n = 3. *p < 0.05 between the control (0 mg/kg) and the indicated treatments in each tissue.
To further test whether SFN promotes the degradation of the UPS function reporter, GFPu, also in mice, we injected the GFPu transgenic (Tg) mice either with vehicle or with SFN and then examined GFPu protein levels in the brain, liver, and heart. The GFPu Tg mouse has previously been described and has shown that it is a reliable tool for monitoring in vivo changes of UPS proteolytic function in virtually all major organs (Kumarapeli et al. 2005). Three days after daily injections, the mice were sacrificed and the GFPu levels in the cerebral cortex, heart, and liver were examined by western blot analysis. As shown in Figs 1C–1H, treatment of the mice with 0.5 mg/kg SFN strikingly reduces GFPu protein levels in all tissues examined. Interestingly, different tissues showed distinct responses to SFN. The cerebral cortex appeared to be more sensitive than other tissues and the GFPu protein level revealed SFN-dose-dependent reduction (Figs. 1C & 1D). It should be noted that a low dose of SFN (0.125 mg/kg) resulted in significant reduction of GFPu protein in the cerebral cortex, but this dose did not caused a similar effect in other types of tissues. To enhance degradation of GFPu in the heart, 0.25 mg/kg SFN is required (Figs. 1E & 1F). In contrast, the UPS in the liver was not as sensitive as in the brain and heart. To significantly enhance the degradation of GFPu, it required at least 0.5 mg/kg SFN (Figs. 1G & 1H). It should also be noted that the GFPu protein levels were closed corresponding to the ubiquitinated/polyubiquitinated protein levels (Figs. 1C, 1E, and 1G). These in vivo data indicate that SFN enhances the UPS function in different tissues, including the brain, when appropriate concentrations of the compound were administered to the animals.
SFN enhances proteasome activity in vivo
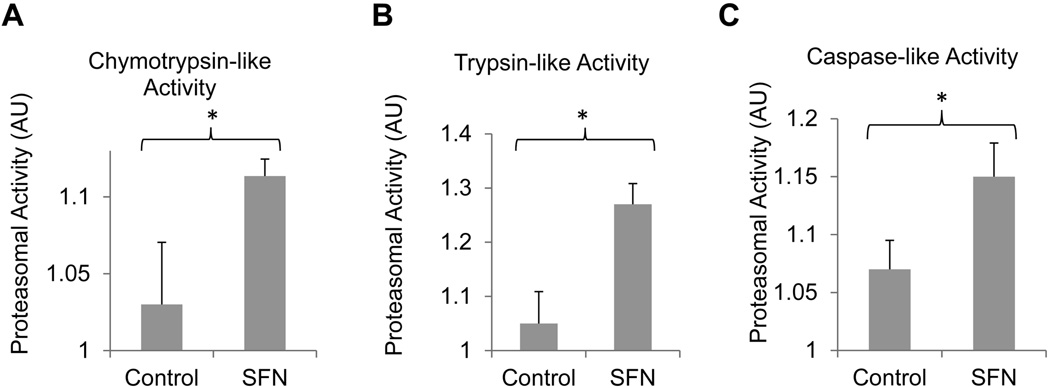
The above experiments show that SFN promotes the degradation of GFPu, the surrogate substrate of the 26S proteasome, suggesting that SFN is a potent enhancer of the UPS. To determine whether SFN also exerts a direct effect on proteasome intrinsic peptidase activities, we further tested the effect of SFN on the peptidase activity of different tissue lysates of the GFPu mice injected with SFN or vehicle. In UPS-mediated proteolysis, peptide cleavage occurs inside the 20S proteasome (Wojcik & DeMartino 2003). Three measurable peptidase activities, including chymotrypsin-like, caspase-like, and trypsin-like activities, have been described in 20S proteasomes, which are carried out by β5-, β2-, and β1-subunits, respectively. Using synthetic flurorogenic peptide substrates, we found that all three peptidase activities in the cerebral cortex were significantly enhanced by SFN following 0.5 mg/kg injection (Figs. 2A, 2B, and 2C).
Fig. 2.
SFN enhances chymotrypsin-like (A), trypsin-like (B), and caspase-like (C) peptidase activities of the proteasome in the brains. The GFPu mice were injected daily with either vehicle or 0.5 mg/kg of SFN. Three days after injections, the mouse cerebral cortex was isolated and subjected to proteasome activity assays. Data are shown as mean ± SD; n = 4. *p < 0.01.
SFN enhances Atg activity in vivo
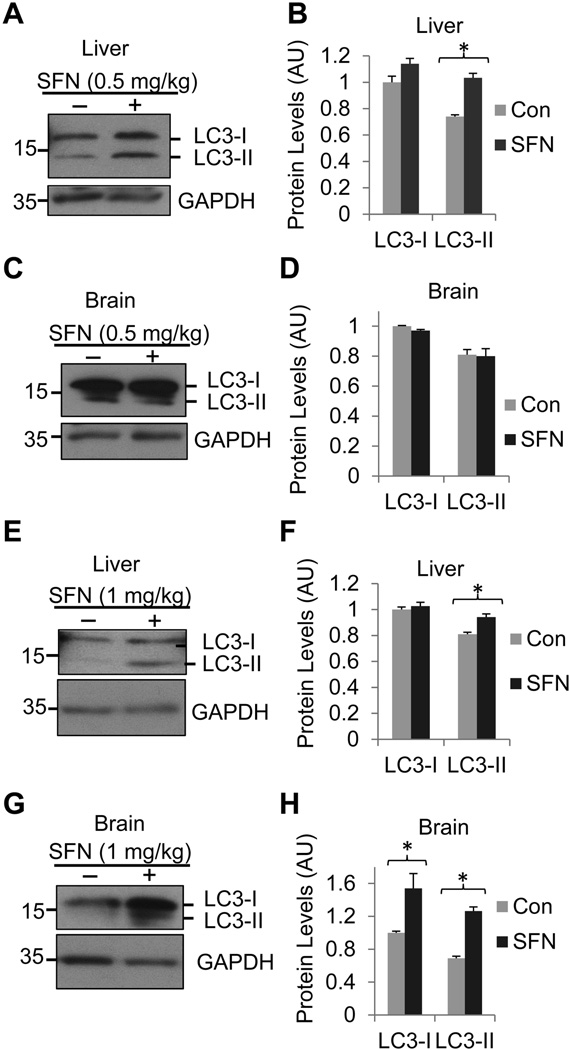
Previous data have shown that SFN activates Atg in human prostate cancer cells (Herman-Antosiewicz et al. 2006). To test this possibility in vivo, we injected mice with 0.5 or 1 mg/kg of SFN daily and the mice were sacrificed 3 days after injections. We examined whether treatment of mice with SFN up-regulates, processes, and recruits the biochemical marker of Atg, microtubule-associated protein 1 light chain 3 (LC3) to autophagosomes of (Kabeya et al. 2000). The LC3 is synthesized as a precursor, which is subsequently truncated to cytosolic LC3-I and then conjugated with phosphatidylethanolamine to become membrane-associated LC3-II (Ogata et al. 2006). As the amount of converted LC3-II protein is correlated to the number of autophagosomes, LC3-II is commonly considered an excellent indication for the activation of the autophagic pathway (Mizushima & Yoshimori 2007). Western blot showed that treatment of mice with 0.5 mg/kg SFN increased LC3-II protein levels in the liver (Figs. 3A & 3B) but not in the brain cortex (Figs. 3C & 3D). When the SFN dose was increased to 1 mg/kg, however, increased LC3-II protein levels were seen in both the liver (Figs. 3E & 3F) and brain cortex (Figs. 3G & 3H). Interestingly, treatment of mice with a high dose of SFN increased both LC3-I and LC3-II protein levels in the brain cortex, indicating that the high concentration of SFN can activate the early cytosolic form of LC3 (LC3-I) and the late membrane-bound form of LC3 (LC3-II) in the brain cortex. These results suggest that SFN enhances autophagy in the brain and non-brain cells.
Fig. 3.
SFN enhances Atg activity in the brain. Mice were injected with either 0.5 (A, B, C, and D) or 1 mg/kg (E, F, G, and H) SFN or vehicle daily. After 3 days, they were sacrificed and both the liver and the brain cortex were isolated for analysis of LC3-I and II proteins. LC3 protein levels were analyzed by western blot analysis using the liver (A, E) or brain (C, G) from mice injected with 0.5 or 1 mg/kg of SFN, respectively. Quantitations of protein levels of A, C, E, and G were shown in B, D, F, and H, respectively. Data are shown as mean ± SD; n = 3. *p < 0.01.
SFN promotes mHtt degradation in a cell culture
The results obtained above suggest that SFN enhances both the UPS and Atg activities and thereby promotes mHtt degradation. To examine this possibility, we conducted a classical cycloheximide chase assay measuring the rate of turnover of the mHtt protein in cells that had been transfected with a plasmid encoding an N-terminal Htt containing 94 CAG repeats (mHtt-94Q) (Dong et al. 2012b). The transfected cells were treated the next day with both SFN and cycloheximide (CHX) or the vehicle (dimethyl sulfoxide, DMSO) for different times as indicated in Figs. 4A & 4B. The mHtt-94Q protein in the cells treated with SFN was degraded more rapidly than that in the cells treated with the vehicle (Figs. 4A & 4B), indicating that SFN promotes mHtt degradation. To determine whether SFN-induced mHtt degradation is through the UPS or Atg pathway, we repeated the experiment in presence of a proteasome inhibitor, MG132, or an Atg inhibitor, 3-methyladenine (3MA). Treatment of the cells with MG132 combined with CHX and SFN for 10 h significantly abolished SFN-enhanced mHtt degradation, whereas the treatment with 3MA did not alter SFN-mediated mHtt degradation (Fig. 4C & 4D), suggesting that SFN-induced mHtt degradation is mainly through the UPS pathway. To further determine whether SFN reduces mHtt protein level in absence of CHX, we treated a previously generated HEK293 cell line stably expressing N-terminal mHtt fragment containing 94Q (Dong et al. 2012a, Dong et al. 2012b) with the same dose of SFN as the CHX chase assay. We observed that in absence of CHX, treating the HD cells with SFN for 13 h also reduced mHtt protein level (Figs. 4E & 4F). Taken together, these data indicate that SFN accelerates mHtt degradation and this effect is largely depends on the UPS pathway.
Fig. 4.
SFN promotes mHtt degradation in cell culture.
A. Cycloheximide chase analysis by western blot of HEK293 cells transiently transfected with a plasmid encoding mHtt containing 94Q and treated with cycloheximide (CHX, 10 µg/ml) plus SFN (10 µM) or CHX plus DMSO for indicated time periods.
B. Quantitation of mHtt94Q protein levels performed in A. A two-way ANOVA (time by treatment) revealed a significant effect of SFN treatment (F(1,30) = 1986, p<0.0001), and a significant effect of time (F(5, 30)=968.6, p<0.0001).
C. SFN-mediated mHtt degradation is mainly through the UPS pathway. HEK293 cells transiently transfected with a plasmid encoding mHtt containing 94Q were treated with cycloheximide (CHX, 10 µg/ml) in presence of DMSO, SFN (10 µM), SFN plus MG132, SFN plus 3MA, or SFN plus the MG132 and 3MA (double inhibitors, DI) for 10 h.
D. Quantitation of mHtt94Q protein levels performed in C. Data were shown as mean ± SD; n = 3. *p < 0.05.
E. SFN treatment reduces mHtt protein level in absence of CHX. HEK293 cells stably expressing 94Q were treated with 10 µM SFN or the equal volume of DMSO (Control) for 13 h.
F. Graph showing mHtt94Q protein levels in E. Data were shown as mean ± SD; n = 3. *p < 0.05.
SFN reduces mHtt-caused toxicity in cell cultures
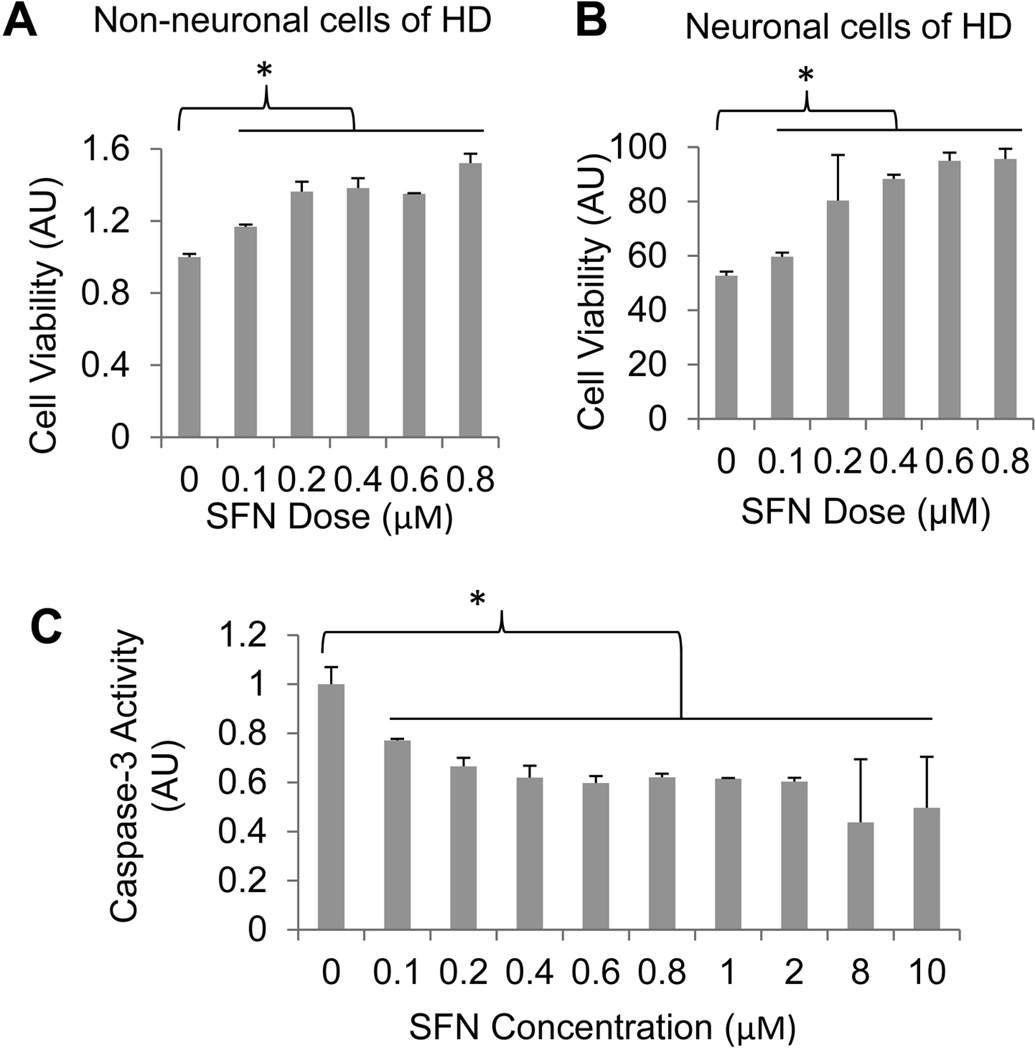
Accumulation of mHtt protein and oxidative stress has been causatively linked to HD (Liang et al. 2011, Jin et al. 2013). The combination of anti-oxidative and protein degradation-enhancing properties of SFN may make the compound a good candidate for use as a therapeutic reagent for treating HD. To test this possibility, we treated a previously generated HEK293 cell line stably expressing N-terminal mHtt fragment containing 94Q with increasing concentrations of SFN. Twenty-four hours after the treatment, the cells were challenged with 100 µM of menadione, an oxidative stress inducer (Caricchio et al. 1999), to potentiate mHtt-caused toxicity by exacerbating oxidative stress (Wang et al. 2006, Dong et al. 2011). As shown in Fig. 5A, treatment of the cells with SFN enhanced cell viability largely in dose-dependent manner. To further examine whether SFN also reduces mHtt caused-toxicity in neuronal cells, we therefore repeated the similar treatments and cell viability assay using a previously established inducible neuronal cell model of HD (Dong et al. 2011). As shown in Fig. 5B, the results we obtained were similar to those obtained using HEK293 cells; namely, treating the HD neurons with SFN enhanced neuronal survival.
Fig. 5.
SFN enhances cell survival in non-neuronal and neuronal cell models of HD. HEK293 cells stably expressing N-terminal mHtt containing 94Q (A) or differentiated neuronal cells stably expressing GFP-tagged N-terminal mHtt containing 74Q (B) were incubated with indicated concentrations of SFN, and 24 h after the incubation cells were treated with 100 µM menadione for 6 h before cell viability was assessed. Data were shown as mean ± SD; n = 3. *p < 0.05. C, Caspase-3-like activity measured from the 94Q HD cells after treating with SFN at the indicated concentrations. Data were shown as mean ± SD; n = 3. *p < 0.01.
Previous studies have indicated that SFN induces apoptosis in different cancer cell lines (Herman-Antosiewicz et al. 2006, Vyas et al. 2013, Gamet-Payrastre et al. 2000). To further examine whether SFN triggers apoptotic cell death in mHtt-expressing cells, we assessed caspase-3 activity following treatment of the HEK293 cells stably expressing 94Q mHtt with different concentrations of SFN as described above. SFN did not activate caspase-3 in the HD cells but on the contrary, it reduced caspase-3 activity when the cells were treated with the indicated concentrations of the compound (Fig. 5C), confirming the observation that SFN protects HD cells.
Discussion
The major discoveries we obtained here are that SFN enhances UPS function and Atg activity in the brain and peripheral tissues, such as the liver, and promotes turnover of a UPS surrogate substrate, GFPu, and ubiquitinated proteins in vivo. Moreover, we also demonstrated, for the first time to our knowledge, that treatment of cells expressing mHtt with SFN augmented the degradation of mHtt and reduced its toxicity in both non-neuronal and neuronal cell cultures. These results raise the possibility that SFN is a potential drug for the treatment of HD and other polyQ expansion-caused neurodegenerative disorders.
The enhanced degradation of ubiquitinated proteins and mHtt and reduced mHtt toxicity observed here are likely synergistic effects of multiple pathways activated by SFN. On one hand, as we demonstrated, all of the three peptidase-like activities of the proteasome were elevated in the brain tissue. This may be at least partially caused by upregulation of the 26S proteasome subunit, PSMB5 (Kwak et al. 2007). On the other hand, as we showed here SFN also activates Atg, which promotes removal of the bulk unwanted substrates. Additionally, SFN activates heat shock transcription factor 1-mediated heat shock response, which may have a direct or indirect effect on the degradation and toxicity of intracellular misfolded proteins (Gan et al. 2010). Finally, SFN is an activator of the transcription factor Nrf2 and thereby induces the phase II antioxidant genes (Chapple et al. 2012). Thus, SFN can reduce free radicals-caused oxidative damage. All of these functions make the compound an attractive candidate for treating HD and other neurodegenerative disorders. The data we have provided here strongly support this possibility.
Another apparent reason for SFN to be a therapeutic reagent for HD is that it can cross the blood-brain barrier. When injected peripherally, we observed enhanced degradation of GFPu and ubiquitinated proteins by SFN in the brain cells, suggesting that the compound can enter the brain. Several studies have shown that peak plasma concentrations of SFN are attained at 1–2 h following its oral or intravenous administration in rodents (Hanlon et al. 2008, Keum et al. 2009). SFN can be detected in the brain and follow similar kinetics as plasma, with highest concentration in the brain at 2 h following oral gavage (Clarke et al. 2011). Moreover, due to the fact that SFN is a naturally-occurring product with a small molecular weight, it is very likely that human tissue cells will tolerate a high dose of SFN, leading to better treatment of HD and other neurodegenerative diseases.
Over the last years, SFN has been frequently tested in a number of clinical trials for cancer. However, it has not been seen in preclinical or clinical tests of the compound in HD. For the next step, it is important to test the effect of SFN in HD animal models, especially the mouse models as a number of HD mouse models that well-replicate many aspects of the major pathological features and symptoms of human HD have been created. Our data present here strongly suggest this necessity. If future studies prove SFN to be effective in treating HD, it will bring hope not only to HD patients but also to other polyQ repeats-caused neurodegenerative disorders. This will provide a new option to the field in utilizing and developing additional new therapies for currently intractable disorders.
Acknowledgments
We thank Dr. Robin Miskimins for critical reading of the manuscript. The animal experiments in this study were approved by the IACUC of the University of South Dakota (USD) and were in compliance with the ARRIVE guidelines. This work was supported by start-up funds from the USD (HW) and HW was supported in part by an grant R15NS071459.
Abbreviations used in this paper
- HD
Huntington’s disease
- Htt
huntingtin
- mHtt
mutant huntingtin
- polyQ
polyglutamine
- UPS
ubiquitin proteasome system
- GFP
green fluorescent protein
- GFPu
GFP UPS reporter
- DMSO
dimethyl sulfoxide
- Atg
autophagy
- SFN
sulforaphane
- Tg
transgenic
- LC3
microtubule-associated protein 1 light chain 3
- 3MA
3-methyladenine
Footnotes
Conflict of interest statement
The authors have declared no conflict of interest.
References
- Bates G, Harper P, Jones L. Huntington's Disease. 3rd edn. Oxford: Oxford University Press; 2002. [Google Scholar]
- Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292:1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- Caricchio R, Kovalenko D, Kaufmann WK, Cohen PL. Apoptosis provoked by the oxidative stress inducer menadione (Vitamin K(3)) is mediated by the Fas/Fas ligand system. Clin Immunol. 1999;93:65–74. doi: 10.1006/clim.1999.4757. [DOI] [PubMed] [Google Scholar]
- Chang DT, Rintoul GL, Pandipati S, Reynolds IJ. Mutant huntingtin aggregates impair mitochondrial movement and trafficking in cortical neurons. Neurobiology of disease. 2006;22:388–400. doi: 10.1016/j.nbd.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Chapple SJ, Siow RC, Mann GE. Crosstalk between Nrf2 and the proteasome: therapeutic potential of Nrf2 inducers in vascular disease and aging. The international journal of biochemistry & cell biology. 2012;44:1315–1320. doi: 10.1016/j.biocel.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Hsu A, Williams DE, Dashwood RH, Stevens JF, Yamamoto M, Ho E. Metabolism and tissue distribution of sulforaphane in Nrf2 knockout and wild-type mice. Pharmaceutical research. 2011;28:3171–3179. doi: 10.1007/s11095-011-0500-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Callegari E, Gloeckner CJ, Ueffing M, Wang H. Mass spectrometric identification of novel posttranslational modification sites in Huntingtin. Proteomics. 2012a;12:2060–2064. doi: 10.1002/pmic.201100380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Callegari EA, Gloeckner CJ, Ueffing M, Wang H. Prothymosin-alpha interacts with mutant huntingtin and suppresses its cytotoxicity in cell culture. The Journal of biological chemistry. 2012b;287:1279–1289. doi: 10.1074/jbc.M111.294280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Ferguson JM, Duling AJ, et al. Modeling pathogenesis of Huntington's disease with inducible neuroprogenitor cells. Cellular and molecular neurobiology. 2011;31:737–747. doi: 10.1007/s10571-011-9679-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong G, Gross K, Qiao F, et al. Calretinin interacts with huntingtin and reduces mutant huntingtin-caused cytotoxicity. Journal of neurochemistry. 2012c;123:437–446. doi: 10.1111/j.1471-4159.2012.07919.x. [DOI] [PubMed] [Google Scholar]
- Fahey JW, Talalay P. Antioxidant functions of sulforaphane: a potent inducer of Phase II detoxication enzymes. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association. 1999;37:973–979. doi: 10.1016/s0278-6915(99)00082-4. [DOI] [PubMed] [Google Scholar]
- Gamet-Payrastre L, Li PF, Lumeau S, Cassar G, Dupont MA, Chevolleau S, Gasc N, Tulliez J, Terce F. Sulforaphane, a naturally occurring isothiocyanate, induces cell cycle arrest and apoptosis in HT29 human colon cancer cells. Cancer research. 2000;60:1426–1433. [PubMed] [Google Scholar]
- Gan N, Wu YC, Brunet M, Garrido C, Chung FL, Dai C, Mi L. Sulforaphane activates heat shock response and enhances proteasome activity through up-regulation of Hsp27. The Journal of biological chemistry. 2010;285:35528–35536. doi: 10.1074/jbc.M110.152686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami A, Dikshit P, Mishra A, Mulherkar S, Nukina N, Jana NR. Oxidative stress promotes mutant huntingtin aggregation and mutant huntingtin-dependent cell death by mimicking proteasomal malfunction. Biochemical and biophysical research communications. 2006;342:184–190. doi: 10.1016/j.bbrc.2006.01.136. [DOI] [PubMed] [Google Scholar]
- Hanlon N, Coldham N, Gielbert A, Kuhnert N, Sauer MJ, King LJ, Ioannides C. Absolute bioavailability and dose-dependent pharmacokinetic behaviour of dietary doses of the chemopreventive isothiocyanate sulforaphane in rat. The British journal of nutrition. 2008;99:559–564. doi: 10.1017/S0007114507824093. [DOI] [PubMed] [Google Scholar]
- Herman-Antosiewicz A, Johnson DE, Singh SV. Sulforaphane causes autophagy to inhibit release of cytochrome C and apoptosis in human prostate cancer cells. Cancer research. 2006;66:5828–5835. doi: 10.1158/0008-5472.CAN-06-0139. [DOI] [PubMed] [Google Scholar]
- Jin YN, Yu YV, Gundemir S, Jo C, Cui M, Tieu K, Johnson GV. Impaired mitochondrial dynamics and Nrf2 signaling contribute to compromised responses to oxidative stress in striatal cells expressing full-length mutant huntingtin. PloS one. 2013;8:e57932. doi: 10.1371/journal.pone.0057932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO journal. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annual review of pharmacology and toxicology. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- Keum YS, Khor TO, Lin W, Shen G, Kwon KH, Barve A, Li W, Kong AN. Pharmacokinetics and pharmacodynamics of broccoli sprouts on the suppression of prostate cancer in transgenic adenocarcinoma of mouse prostate (TRAMP) mice: implication of induction of Nrf2, HO-1 and apoptosis and the suppression of Akt-dependent kinase pathway. Pharmaceutical research. 2009;26:2324–2331. doi: 10.1007/s11095-009-9948-5. [DOI] [PubMed] [Google Scholar]
- Kim HV, Kim HY, Ehrlich HY, Choi SY, Kim DJ, Kim Y. Amelioration of Alzheimer's disease by neuroprotective effect of sulforaphane in animal model. Amyloid : the international journal of experimental and clinical investigation : the official journal of the International Society of Amyloidosis. 2013;20:7–12. doi: 10.3109/13506129.2012.751367. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Monitoring activity and inhibition of 26S proteasomes with fluorogenic peptide substrates. Methods in enzymology. 2005;398:364–378. doi: 10.1016/S0076-6879(05)98030-0. [DOI] [PubMed] [Google Scholar]
- Kumarapeli AR, Horak KM, Glasford JW, Li J, Chen Q, Liu J, Zheng H, Wang X. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2005;19:2051–2053. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- Kwak MK, Cho JM, Huang B, Shin S, Kensler TW. Role of increased expression of the proteasome in the protective effects of sulforaphane against hydrogen peroxide-mediated cytotoxicity in murine neuroblastoma cells. Free radical biology & medicine. 2007;43:809–817. doi: 10.1016/j.freeradbiomed.2007.05.029. [DOI] [PubMed] [Google Scholar]
- Li J, Powell SR, Wang X. Enhancement of proteasome function by PA28α overexpression protects against oxidative stress. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25:883–893. doi: 10.1096/fj.10-160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li H, Li XJ. Intracellular degradation of misfolded proteins in polyglutamine neurodegenerative diseases. Brain Res Rev. 2008;59:245–252. doi: 10.1016/j.brainresrev.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Li S. Proteasomal dysfunction in aging and Huntington disease. Neurobiology of disease. 2011;43:4–8. doi: 10.1016/j.nbd.2010.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Q, Ouyang X, Schneider L, Zhang J. Reduction of mutant huntingtin accumulation and toxicity by lysosomal cathepsins D and B in neurons. Molecular neurodegeneration. 2011;6:37. doi: 10.1186/1750-1326-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zheng H, Tang M, Ryu YC, Wang X. A therapeutic dose of doxorubicin activates ubiquitin-proteasome system-mediated proteolysis by acting on both the ubiquitination apparatus and proteasome. American journal of physiology. Heart and circulatory physiology. 2008;295:H2541–H2550. doi: 10.1152/ajpheart.01052.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3:542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- Morroni F, Tarozzi A, Sita G, Bolondi C, Zolezzi Moraga JM, Cantelli-Forti G, Hrelia P. Neuroprotective effect of sulforaphane in 6-hydroxydopamine-lesioned mouse model of Parkinson's disease. Neurotoxicology. 2013;36C:63–71. doi: 10.1016/j.neuro.2013.03.004. [DOI] [PubMed] [Google Scholar]
- Ogata M, Hino S, Saito A, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Molecular and cellular biology. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravikumar B, Vacher C, Berger Z, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature genetics. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Seo H, Sonntag KC, Isacson O. Generalized brain and skin proteasome inhibition in Huntington's disease. Annals of neurology. 2004;56:319–328. doi: 10.1002/ana.20207. [DOI] [PubMed] [Google Scholar]
- Seo H, Sonntag KC, Kim W, Cattaneo E, Isacson O. Proteasome activator enhances survival of huntington's disease neuronal model cells. PloS one. 2007;2:e238. doi: 10.1371/journal.pone.0000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas AR, Hahm ER, Arlotti JA, Watkins S, Stolz DB, Desai D, Amin S, Singh SV. Chemoprevention of prostate cancer by d,l-sulforaphane is augmented by pharmacological inhibition of autophagy. Cancer research. 2013;73:5985–5995. doi: 10.1158/0008-5472.CAN-13-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Human molecular genetics. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- Wojcik C, DeMartino GN. Intracellular localization of proteasomes. The international journal of biochemistry & cell biology. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Talalay P, Cho CG, Posner GH. A major inducer of anticarcinogenic protective enzymes from broccoli: isolation and elucidation of structure. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:2399–2403. doi: 10.1073/pnas.89.6.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Orr HT. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–247. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]