Abstract
Personalized medicine holds great promise for cancer treatment, with the potential to address challenges associated with drug sensitivity and interpatient variability. Circulating tumor cells (CTC) can be useful for screening cancer drugs as they may reflect the severity and heterogeneity of primary tumors. Here we present a platform for rapidly evaluating individualized drug susceptibility. Treatment efficacy is evaluated directly in blood, employing a relevant environment for drug administration, and assessed by comparison of CTC counts in treated and control samples. Multiple drugs at varying concentrations are evaluated simultaneously to predict an appropriate therapy for individual patients.
Keywords: personalized medicine, circulating tumor cell, chemotherapy, drug resistance
1. Introduction
It is increasingly apparent that the most effective treatment for a cancer patient is a personalized approach based on predictive criteria for that individual. Traditional practice to achieve this goal has been to identify predictors of sensitivity or resistance in malignant cells. For example, it has been shown that panitumumab can be an effective therapy for colorectal cancer patients, but only in patients without KRAS mutation, which renders the treatment ineffective [1]. Thus, patients are screened for mutated KRAS prior to pantumumab treatment. Patients with non-small-cell lung cancer are evaluated for specifically mutated EGFR prior to being placed on gefitinib [2]. However, for more general chemotherapeutics, such as taxanes, no single mutation or marker has been identified that will serve as a reliable predictor of patient response. Chemotherapeutic resistance, both intrinsic and acquired, is a significant problem and is believed to result in failure in more than 90% of patients with metastatic disease [3]. In an attempt to determine patient-specific sensitivity to cytotoxic and cytostatic agents, studies have been conducted wherein tumor cells are biopsied and treated ex vivo. Unfortunately no significant benefit has been found in these types of assays because sensitivity ex vivo does not necessarily translate to a similar response in vivo [4]. This is likely due in part to spatial heterogeneity within tumors and the fact that biopsies only sample a small section of a tumor [5; 6], and in part a consequence of the environment in which the cells are treated [7].
In recent years much interest has been focused on circulating tumor cells (CTC) [8]. Many studies have found that CTC appear early in the disease, and their prevalence in blood correlates with disease severity [9; 10; 11; 12]. Clinicians are beginning to view CTC isolated from blood draws as a ‘fluid biopsy,’ something of a snapshot of the current state of a dynamic tumor, and CTC are believed to reflect in some way the breadth of tumor heterogeneity [13]. Indeed, the case has been made that CTC are the relevant cancer cell subpopulation to target for therapy based on the fact that 90% of cancer deaths are due to metastasis [14]. In addition, the circulatory system, within which cancer cells are termed CTC, is the primary route of metastasis [15]. As such, CTC are being investigated on a patient-to-patient level for characterization purposes, such as epithelial-to-mesenchymal (EMT) state [16] and detection of surface markers that correlate with specific drug response [17].
We recently reported a technique for the isolation of CTC from patient blood in a relatively simple device using off-the-shelf components [18; 19]. The device is modeled on an inflamed postcapillary venule and is functionalized with recombinant human E-selectin to rapidly bind flowing cells and anti-EpCAM antibodies to firmly adhere cancer cells. It has been suggested that E-selectin plays a role in metastasis, specifically in the extravasation of metastatic cells [20; 21; 22]. In this paper we present a technique to rapidly screen patient samples for sensitivity to multiple chemotherapeutics in a relevant setting. To accomplish this, blood samples from a patient diagnosed with metastatic cancer are split into multiple aliquots with chemotherapeutics introduced at clinically relevant dosages to treat the CTC in situ. Subsequently, the CTC are isolated from the paired aliquots and enumerated. Reductions in CTC count are interpreted as drug sensitivities. This assumption was validated using drug-sensitive cell lines spiked into normal whole blood. It is concluded that one may successfully detect significant CTC count reductions using this approach, providing a platform upon which to make informed therapeutic decisions. This technique has the potential for additional use as a companion tool to detect acquired resistance throughout treatment.
2. Materials and Methods
2.1 Cell culture
BT20 and PC3 cells were purchased from the American Type Culture Collection (ATCC, Manassas, VA). BT20 cells were grown in Eagle’s Modified Medium (ATCC) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) and 1% penicillin-streptomycin (Lonza, Basel, Switzerland). PC3 cells were grown in RPMI 1640 media (VWR, Randor, PA) supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were maintained at 37°C and 5% CO2.
2.2 Antibodies and reagents
Anti-EpCAM (clone 158210), anti-EpCAM-FITC (clone 158206) antibodies, mouse IgG2b-FITC isotype control, and recombinant human-E-selectin were purchased from R&D Systems (Minneapolis, MN). Anti-CD45-APC (clone HI30) antibody, anti-sialyl Lewisx antibody (clone CSLEX1), mouse IgG1 isotype control, and Annexin V-APC kit were obtained from BD Biosciences (San Jose, CA). Anti-EpCAM-FITC (clone HEA-125) was obtained from Miltenyi Biotec (Auburn, CA). Anti-mouse IgG1-Alexa488 secondary antibody was purchased from Life Technologies (Grand Island, NY). Halloysite nanotubes were a gift from NaturalNano (Rochester, NY). Ficoll-Paque was purchased from GE Healthcare (Waukesha, WI). Erythrocyte lysis buffer was obtained from Qaigen (Germantown, MD). Docetaxel, mitoxantrone, and calcium carbonate were purchased from Sigma Aldrich (St. Louis, MO). Doxorubicin was purchased from Sellek-Pfizer (Houston, TX). ViaCount Viability Kit was purchased from Millipore (Billerica, MA). Hank’s balanced salt solution (HBSS), phosphate-buffered saline (PBS), PBS supplemented with calcium and magnesium, and trypsin were purchased from Life Technologies (Grand Island, NY). Paraformaldehyde was acquired from Electron Microscopy Sciences (Hatfield, PA). DAPI was obtained from Vector Laboratories (Burlingame, CA). Bovine serum albumin (BSA) was purchased from Sigma Aldrich (St. Louis, MO). Dimethyl sulfoxide (DMSO) was obtained from Avantor Performance Materials Inc. (Center Valley, PA).
2.3 Preparation of selectin-functionalized microtubes
Selectin-functionalized microtubes for cancer cell isolation were prepared as previously described [18,19]. Microrenathane tubing was washed with ethanol and distilled water, then coated with poly-L-lysine (1:250) and 6.6 wt% halloysite nanotubes. The tubes were subsequently washed with distilled water and allowed to cure overnight at RT. The halloysite-coated microtubes were then perfused with 20 ug/mL Protein-G and allowed to incubate for 2 h at RT. A solution of 10 ug/mL E-selectin-Fc chimera and 50 ug/mL anti-EpCAM antibody was then pulled into the microtubes. The tubes were treated with this solution for 2 h at RT, and then blocked with 5% milk for 1 h at RT.
2.4 Determination of false positive rate and capture efficiency
For the determination of false positive rates, 10 mL of whole blood was drawn from four healthy volunteers after informed consent and split into matched 5 mL samples. One sample of each matched pair was treated with 15 ug/mL docetaxel, and the other sample was treated with vehicle control. Samples were placed in BSA-blocked test tubes and placed on a rocker at 37°C for 24 h. Buffy coats were isolated by Ficoll density centrifugation, washed, and suspended in calcium-saturated PBS. Samples were processed and stained in an identical manner to that used for CTC capture. For the determination of capture efficiency, 5 mL of whole blood was drawn from four healthy volunteers and the buffy coats isolated. The buffy coats were washed and suspended in calcium-saturated PBS. 1000 BT20 cells were added to each sample and immediately processed through the CTC capture device. Staining was carried out in an identical manner as that used for CTC capture, described below.
2.5 Spiking of cancer cell line cells into blood
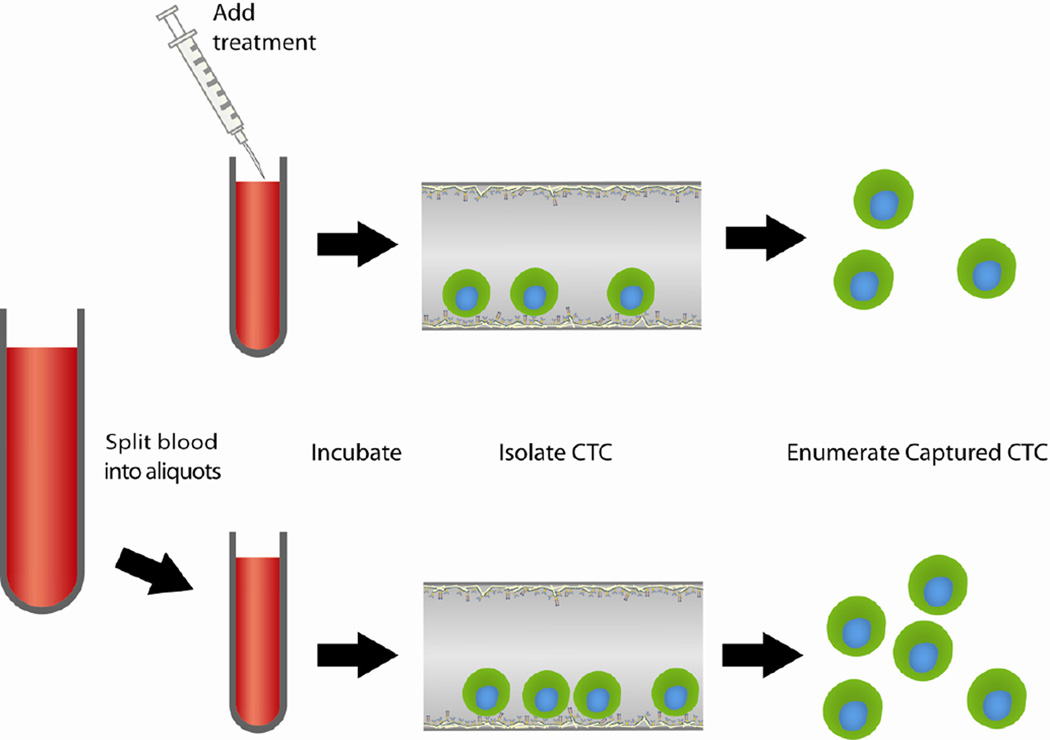
Cancer cell line cells were spiked in blood, treated with chemotherapeutic drugs, and then isolated. This process is depicted schematically in Fig. 1. Peripheral blood was drawn from healthy volunteers after informed consent and transferred to 8 mL polystyrene round-bottomed tubes (BD Biosciences) in which the interior lumen had been blocked with 3% BSA for 1 h at room temperature. 50,000 breast cancer (BT20) or prostate cancer (PC3) cells were added to 5 mL of blood. The spiked blood was then treated with vehicle control (dimethylsulfoxide, DMSO) or one of three drug dosages based on published pharmacokinetic data (20% of peak plasma concentration (PPC), 100% PPC, and 300% PPC). Breast cancer spiked blood was treated with docetaxel (1 ug/mL, 5 ug/mL, 15 ug/mL) or doxorubicin (0.2 ug/mL, 1 ug/mL, 3 ug/mL); prostate cancer spiked blood was treated with docetaxel or mitoxantrone (0.1 ug/mL, 0.5 ug/mL, 1.5 ug/mL). Peak plasma concentrations are known from previous pharmacokinetic studies [18–20]. Samples were incubated for 24 h at 37°C on a BioRad UltraRocker rocking platform (Hercules, CA).
Figure 1.
Schematic of CTC analysis protocol.
2.6 Cell isolation and enumeration from spiked blood
Buffy coat was extracted from spiked blood using a Ficoll density centrifugation as previously described [18]. Briefly, buffy coat was washed in HBSS and any remaining red blood cells were lysed with erythrocyte lysis buffer for 10 min at room temperature (RT). Cells were washed with HBSS and resuspended in 2 mL of flow buffer. Flow buffer was prepared by saturating PBS containing Ca2+ and Mg2+ with CaCO3, followed by sterile filtration through a 0.2 µm PFTE syringe filter (Millipore). Cells were perfused through the selectin-functionalized microtube device at a shear stress of 2 dyn/cm2. After flow, the microtube devices were washed with cell-free flow buffer, and adherent cells were removed from the tube by introducing trypsin for 10 min at RT. The recovered cells were plated onto glass bottom petri dishes (Grenier Bioone, Frickenhausen, Germany) and allowed to recover in media supplemented with 30% FBS for 4 h.
Cells were fixed in 4% paraformaldehyde for 45 min at RT. Plates were incubated with anti-EpCAM antibody conjugated to FITC diluted 1:100 in PBS for 1 h at RT followed by incubation with anti-CD45-APC antibody diluted 1:100 for 45 min at RT. DAPI was added and the plates were imaged using an Olympus IX81 fluorescence microscope (Center Valley, PA) or Zeiss LSM710 confocal microscope (Oberkochen, Germany) within the Life Science Core Facility at Cornell University. Cell counts were based on EpCAM and CD45 expression, nucleus size and shape, and cell size and morphology. A CTC was taken as any cell that met the following requirements: greater than 8 um in size, nonsymmetrical nucleus, positive for EpCAM, negative for CD45. Fluorescent micrographs were taken at 20 randomly selected locations within each well, and total cell counts estimated based on the total well area [18].
Processed cells that were not captured in the tube were collected, washed with PBS, and incubated with anti-EpCAM-FITC (clone 158206) for 1 h at RT. Stained cells were subsequently washed and stained with annexin-V and propidium iodide according to manufacturer instructions. Quantification was carried out using a Millipore Guava Easycyte flow cytometer.
2.7 Patient sample isolation
Two tubes of peripheral whole blood (7.5 mL per tube) was collected from patients diagnosed with stage IV cancer by BioCytics Inc. at Carolina BioOncology Institute, PLLC, after informed consent. Samples were analyzed from 3 breast cancer patients (Br1 through Br3), 2 prostate cancer patients (Pr1 and Pr2), one renal cancer patient (Re1), and one colon cancer patient (Co1). Samples were shipped overnight to Cornell University where they were split into 3 2.5 mL samples and treated with vehicle control, subclinical (20% PPC), or clinical dosages (100% PPC) of drug. Drugs were selected based on the cancer type. Prostate samples were treated with docetaxel and mitoxantrone; breast, colon, and renal samples were treated with docetaxel and doxorubicin. Samples were processed and enumerated in the precise manner as in cell spiking experiments as described above.
2.8 EpCAM and sialyl Lewisx expression following drug treatment
In order to determine whether the reduction in captured cells was from cell death or loss of adhesion ability, the expression of EpCAM and sialyl Lewisx, a selectin ligand moiety, was measured after drug treatment. BT20 cells were plated on 24 well plates. Cells were treated with the same dosages and drugs as in blood spiking experiments. The plates were incubated for 4 h at 37°C. The cells were released from the plates with enzyme-free cell dissociation buffer (Life Technologies). Cells were stained with a 1:100 dilution of anti-EpCAM conjugated with FITC (clone HEA-125), 10µg/mL anti-sLex for 30 minutes on ice. Cells were washed twice with buffer and analyzed using a flow cytometer.
2.9 Cell viability
Cells were plated on 24 well plates and treated with drug at the same concentrations used in the isolation studies for 24 h at 37°C. Cells were released from the plate with trypsin and washed with buffer. Cells were then diluted 1:10 in ViaCount viability reagent and incubated for 10 min at RT, according to the manufacturer instructions. The samples were then processed on a flow cytometer using built-in ViaCount software.
2.10 Statistics
All graphical error bars represent standard error of the mean. Significance was determined by performing an unpaired two-tailed t-test with α=0.05 in GraphPad Prism.
3. Results
3.1 BT20 and PC3 cells showed dose dependent susceptibility to chemotherapeutic drugs in vitro
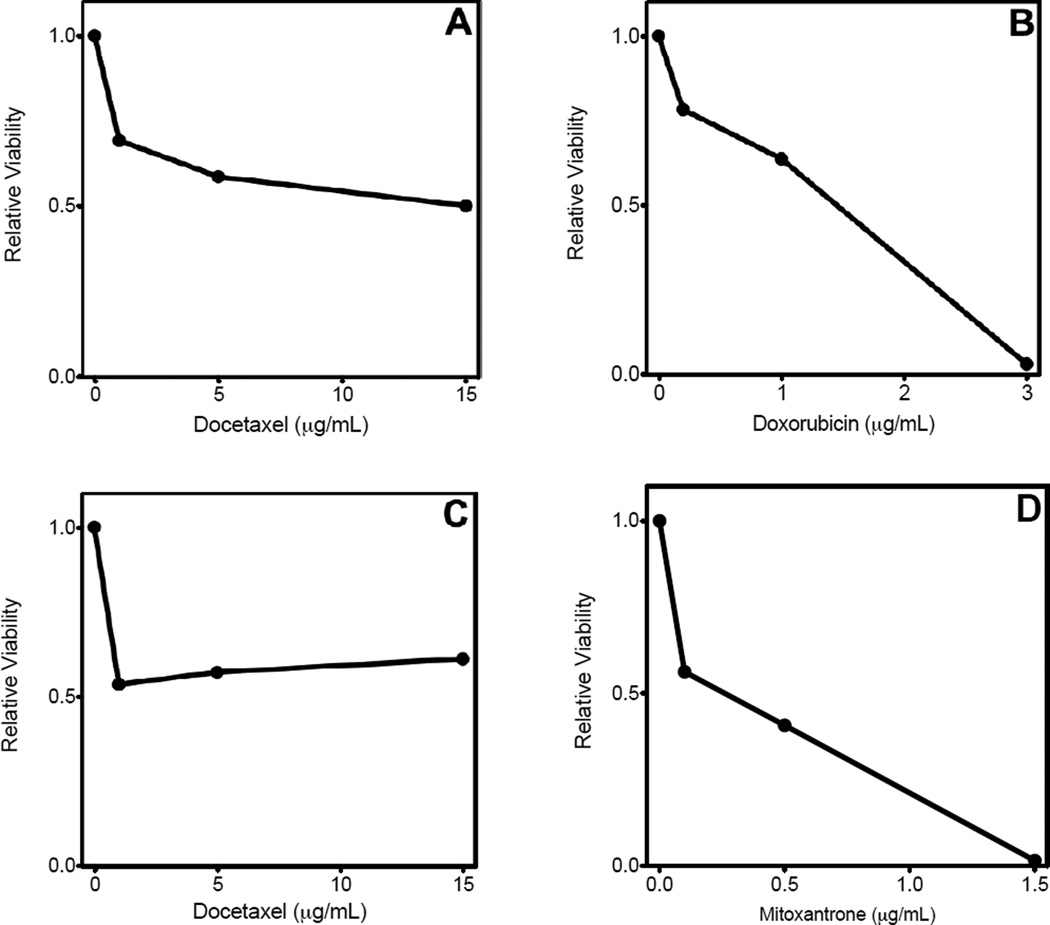
Chemotherapeutic drugs of interest (docetaxel, doxorubicin, mitoxantrone) were tested for their efficacy in vitro prior to testing the drugs in situ in whole blood (Fig. 2). Data are expressed as the number of viable cells relative to the untreated sample. BT20 showed dose-dependence, and this effect reached a plateau at ~50% viability with docetaxel. A similar effect occurred with docetaxel on PC3. Extended dose dependence was seen with doxorubicin and BT20 as well as with mitoxantrone and PC3, where viability was reduced to 3 and 1.5%, respectively.
Figure 2.
Breast and prostate cancer cell lines are sensitive to docetaxel, doxorubicin, and mitoxantrone in vitro. Results are presented as the ratio of viable cells following 24 h of drug administration to the number of viable cells in the control sample. (A) BT20 cells treated with docetaxel. (B) BT20 cells treated with doxorubicin. (C) PC3 cells treated with docetaxel. (D) PC3 cells treated with mitoxantrone. Figures are representative of two independent experiments.
3.2 Spiked BT20 cells are captured at high efficiency and identified with high specificity
1000 BT20 cells were spiked into buffy coat samples from four healthy volunteers and the mean recovery determined to be 82.0±9.4% (mean±SEM, Supplemental Fig. 1). Buffy coat samples were processed in an identical manner without spiked cells to determine false positive rates. The mean number of positively stained cells recovered from donor samples was 16.5±9.4 cells per donor with no drug treatment, and 0 cells following treatment with 15 ug/mL docetaxel in matched samples.
3.3 BT20 and PC3 cells showed drug dependent susceptibility to chemotherapeutic drugs in whole blood
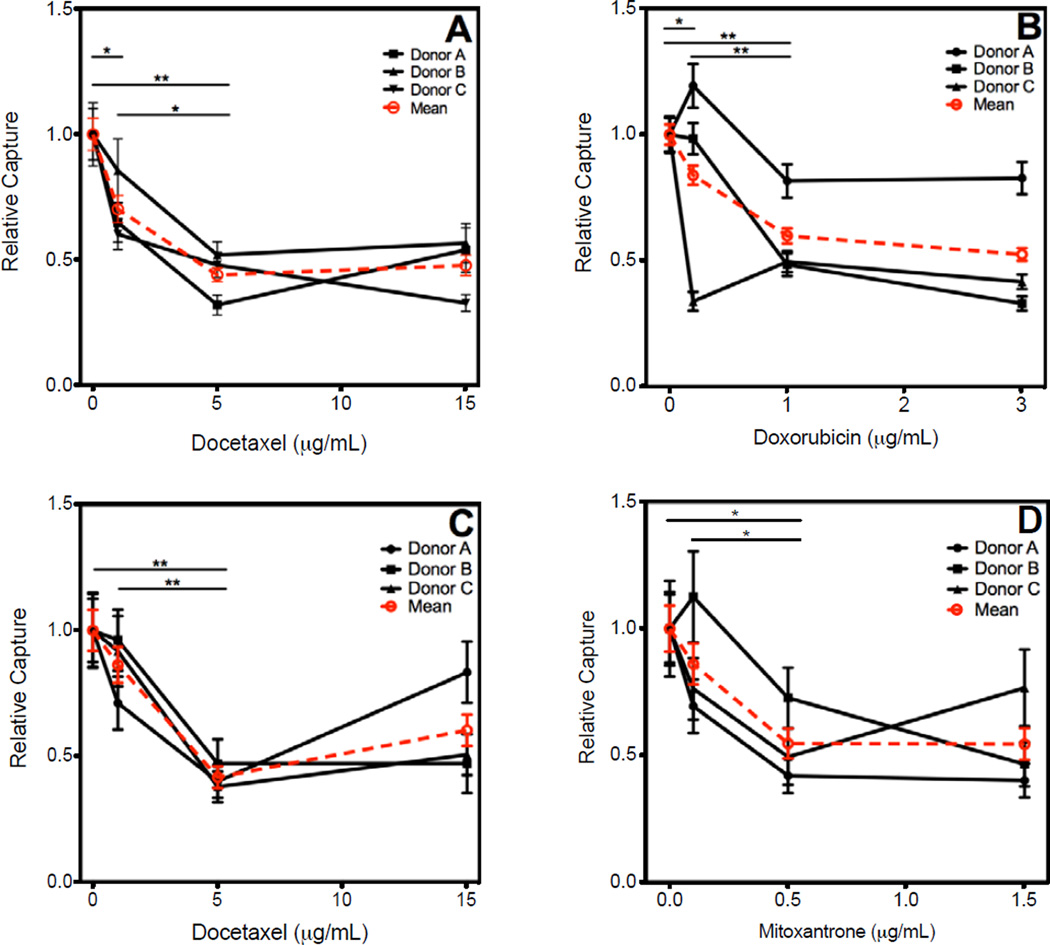
50,000 BT20 or PC3 cells were spiked into whole blood, treated with appropriate chemotherapeutic drug, and isolated as described above. The clinical dosage of each drug was taken to be the maximum plasma concentration determined by previous pharmacokinetic studies [18–20]. BT20 cells were treated with docetaxel (1 ug/mL, 5 ug/mL, 15 ug/mL) and doxorubicin (0.2 ug/mL, 1 ug/mL, 3 ug/mL). Cell counts of BT20 treated with docetaxel were reduced to 70.2±5.4% (mean±SEM), 43.9±2.7%, and 47.7±4.1% of the untreated sample. When treated with doxorubicin, cell counts decreased to 83.9±3.8%, 59.9±3.0%, and 52.4±2.5%, with respect to the untreated control. PC3 cells were treated with docetaxel and mitoxantrone (0.1 ug/mL, 0.5 ug/mL, 1.5 ug/mL). Docetaxel treatment of PC3 cells reduced the cell count to 86.3±7.1%, 41.7±4.4%, and 60.3±6.2 of control, while mitoxantrone treatment counts were 86.1±8.0%, 54.7±5.9%, and 54.5±6.2% of the untreated control (Fig. 3, Table I).
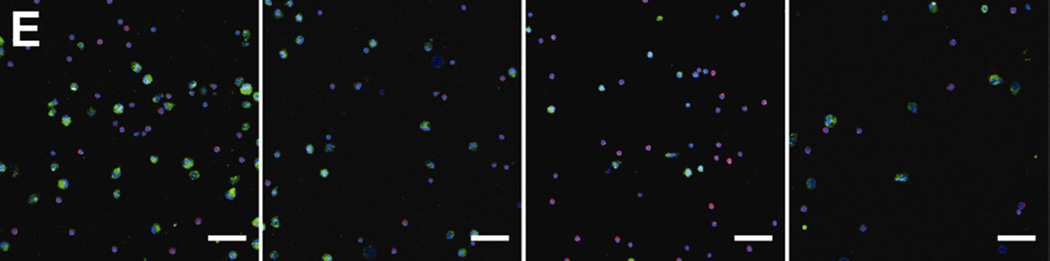
Figure 3.
Breast and prostate cancer cell lines were spiked into 5 mL whole blood and treated with various doses of drugs. Following 24 h incubation, cancer cells were isolated from the blood and enumerated. Results are presented with individual donor data represented by black lines and the mean capture by a red dotted line. (A) BT20 cells treated with docetaxel. (B) BT20 cells treated with doxorubicin. (C) PC3 cells treated with docetaxel. (D) PC3 cells treated with mitoxantrone. (E) Representative micrographs of PC3 cells captured from blood samples treated with docetaxel. Cells were stained for EpCAM (green), CD45 (red), and nucleus (DAPI). Error bars represent standard error of the mean. * P < 0.05, ** P < 0.01, *** P < 0.001; scale bar = 50 µm.
Table I.
Experimental data detailing the number of cells captured from cell spiking in whole blood. PPC = 5 ug/mL docetaxel (DT), 1 ug/mL doxorubicin (DOX), and 0.5 ug/mL mitoxantrone (MTX).
| Cell Line |
Treatment | Donor | Control | 20% PPC | 100% PPC | 300% PPC |
|---|---|---|---|---|---|---|
| BT20 | DT | A | 1522 ± 136 | 987 ± 81 | 487 ± 42 | 820 ± 114 |
| BT20 | DT | B | 2597 ± 186 | 2222 ± 289 | 1347 ± 97 | 1469 ± 173 |
| BT20 | DT | C | 1906 ± 139 | 1147 ± 82 | 914 ± 67 | 624 ± 45 |
| BT20 | DOX | A | 2276 ± 116 | 2717 ± 142 | 1857 ± 118 | 1883 ± 110 |
| BT20 | DOX | B | 2539 ± 121 | 2499 ± 103 | 1228 ± 100 | 837 ± 59 |
| BT20 | DOX | C | 1521 ± 72 | 514 ± 50 | 754 ± 51 | 632 ± 33 |
| PC3 | DT | A | 1251 ± 112 | 890 ± 105 | 500 ± 68 | 1043 ± 120 |
| PC3 | DT | B | 4260 ± 437 | 4095 ± 300 | 2006 ± 355 | 2006 ± 455 |
| PC3 | DT | C | 3992 ± 422 | 3662 ± 400 | 1512 ± 184 | 2020 ± 241 |
| PC3 | MTX | A | 6019 ± 572 | 4178 ± 494 | 2529 ± 332 | 2419 ± 332 |
| PC3 | MTX | B | 2419 ± 247 | 2721 ± 332 | 1759 ± 224 | 1127 ± 176 |
| PC3 | MTX | C | 7778 ± 1039 | 5937 ± 518 | 3848 ± 690 | 5964 ± 861 |
To confirm that the uncaptured cells were indeed rendered not viable rather than just non-adhesive, the cells from the syringe that did not stick to the tube were stained with annexin-V and propidium iodide. No significant number of viable EpCAM-positive cells were observed in any of the samples studied (data not shown).
3.4 Chemotherapeutic drug treatment did not cause loss of EpCAM or sialyl Lewisx expression
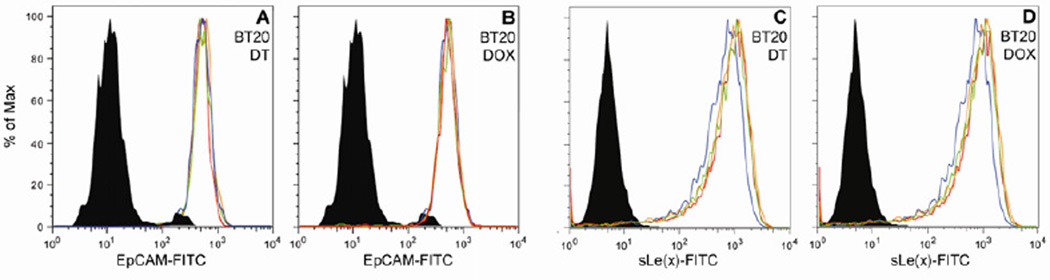
It was investigated whether the reduction of isolated cells as a result of drug treatment was due to drug efficacy or to simply loss of adhesion markers. To address this, the surface expression of EpCAM and sialyl Lewisx (sLe(x)) on BT20 cells was tested following drug treatment by flow cytometry (Fig. 4). No significant change in expression was seen post-treatment for any of the drug concentrations. This suggests that the reduced cell counts in drug treated samples were due to reduction in the number of viable cells rather than a loss of adhesion affinity per se.
Figure 4.
EpCAM and sialyl Lewisx (sLe(x)) expression of BT20 cells did not change following treatment with docetaxel or doxorubicin, as evaluated by flow cytometry. Data is presented in histograms wherein the black shaded region represents isotype control, the blue line is the control untreated sample, the red line is the 20% PPC, the orange line is 100% PPC, and the green line is 300% PPC. (A) EpCAM expression on cells treated with docetaxel (A) and doxorubicin (B) sLe(x) expression on cells treated with docetaxel (C) and doxorubicin (D). Figures are representative of three independent experiments.
3.5 Primary cancer blood samples show heterogeneous susceptibility to chemotherapeutic drugs
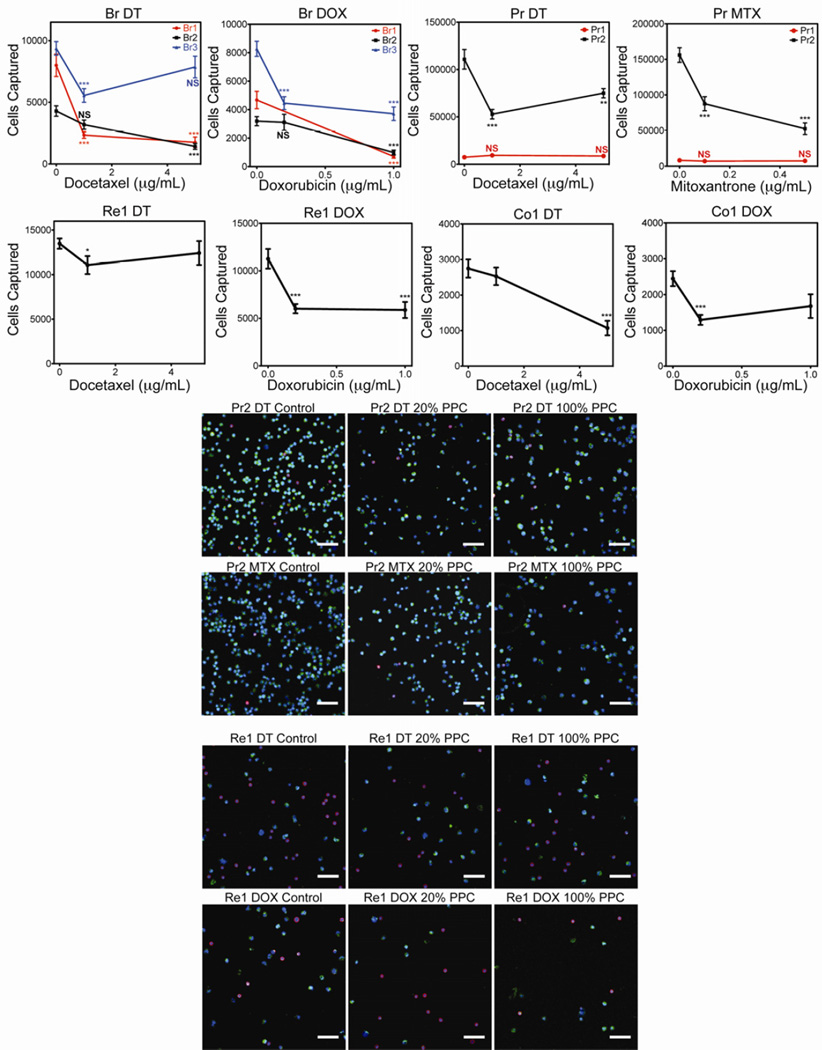
To investigate the relevance of this platform for clinical use, we tested primary blood samples from 7 cancer patients (3 breast, 2 prostate, 1 colon, 1 renal). Subclinical and clinical dosages were tested. Breast, colon, and renal blood samples were treated with docetaxel and doxorubicin, while prostate cancer blood was treated with docetaxel and mitoxantrone (Fig 5; Table II.). Overall, drug susceptibility for at least one of the drugs tested in 6 of 7 patients was detected. The CTC from 3 patients were susceptible to only one of the drugs tested (Co1, Re1, Br3) while the CTC from another 3 patients were susceptible to both (Br1, Br2, Pr2). The remaining patient (Pr1) was not susceptible to either drug tested.
Figure 5.
Patient samples were collected from three breast cancer patients (Br1, Br2, and Br3), two prostate cancer patients (Pr1 and Pr2), one renal and one colon cancer patient (Re1 and Co1, respectively). Each tube of whole blood was split into three aliquots and treated with vehicle control, 20% PPC, or 100% PPC of the appropriate drug. DT = docetaxel, DOX = doxorubicin, MTX = mitoxantrone. Error bars represent standard error of the mean. * P < 0.05, ** P < 0.01, *** P < 0.001; scale bar=50 um. Representative micrographs of CTC capture from patient Pr2 and Re1 show the reduction in CTC count following drug treatment. Scale bars = 50 µm.
Table II.
Experimental data the number of cells captured from cancer patient blood samples. PPC = 5 ug/mL docetaxel (DT), 1 ug/mL doxorubicin (DOX), and 0.5 ug/mL mitoxantrone (MTX).
| Donor | Treatment | Control | 20% PPC | 100% PPC |
|---|---|---|---|---|
| Br1 | DT | 7998 ± 905 | 2336 ± 255 | 1759 ± 423 |
| Br1 | DOX | 4672 ± 609 | -- -- | 715 ± 120 |
| Br2 | DT | 4288 ± 423 | 3188 ± 365 | 1429 ± 241 |
| Br2 | DOX | 3188 ± 318 | 3106 ± 563 | 989 ± 167 |
| Br3 | DT | 9372 ± 550 | 5552 ± 550 | 7861 ± 879 |
| Br3 | DOX | 8273 ± 533 | 4453 ± 449 | 3710 ± 473 |
| Pr1 | DT | 7476 ± 524 | 9455 ± 1093 | 8768 ± 1070 |
| Pr1 | MTX | 8300 ± 603 | 7146 ± 722 | 7531 ± 886 |
| Pr2 | DT | 110765 ± 10336 | 52826 ± 5043 | 74952 ± 4918 |
| Pr2 | MTX | 156087 ± 10276 | 87622 ± 9860 | 52606 ± 8115 |
| Re1 | DT | 13486 ± 576 | 11076 ± 1011 | 12423 ± 1345 |
| Re1 | DOX | 11269 ± 1034 | 6019 ± 486 | 5882 ± 843 |
| Co1 | DT | 2749 ± 255 | 2529 ± 247 | 1072 ± 205 |
| Co1 | DOX | 2446 ± 209 | 1292 ± 140 | 1677 ± 332 |
4. Discussion
In this paper a novel platform is presented for the prediction of cancer drug efficacy on a patient-to-patient basis, in a manner suitable for pre-screening prior to systemic administration. This platform was first characterized by spiking breast and prostate cancer cell lines at known quantities into healthy blood, creating model samples of blood containing cancer cells with well-defined susceptibilities. Based on studies of drug efficacy on these cell lines in media (Fig. 2), we were able to recapitulate the therapeutic effect in whole blood (Fig. 3). It is interesting to note that the effect of doxorubicin and mitoxantrone at their highest dosages was to eliminate nearly all cancer cells in media, however in whole blood there was no significant increase in cell elimination in response to the clinical dosage. The observed limit of efficacy to about 50% viability is likely due to various factors present in the milieu of whole blood. This underscores another advantage of our system, specifically that drug efficacy may be tested in the same environment in which it is actually administered. The dose dependence of treatment observed also demonstrates that it may be possible to identify patients that would respond to subclinical dosages at a level of efficiency equal to the maximum dosage, ameliorating detrimental side effects associated with chemotherapeutic toxicity.
A high degree of spiked cancer cell loss was observed following incubation of blood samples for 24 h on a rocker. The observed capture efficiency of 82% (Supplemental Fig. 1) strongly suggests that cell loss is due to cell death. Loss of cell viability is most likely due in part to the fact that the test tubes were thoroughly blocked with BSA and the motion of the blood from the rocker prohibited cell adhesion, contributing to in cell death via anoikis [23; 24]. Further cell death is likely the result of inhospitable factors within the whole blood collected from healthy volunteers, which would explain the relatively high degree of variability between donors (Table I). Nonetheless, we were able to detect a therapeutic reduction in cell number, which is significant due to the fact that all comparative samples were matched. This is not expected to be the case for clinical samples since the cancer cells are not foreign transplants from a different donor but native to the patient, and, additionally, primary cancer cells have been shown to avoid anoikis by various mechanisms [24; 25], and can escape immune activity [26; 27; 28].
Significant quantities of CTC were detected in blood samples of 7 patients diagnosed with metastatic cancer (Fig. 5). Of these, 6 showed a marked reduction in CTC count following enumeration, and of these 6 samples, three of them showed CTC reduction in response to one of the two drugs tested. The fact that 3 out of 7 of the samples responded differently to different drugs when treated in an identical manner otherwise suggests that these results are in fact due to sensitivities of the CTC. While all of the patient samples analyzed showed distinctly high CTC counts, one patient had exceptionally high counts: Pr2. It is interesting to note that Pr2, which showed much higher CTC counts than Pr1, also had a much higher PSA level of 2,149 ng/mL compared to 643 ng/mL for Pr1. It is important to note that this study was designed to assay drug sensitivity. Reduction in CTC capture is a likely indication of drug sensitivity in the patient, however negative results are not conclusive of drug resistance in the patient. It remains to be seen if our technique provides a true predictor of therapeutic response of primary tumor and metastatic lesions. Nevertheless, the assay is intended to be carried out in a relevant biological environment rather than an engineered environment, which may be a necessary step to development of a successful predictive clinical tool.
Clinical trials have been performed and more are in progress that monitor CTC count throughout the treatment of different cohorts of patients. It has been shown that CTC count is a reliable predictor of response and relapse [29; 30]. The combination of these clinical trial findings with the suggestion that CTC may be the most deadly subpopulation of cancer cells (in that they propagate metastasis) makes CTC a particularly promising substrate for the development of personalized medicine determination in the clinic [31]. The assay developed here has the potential to be used in a number of ways. Patient cohorts could be selected based on drug sensitivity pre-screening. Alternatively, acquired resistance to chemotherapeutics can be monitored throughout the progress of clinical trials. Furthermore, as we have shown here for the administration of docetaxel and doxorubicin to renal and colon cancers, this platform allows for rapid screening of drugs approved for some cancers but remaining to be evaluated for others. This is particularly useful considering recent observations by the Cancer Genome Atlas Research Network that cancers from different tissues can have strikingly similar genetic signatures [32; 33].
In conclusion, we have developed a novel platform for screening drug efficacies of chemotherapeutics using CTC enumeration as a diagnostic output as a predictor for drug susceptibility in individual patients. BT20 and PC3 cells were spiked into whole blood and treated with the purpose of validating this technique. The assay is carried out in a rapid procedure that, in a clinical setting, could predict a patient’s sensitivity in a single day. Two doses of two therapeutic agents were assayed simultaneously in this study; scale up to test more drugs is limited only by the quantity of blood that can be drawn from a patient and the required sample volume per test. Additionally, this technique is not limited to the isolation technique used in this paper; it can be adapted to any CTC detection or isolation method.
Supplementary Material
To determine capture efficiency, 1,000 BT20 cells were spiked into the buffy coats isolated from four healthy donors. Samples were processed through the microtube device and BT20 were enumerated in an identical manner as that used for cell line spiking and CTC isolation experiments. Dotted line shows 100% recovery. Error bars represent SEM.
Acknowledgements
This material was funded by National Institutes of Health Grant No. CA143876 (M.R.K.) and is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-0707428 (A.D.H.) and DGE-1144153 (J.R.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no conflicts of interest in this work.
References
- 1.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 2.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 3.Longley DB, Johnston PG. Molecular mechanisms of drug resistance. J Pathol. 2005;205:275–292. doi: 10.1002/path.1706. [DOI] [PubMed] [Google Scholar]
- 4.Samson DJ, Seidenfeld J, Ziegler K, Aronson N. Chemotherapy sensitivity and resistance assays: A systematic review. Journal of Clinical Oncology. 2004;22:3618–3630. doi: 10.1200/JCO.2004.04.077. [DOI] [PubMed] [Google Scholar]
- 5.Gonzalez-Garcia I, Sole RV, Costa J. Metapopulation dynamics and spatial heterogeneity in cancer. Proc Natl Acad Sci U S A. 2002;99:13085–13089. doi: 10.1073/pnas.202139299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, Halushka MK, Sukumar S, Parker LM, Anderson KS, Harris LN, Garber JE, Richardson AL, Schnitt SJ, Nikolsky Y, Gelman RS, Polyak K. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 7.Sun Y, Campisi J, Higano C, Beer TM, Porter P, Coleman I, True L, Nelson PS. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012;18:1359–1368. doi: 10.1038/nm.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hughes AD, King MR. Nanobiotechnology for the capture and manipulation of circulating tumor cells. Wires Nanomed Nanobi. 2012;4:291–309. doi: 10.1002/wnan.168. [DOI] [PubMed] [Google Scholar]
- 9.De Giorgi U, Mego M, Rohren EM, Liu P, Handy BC, Reuben JM, Macapinlac HA, Hortobagyi GN, Cristofanilli M, Ueno NT. 18F-FDG PET/CT findings and circulating tumor cell counts in the monitoring of systemic therapies for bone metastases from breast cancer. J Nucl Med. 2010;51:1213–1218. doi: 10.2967/jnumed.110.076455. [DOI] [PubMed] [Google Scholar]
- 10.Maheswaran S, Haber DA. Circulating tumor cells: a window into cancer biology and metastasis. Curr Opin Genet Dev. 2010;20:96–99. doi: 10.1016/j.gde.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10:6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 12.Miller MC, Doyle GV, Terstappen LW. Significance of Circulating Tumor Cells Detected by the CellSearch System in Patients with Metastatic Breast Colorectal and Prostate Cancer. J Oncol. 2010;2010:617421. doi: 10.1155/2010/617421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davnall F, Yip CS, Ljungqvist G, Selmi M, Ng F, Sanghera B, Ganeshan B, Miles KA, Cook GJ, Goh V. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012;3:573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wittekind C, Neid M. Cancer invasion and metastasis. Oncology. 2005;69(Suppl 1):14–16. doi: 10.1159/000086626. [DOI] [PubMed] [Google Scholar]
- 15.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 16.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyamoto DT, Lee RJ, Stott SL, Ting DT, Wittner BS, Ulman M, Smas ME, Lord JB, Brannigan BW, Trautwein J, Bander NH, Wu CL, Sequist LV, Smith MR, Ramaswamy S, Toner M, Maheswaran S, Haber DA. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes AD, Mattison J, Western LT, Powderly JD, Greene BT, King MR. Microtube device for selectin-mediated capture of viable circulating tumor cells from blood. Clin Chem. 2012;58:846–853. doi: 10.1373/clinchem.2011.176669. [DOI] [PubMed] [Google Scholar]
- 19.Hughes AD, Mattison J, Powderly JD, Greene BT, King MR. Rapid isolation of viable circulating tumor cells from patient blood samples. J Vis Exp. 2012:e4248. doi: 10.3791/4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hiratsuka S, Goel S, Kamoun WS, Maru Y, Fukumura D, Duda DG, Jain RK. Endothelial focal adhesion kinase mediates cancer cell homing to discrete regions of the lungs via E-selectin up-regulation. Proc Natl Acad Sci U S A. 2011;108:3725–3730. doi: 10.1073/pnas.1100446108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orr FW, Wang HH, Lafrenie RM, Scherbarth S, Nance DM. Interactions between cancer cells and the endothelium in metastasis. J Pathol. 2000;190:310–329. doi: 10.1002/(SICI)1096-9896(200002)190:3<310::AID-PATH525>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 22.Geng Y, Marshall JR, King MR. Glycomechanics of the metastatic cascade: tumor cell-endothelial cell interactions in the circulation. Ann Biomed Eng. 2012;40:790–805. doi: 10.1007/s10439-011-0463-6. [DOI] [PubMed] [Google Scholar]
- 23.Fiucci G, Ravid D, Reich R, Liscovitch M. Caveolin-1 inhibits anchorage-independent growth, anoikis and invasiveness in MCF-7 human breast cancer cells. Oncogene. 2002;21:2365–2375. doi: 10.1038/sj.onc.1205300. [DOI] [PubMed] [Google Scholar]
- 24.Jiang C, Wang Z, Ganther H, Lu J. Caspases as key executors of methyl selenium-induced apoptosis (anoikis) of DU-145 prostate cancer cells. Cancer Res. 2001;61:3062–3070. [PubMed] [Google Scholar]
- 25.Berezovskaya O, Schimmer AD, Glinskii AB, Pinilla C, Hoffman RM, Reed JC, Glinsky GV. Increased expression of apoptosis inhibitor protein XIAP contributes to anoikis resistance of circulating human prostate cancer metastasis precursor cells. Cancer Res. 2005;65:2378–2386. doi: 10.1158/0008-5472.CAN-04-2649. [DOI] [PubMed] [Google Scholar]
- 26.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 27.Kim R, Emi M, Tanabe K, Arihiro K. Tumor-driven evolution of immunosuppressive networks during malignant progression. Cancer Res. 2006;66:5527–5536. doi: 10.1158/0008-5472.CAN-05-4128. [DOI] [PubMed] [Google Scholar]
- 28.Pedroza-Gonzalez A, Verhoef C, Ijzermans JN, Peppelenbosch MP, Kwekkeboom J, Verheij J, Janssen HL, Sprengers D. Activated tumor-infiltrating CD4+ regulatory T cells restrain antitumor immunity in patients with primary or metastatic liver cancer. Hepatology. 2013;57:183–194. doi: 10.1002/hep.26013. [DOI] [PubMed] [Google Scholar]
- 29.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 30.Bidard F, Fehm T, Ignatiadis M, Smerage J, Alix-Panabieres C, Janni W, Messina C, Paoletti C, Muller V, Hayes D, Piccart M, Pierga J-Y. Clinical application of circulating tumor cells in breast cancer: overview of the current interventional trials. Cancer Metastasis Rev. 2013;32:179–188. doi: 10.1007/s10555-012-9398-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greene BT, Hughes AD, King MR. Circulating tumor cells: the substrate of personalized medicine? Front Oncol. 2012;2:69. doi: 10.3389/fonc.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, Yau C, Laird PW, Ding L, Zhang W, Mills GB, Kucherlapati R, Mardis ER, Levine DA. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genomic and Epigenomic Landscapes of Adult De Novo Acute Myeloid Leukemia. New England Journal of Medicine. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
To determine capture efficiency, 1,000 BT20 cells were spiked into the buffy coats isolated from four healthy donors. Samples were processed through the microtube device and BT20 were enumerated in an identical manner as that used for cell line spiking and CTC isolation experiments. Dotted line shows 100% recovery. Error bars represent SEM.