Abstract
Key message
We report the first complete set of alien addition lines of G. hirsutum . The characterized lines can be used to introduce valuable traits from G. australe into cultivated cotton.
Abstract
Gossypium australe is a diploid wild cotton species (2n = 26, GG) native to Australia that possesses valuable characteristics unavailable in the cultivated cotton gene pool, such as delayed pigment gland morphogenesis in the seed and resistances to pests and diseases. However, it is very difficult to directly transfer favorable traits into cultivated cotton through conventional gene recombination due to the absence of pairing and crossover between chromosomes of G. australe and Gossypium hirsutum (2n = 52, AADD). To enhance the transfer of favorable genes from wild species into cultivated cotton, we developed a set of hirsutum–australe monosomic alien chromosome addition lines (MAAL) using a combination of morphological survey, microsatellite marker-assisted selection, and molecular cytogenetic analysis. The amphidiploid (2n = 78, AADDGG) of G. australe and G. hirsutum was consecutively backcrossed with upland cotton to develop alien addition lines of individual G. australe chromosomes in G. hirsutum. From these backcross progeny, we generated the first complete set of chromosome addition lines in cotton; 11 of 13 lines are monosomic additions, and chromosomes 7Ga and 13Ga are multiple additions. MAALs of 1Ga and 11Ga were the first to be isolated. The chromosome addition lines can be employed as bridges for the transfer of desired genes from G. australe into G. hirsutum, as well as for gene assignment, isolation of chromosome-specific probes, flow sorting and microdissection of chromosome, development of chromosome-specific ‘‘paints’’ for fluorochrome-labeled DNA fragments, physical mapping, and selective isolation and mapping of cDNAs for a particular G. australe chromosome.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-014-2283-1) contains supplementary material, which is available to authorized users.
Introduction
Cotton, Gossypium spp., is the most important natural textile fiber, the second best potential source of plant protein and the fifth largest oilseed crop in the world (Bi et al. 1999a, b). The genus Gossypium consists of approximately 45 diploid (A–G and K genomes, 2n = 2x = 26) and five tetraploid species (AD genome, 2n = 4x = 52) (Fryxell 1992). All of these species have been classified into primary, secondary, and tertiary germplasm pools (Stewart 1995) on the basis of the relative genetic accessibility and utility of species to cotton improvement efforts (Harlan and de Wart 1971), being a valuable reservoir of agronomically useful genes and genes for resistance to pests and diseases (Endrizzi et al. 1985; Stewart 1995).
G. australe F. Mueller, a wild diploid cotton species (2n = 2x = 26, GG) native to Australia, possesses numerous economically valuable characteristics such as delayed pigment gland morphogenesis, which would be conducive to production of seeds with very low levels of gossypol as a potential source of food and feed for both human and animal consumption, resistance to pest insects (aphids and mites) and diseases (Fusarium and Verticillium wilts), and tolerance to abiotic stresses (drought), that would be useful if transferred into the most important tetraploid cultivated species, G. hirsutum L. (2n = 4x = 52, AADD). G. australe, however, belongs to the tertiary gene pool of Gossypium. These species present the greatest challenge to breeders for utilization in cotton improvement, because differences in chromosome number and structure restrict homoeologous chromosome recombination and limit introgression across genomes due to the failure of corresponding chromosomes to pair at meiosis in the interspecific hybrid with G. hirsutum. Thus, the transfer of genes from G. australe directly into G. hirsutum cannot be accomplished by conventional breeding methods such as backcrossing. One strategy is to produce monosomic alien addition lines (MAALs, 2n = 4x = 52 + II) by repeated backcrossing to an allohexaploid (2n = 6x = 78) of the G. hirsutum × G. australe F1 (2n = 3x = 39) by chromosome doubling following colchicine treatment, which can be used as a bridge to transfer desired genes from G. australe into G. hirsutum (Stewart, 1995). Over the past two decades, MAALs have been widely available in wheat (Friebe et al. 2000; Kishii et al. 2004; Wang et al. 2001; Kong et al. 2008), rice (Multani et al. 2003), potato (Ali et al. 2001), cucumber (Chen et al. 2004), tobacco (Chen et al. 2002), oat (Kynast et al. 2001), sugar beet (Reamon-Ramos and Wricke 1992; Gao et al. 2001), and rapeseed (Srinivasan et al. 1998; Budahn et al. 2008). The system has been used in numerous studies, such as chromosome pairing (Cifuentes and Benavente 2009; Molnár and Molnár-Láng 2010), recombination (Ji and Chetelat 2003; Pertuzé et al. 2003), gene transfer (Peterka et al. 2004; Fu et al. 2012; Chen et al. 1992), gene mapping (Geleta et al. 2012; Chen et al. 1992), gene tagging, genome structure, evolution, microdissection, and microcloning for chromosome-specific library construction (Shim et al. 2010; Kynast et al. 2004; Fang et al. 2004; Jiang et al. 2005; Bento et al. 2010). The application of molecular biological techniques using such stocks led to development of the field of molecular cytogenetics, allowing specific DNA sequences to be mapped to a physical chromosomal location.
In cotton, several previous reports on efforts to isolate alien addition lines of diploid Gossypium species in G. hirsutum were based mainly on classical cytogenetic analyses combined with morphological observation (Hau 1981; Rooney and Stelly 1991; Mergeai 1992). Due to the unreliability and time-consuming nature of these techniques, however, there was no complete set of diploid species MAALs in G. hirsutum reported by the end of twentieth century. With the advent of molecular genetic markers and molecular cytogenetic techniques such as genomic in situ hybridization (GISH) in cotton, and the construction of cotton molecular genetic maps, cotton researchers have new tools to identify and characterize MAALs for transferring wild traits of interest. The above techniques have facilitated the isolation of MAALs. Ahoton et al. (2003) isolated six of the possible 13 MAALs carrying G. australe chromosomes in a G. hirsutum background. Sarr et al. (2011) also identified five new MAALs of G. australe in G. hirsutum using simple sequence repeat (SSR) and GISH analysis based on the results of Ahoton et al. (2003). Zhou et al. (2004) also isolated two MAALs of G. somalense in G. hirsutum using random amplified polymorphic DNA (RAPD) and classical cytogenetic analysis.
In order to develop a complete set of monosomic alien chromosome addition lines from G. australe into G. hirsutum, we backcrossed the interspecific hexaploid hybrid of G. hirsutum × G. australe to G. hirsutum, and pentaploid plants (i.e., BC1) were obtained for a subsequent backcross with G. hirsutum. For the BC2, BC3, BC4, and BC5 progeny, GISH was employed to identify chromosome(s) from G. austral. Furthermore, a set of G. australe-specific SSR markers were developed and used to identify G. australe chromosome(s) that were present in the G. hirsutum background. As a result of this work, eleven MAALs, one double monosomic alien addition line (DMAAL), and one triple monosomic alien addition line (TMAAL) were identified and characterized.
Materials and methods
Plant materials
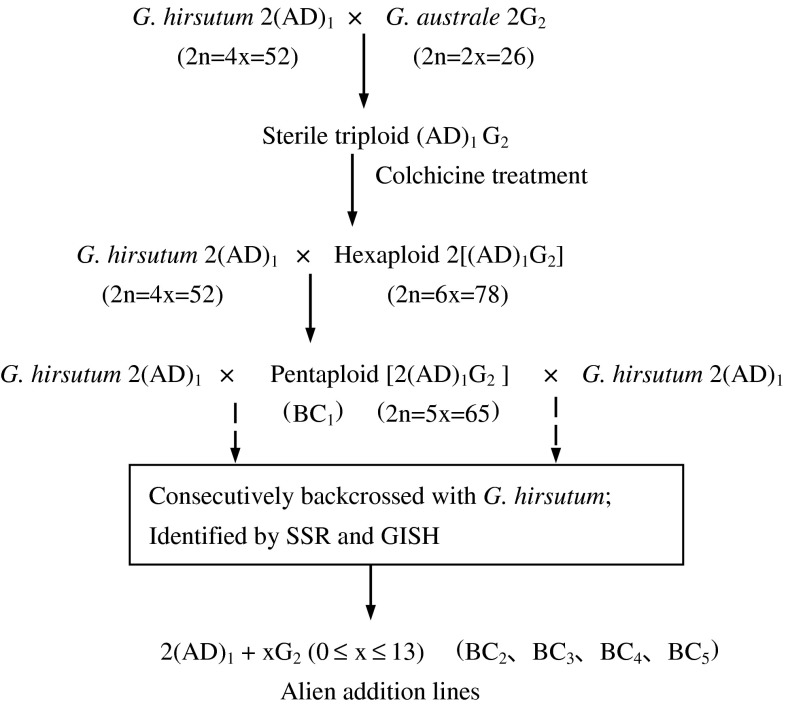
An allohexaploid cotton line was obtained from the chromosome-doubled triploid F1 of the cross G. hirsutum × G. australe. The allohexaploid (kindly provided by Dr. Curt Brubaker; Brubaker and Brown 2003) used as the paternal parent was directly crossed with G. hirsutum acc. TM-1, and seven pentaploid seeds were obtained in 2007; the six pentaploid plants obtained were grown at Jiangpu Breeding Station of Nanjing Agricultural University (JBS/NAU). When they flowered, all six pentaploid plants were backcrossed with TM-1 at the Pailou Experimental Station of Nanjing Agricultural University (PES/NAU); the plant hormone GA3 was applied to the flower base for protection from shedding when crossing. All cottonseeds obtained were sown in nursery pots and the seedlings were transplanted into clay pots in the field. In winter, all plants were moved into a glasshouse at PES/NAU. The scheme for developing the monosomic alien addition lines is shown in Fig. 1.
Fig. 1.
Scheme for the development of alien chromosome addition lines of G. australe in G. hirsutum
DNA extraction and SSR primer pair screening
Total genomic DNA was extracted from young leaves of the two parents, G. hirsutum and G. australe, the interspecific hexaploid, and individuals of the BC1, BC2, BC3, BC4, and BC5 generations as described by Paterson et al. (1993) with some modifications.
Based on the high-density tetraploid cotton linkage map constructed in our institute (Guo et al. 2007), 2,168 microsatellite (simple sequence repeat, SSR) primer pairs were selected to screen for polymorphisms between the two parental species, G. hirsutum and G. australe. All SSR primer information used in this work can be downloaded from http://www.cottonmarker.org. SSR-PCR amplifications were performed using a Peltier Thermal Cycler-EDC-810 (Eastwin, Hong Kong), and electrophoresis of the products was performed as described by Zhang et al. (2000, 2002). G. australe-specific marker primers were further employed to identify chromosomes from G. australe.
MAAL nomenclature
Since previous studies have mapped numerous SSR markers onto G. hirsutum chromosomes (Guo et al. 2007), and these SSR markers are cross-transferable in Gossypium species (Guo et al. 2006), they can be used to reveal the homoeologous relationships between G. australe and G. hirsutum chromosomes, as validated by Ahoton et al. (2003). In order to employ a consistent nomenclature as is used for wheat chromosomes (Sears 1954), the thirteen chromosomes of G. australe will be designated 1Ga–13Ga, based on SSR marker distributions on chromosomes in the Dt-subgenome of tetraploid map. There are very few chromosomal structural changes in the Dt-subgenome of cultivated tetraploid cotton. In particular, no chromosome translocations have occurred throughout evolution (Gerstel and Sarvella 1956); thus, the chromosome codes 1–13 in G. australe should correspond to the homoeologous chromosome designations in the Dt-subgenome of tetraploid cotton. The thirteen MAALs will be named MAAL-1Ga to MAAL-13Ga, Ga indicates that G. australe belongs to the G-genome species in Gossypium, and the superscript letter ‘a’ refers to the initial letter in the species name australe.
Chromosome preparation
Root tips from each individual in every BC generation were harvested from germinated seeds and pretreated in 25 μg/mL cycloheximide at room temperature for 2 h to accumulate metaphase cells. Root tips were then fixed in a solution of ethanol:acetic acid (3:1 v/v). After fixation, root tips were rinsed in distilled water and then macerated in 4 % cellulase and 1 % pectinase at 37 °C for 1.5 h and squashed in a drop of 45 % acetic acid. Slides with more than 20 good images of well-spread somatic chromosomes at metaphase were prepared and then stored at −70 °C until use. Two slides were prepared and about 20 cells were examined for each individual. The coverslips were removed from the slides, and the root tips were dehydrated through an ethanol series (70, 90, and 100 %; 5 min each) prior to use in fluorescence in situ hybridization (FISH). Meiotic chromosome spreads were prepared as per Wang et al. (2006) with several modifications. Upon the removal of the calyx and corolla, floral buds from each individual of MAALs were fixed in ethanol–acetic acid (3:1) fixative for 2–24 h at 4 °C. Next, the buds were screened for metaphase I, and several anthers from the selected bud were placed on an ethanol-washed glass slide with a drop of 45 % acetic acid (v/v), freed of debris, and squashed.
Fluorescence in situ hybridization (FISH)
Genomic DNA extracted from G. australe was labeled with digoxigenin-11-dUTP and biotin-16-dUTP (Roche Diagnostics, Mannheim, Germany) by nick translation. The sizes of the labeled DNA probe fragments were between 200 and 500 bp. Fluorescence in situ hybridization was carried out as described by Wang et al. (2006) with some modifications. Chromosomal DNA was denatured by placing the slides in 50 mL 70 % formamide, 2 × SSC at 72 °C for 2.5 min, and then immediately dehydrating them in an ethanol series at −20 °C followed by air-drying. Fifteen microliters of a mixture containing 25–50 ng labeled DNA, 50 % (w/v) dextran sulfate, 10-μg sheared salmon sperm DNA, an appropriate amount of sheared cotton DNA as blocking DNA (probe: blocking DNA = 1:100), and 1.5 μL 20 × SSC, was denatured at 97 °C for 10 min, chilled on ice, annealed at 37 °C for 1 h, and then applied to a dry slide. Following overnight incubation at 37 °C, the coverslips were removed and the slides were washed at increased stringency by rinsing at 43 °C in: 2 × SSC for 5 min (twice); 2 × SSC, 60 % formamide for 13 min; twice more in 2 × SSC for 5 min; and finally in 1× PBS for 5 min. Probes were detected with 20 μg/mL rhodamine-conjugated anti-digoxigenin antibody and avidin–fluorescein (Roche Diagnostics). Slides were stained in 4′,6-diamidino-2-phenylindole (DAPI) (Roche Diagnostics) for 10 min at room temperature, and anti-fade (Vector, USA) was applied under the coverslip. Slides were examined and more than 20 images of well-spread somatic chromosomes at metaphase stage were obtained for each individual using an Olympus BX51 fluorescence microscope. Chromosome and FISH signal images were captured using an Evolution VF CCD camera (Media Cybernetics, Bethesda, MD, USA) and merged using Image-Pro Express software (Media Cybernetics, Bethesda, MD, USA).
Evaluation of traits
The seed index was determined by weighing 100 cottonseeds collected from open bolls of TM-1 (control) and MAAL-4Ga. For the measurement of leaf chlorophyll content of TM-1 (control) and MAAL-8Ga, the youngest fully expanded leaves were collected to extract chlorophyll as described by Bao and Leng (2005). The chlorophyll a and chlorophyll b contents were measured as described by Wang and Zhang (2012) with three experimental and three biological replicates. The shape and size of the bolls were observed at 40 days post-anthesis. Fully expanded leaves from the same position on the MAALs and the control (TM-1) plants were investigated for shape and size. Floral morphological traits were observed at the time of flowering.
Results
Development of set of putative G. australe chromosome-specific SSR primers pairs
We used a total of 2,168 SSR primer pairs/combinations selected from the linkage maps of the G. hirsutum and G. barbadense genomes constructed in our institute (Guo et al. 2007) to screen for genetic polymorphisms between G. hirsutum and G. australe. The results indicated that approximately 66.2 % (1435/2168) of the SSRs detected polymorphisms between these two species. It is well known that translocations have occurred between chromosomes in the At-subgenome of the tetraploids, as well as At-subgenome and A-genome species (Gerstel and Sarvella 1956; Guo et al. 2007), while very few incidences of chromosomal structural changes have occurred in the Dt-subgenome between genomes during tetraploid formation. Hence, we selected and used only unambiguously polymorphic SSR primers in the Dt-subgenome of the tetraploids. In this study, after removing markers clustered on the chromosomes, a total of 160 G. australe-specific marker alleles that were almost evenly distributed on each Dt-subgenome chromosome were obtained to monitor chromosomes of G. australe in G. hirsutum. For each chromosome, there were 10–18 SSR primer pairs/combinations obtained based on the homoeological relationships that exist between G. australe and G. hirsutum chromosomes. These SSR markers covered from 80.5 to 97.3 % of each G. australe chromosome with a density of 6.7–15.0 cM (Table 1; Fig. 2, Fig. 1S). The G. australe-specific SSR markers have been validated in the development of hirsutum–australe alien chromosome addition lines for monitoring the numbers and identities of chromosomes of G. australe in combination with GISH.
Table 1.
SSR primers used to screen for G. australe chromosomes in the alien addition lines
| Chr | 1G | 2G | 3G | 4G | 5G | 6G | 7G | 8G | 9G | 10G | 11G | 12G | 13G |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAU458-230 | NAU3733-490 | shin-250 | NAU6992-200 | NAU3095-210 | BNL3359-200 | NAU5325-180 | BNL2597-220 | BNL686-150 | MUSS266-180 | NAU5461-210 | NAU3032-390 | NAU3685-120 | |
| NAU3346-280 | JESPR-156-90 | TMK08-190 | BNL4030-130 | NAU4907-350 | BNL2569-680 | BNL1694-240 | NAU3010-160 | NAU2739-190 | STV031-160 | dPL0522-195 | NAU3862-270 | BNL193-100 | |
| NAU3714-170 | STV030-135 | BNL3955-150 | JESPR-65-130 | NAU2232-450 | BNL3103-190 | BNL1604-110 | NAU5130-330 | NAU6701-390 | NAU2549-260 | BNL1034-200 | BNL3537-170 | NAU7471-160 | |
| NAU3056-630 | NAU4009-160 | NAU5333-150 | dPL0390-230 | JESPR236-400 | NAU2397-240 | TMB09-280 | dPL0862-220 | NAU6517-300 | NAU6720-150 | Gh508-200 | NAU4914-200 | dPL0286-220 | |
| NAU5172-180 | NAU2633-240 | JESPR101-160 | NAU3508-250 | BNL3535-145 | dc40121-180 | cot030-200 | shin0384-200 | NAU6751-135 | NAU4973-170 | Gh83-140 | NAU1558-380 | cgr5390-340 | |
| MUCS322-450 | NAU7342-270 | NAU1028-120 | MUCS271-360 | dPL0137-140 | BNL3806-160 | NAU2620-320 | cgr5503-120 | NAU5508-160 | Gh424-95 | NAU5436-370 | BNL341-160 | BNL1079-200 | |
| BNL2646-120 | Gh51-130 d | NAU6129-280 | BNL358-250 | BNL390-210 | Gh537-160 | NAU4956-280 | Gh573-145 | NAU2709-200 | TMF09-240 | BNL2632-340 | NAU7594-500 | BNL3280-200 | |
| NAU4073-180 | NAU6692-270 | JESPR-304-140 | cgr5566-155 | NAU6406-230 | JESPR229-115 | cgr5149-120 | cgr5202-150 | JESPR208-160 | NAU0922-220 | cgr5233-430 | cgr5787-120 | NAU3203-160 | |
| NAU2814-230 | NAU4022-200 | NAU2859-270 | NAU3437-250 | NAU5486-270 | dPL0702-210 | NAU2597-190 | NAU3904-240 | JESPR110-170 | NAU1280-130 | Gh434-150 | NAU3442-320 | BNL2652-280 | |
| cgr5309-140 | NAU3573-800 | NAU2898-230 | NAU7290-200 | Gh109-120 | NAU2641-180 | cgr5181-120 | JESPR291-170 | NAU3414-240 | NAU1169-300 | cgr5148-140 | NAU1119-220 | NAU6724-370 | |
| dc40046-240 | JESPR-220-150 | cgr5117-130 | MUSS519-160 | cgr6572-330 | STV031-370 | CIR414-160 | BNL2578-250 | dPL0864-200 | |||||
| NAU6728-330 | Gh229-130 | NAU3502-240 | NAU3414-250 | NAU2602-310 | NAU4925-600 | BNL2571-360 | |||||||
| cgr5510-150 | NAU864-170 | Gh132-170 | NAU3271-200 | NAU3861-450 | |||||||||
| cgr5803-120 | NAU486-230 | NAU3843-280 | |||||||||||
| cgr6240-240 | NAU429-220 | ||||||||||||
| Gh182-190 | NAU6658-180 | ||||||||||||
| JESPR122-320 | |||||||||||||
| NAU3631-270 | |||||||||||||
| Total | 10 | 12 | 10 | 11 | 18 | 12 | 10 | 10 | 13 | 11 | 16 | 13 | 14 |
| Position | 0.0–107.3 | 5.6–110.4 | 14.0–119.5 | 3.5–105.9 | 10.9–185.4 | 8.5–123.9 | 2.4–121.1 | 0.0–149.9 | 0.0–143.9 | 0–112.4 | 5.8–157.7 | 15.3–126.2 | 8.8–103.1 |
| Mean densitya | 10.7 | 8.7 | 10.6 | 9.3 | 9.7 | 9.6 | 11.9 | 15 | 11.1 | 10.2 | 9.5 | 8.5 | 6.7 |
| GDC (cM)b | 107.3 | 104.8 | 105.5 | 102.4 | 174.5 | 115.4 | 118.7 | 149.9 | 143.9 | 112.4 | 151.9 | 110.9 | 94.3 |
| PCC (%)c | 85.3 | 93.5 | 83.6 | 80.2 | 91.9 | 83.7 | 91.7 | 90.3 | 95.2 | 97.3 | 83.9 | 83.0 | 80.5 |
a Genetic distance (cM) between two adjacent markers on a chromosome
b GDC genetic distance coverage (cM)
c PCC percentage of chromosome covered by markers (%)
d Boldface type indicates the molecular markers that are deleted in the MAALs
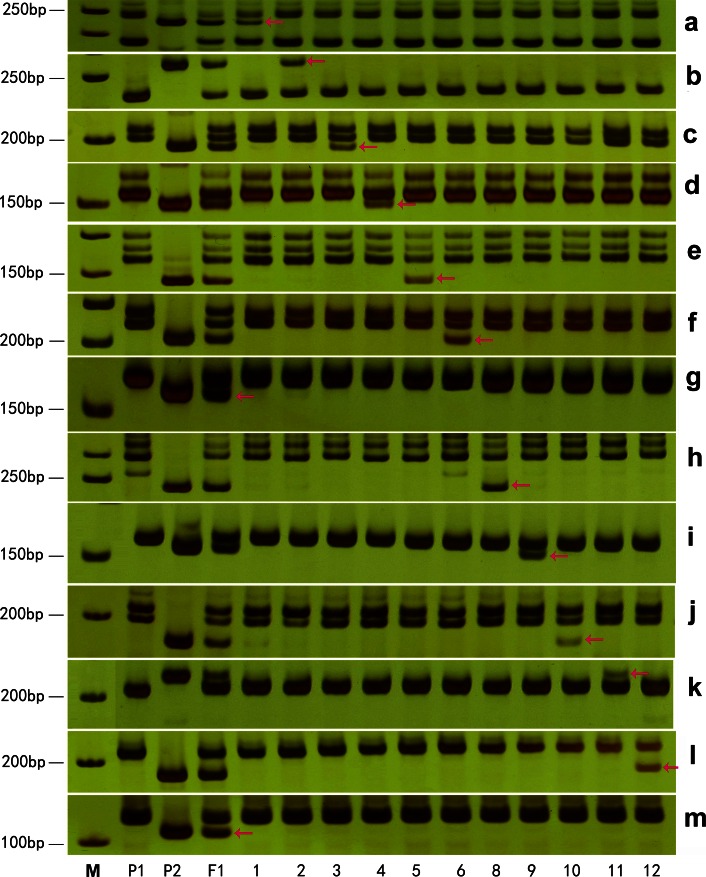
Fig. 2.
A set of G. australe-specific SSR markers were used to identify individual alien chromosomes of G. australe in G. hirsutum. a–m The G. australe-specific amplicons were obtained using 13 individual chromosome-specific primer pairs for markers; NAU5138, NAU6692, NAU805, NAU2120, Cgr5510, dPL0811, dPL0048, NAU6616, NAU3763, NAU4973, NAU2141, dPL0379, and BNL193. The chromosomes correspond to D1–D13 in cultivated tetraploid cotton. P1 G. hirsutum, P2 G. australe, F 1 the hexaploid of G. hirsutum and G. australe; 1–6 and 8–12 show that each of these plants possesses a single different individual chromosome from G. australe, corresponding to 1Ga to 6Ga, and 8Ga to 12Ga, but not 7Ga or 13Ga. M molecular size marker (50 bp ladder). Arrows (red) indicate chromosome-specific markers for G. australe
Transmission rates of alien chromosomes of G. australe in G. hirsutum
Using the pentaploid F1 of G. hirsutum × G. australe (2n = AADDGG = 78) × G. hirsutum as the maternal parent consecutively backcrossed with G. hirsutum, we found that different G. australe chromosomes had diverse transmission rates during backcrossing into the upland cotton background, and that the number of alien chromosomes drastically decreased in successive backcross generations. These results are described in detail as follows: the BC2 population consisted of 63 individuals, and the number of alien chromosomes transmitted from the BC1 (pentaploid) ranged between 1 and 10. Most plants (85.71 %) carried 1–6 alien chromosomes, and only 14.29 % of plants carried 7–10 alien chromosomes. The alien chromosome with the highest incidence was 10Ga, which was in 84.13 % of the BC2 progeny, followed by 8Ga at 42.86 %; 3Ga, 5Ga, and 12Ga at 38.10 %; 2Ga and 4Ga at 31.75 %; 11Ga at 23.81 %; and 1Ga and 6Ga at 20.63 %. The lowest incidence was 11.11 % for both 7Ga and 9Ga. The first four isolated MAALs carried G. australe chromosomes 2Ga, 5Ga, 10Ga, and 11Ga. The remaining 55 plants carried multiple alien chromosomes (Table 1S).
The above multiple alien chromosome addition lines in the BC2 generation were further backcrossed to G. hirsutum (TM-1), and a total of 164 plants were characterized by PCR and GISH. Of these, 17 (10.37 %) plants had no alien chromosomes. The number of alien chromosomes transmitted from the BC2 were 1–5. Most plants (40.24 %) carried only one alien chromosome, followed by two (21.95 %), three (12.20 %), four (12.20 %), and five (3.05 %) G. australe chromosomes. The highest incidence of an alien chromosome was 85.71 % for 10Ga (similar to transmission from the BC1), followed by 27.21 % for 12Ga. Transmission of the other chromosomes was drastically reduced to <20 %: 19.73 % for 8Ga, 19.05 % for 3Ga, and 12.24 % for 6Ga; transmission was <10 % for 1Ga, 2Ga, 4Ga, 5Ga, 7Ga, 9Ga, 11Ga, and 13Ga. In the BC3, we isolated five new MAALs for 1Ga, 3Ga, 6Ga, 8Ga, and 12Ga. MAAL-10Ga was isolated again with a rate as high as 77.27 %. The others had a very low rate of isolation (Table 2S). The remaining 81 plants were multiple alien addition lines.
The above 81 plants from the BC3 generation were further backcrossed with G. hirsutum (TM-1 as paternal parent), and a total of 237 progeny plants were characterized by PCR and GISH. Of these, 18 (7.59 %) plants had no alien chromosomes. The numbers of alien chromosomes transmitted from the BC3 were 1–4. The majority of plants (57.38 %) carried only one alien chromosome, followed by two (27.43 %), three (6.75 %), and four (0.84 %) chromosomes. The highest incidence for an alien chromosome was 91.32 %, again for 10Ga, followed by 23.74 % for 12Ga and 13.24 % for 3Ga. Transmission of the other chromosomes decreased drastically to <5 %: 1.83 % for 2Ga and 4Ga, 1.37 % for 5Ga and 9Ga, 4.11 % for 6Ga and 13Ga, and 4.57 % for 8Ga; there were no plants carrying 1Ga, 7Ga, or 11Ga. In the BC4 generation, we isolated only one new MAAL, for 9Ga, at the rate of 0.74 %. MAAL-10Ga was isolated again with a rate as high as 89.71 %. The other chromosomes had a very low rate of isolation (Table 3S). The remaining 83 plants were multiple addition lines.
The 83 plants of the BC4 generation were again backcrossed to G. hirsutum (TM-1 as paternal parent), and 32 plants in total were characterized by PCR and GISH. Of these, 15 (46.88 %) plants had no alien chromosomes. The numbers of alien chromosomes transmitted from the BC4 were 1–4. The majority of plants (34.38 %) carried only one alien chromosome, followed by two (15.63 %) and three (3.13 %) chromosomes. The highest incidence for an alien chromosome was 82.35 %, again for 10Ga as in the preceding BC generations, followed by 35.29 % for 4Ga, 11.76 % for 5Ga, and 5.88 % for both 3Ga and 13Ga. In the BC5 generation, only one new MAAL, for chromosome 4Ga, was isolated at the rate of 9.09 %. This was the same as the rates for 3Ga and 5Ga, with 10Ga transmitted at the highest rate of 72.73 % (Table 4S).
In total, the number of progeny in all backcross generations (BC2 to BC5) consisted of 496 individuals, and the numbers of alien chromosomes carried by these plants ranged from 1 to 10. Most of the plants (44.56 %) carried only a single alien chromosome, followed by two (23.19 %), and three or four (combined 16.13 %) chromosomes. A few plants (6.05 %) carried between 5 and 10 alien chromosomes. The highest incidence for any alien chromosome was for 10Ga with a rate as high as 79.03 %, followed by 12Ga at 22.18 %; 3Ga at 16.53 %, 8Ga at 13.31 %, 6Ga at 8.06 %, 5Ga at 7.86 %, 2Ga and 4Ga both at 7.66 %; 13Ga at 6.65 %, 1Ga at 3.63 %, 9Ga and 11Ga both at 3.43 %, and 7Ga with the lowest rate of 2.02 %. Of these, with the exception of 7Ga and 13Ga being multiple addition lines, we isolated 11 monosomic addition lines for G. hirsutum. The relative order of occurrence of monosomic chromosome additions isolated, from high to low, was 10Ga, 8Ga, 3Ga, 6Ga, 5Ga, 12Ga, 2Ga, 11Ga, 1Ga, 4Ga, and 9Ga, (82.81, 5.43, 3.17, 2.26, 1.81, 1.36, 0.90, 0.90, 0.45, 0.45, and 0.45 %, respectively) (Table 2).
Table 2.
Incidence of alien (G. australe) chromosomes in G. hirsutum × G. australe in the BC2 to BC5 generations
| Chromosome number | 1G | 2G | 3G | 4G | 5G | 6G | 7G | 8G | 9G | 10G | 11G | 12G | 13G | No. of individuals |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 52 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50 |
| 52 + 1 | 1 | 2 | 7 | 1 | 4 | 5 | 0 | 12 | 1 | 183 | 2 | 3 | 0 | 221 |
| 52 + 2 | 0 | 5 | 23 | 11 | 5 | 8 | 0 | 12 | 4 | 102 | 1 | 54 | 5 | 115 |
| 52 + 3 | 2 | 5 | 22 | 5 | 7 | 9 | 1 | 11 | 3 | 46 | 1 | 23 | 9 | 48 |
| 52 + 4 | 7 | 9 | 11 | 7 | 6 | 9 | 2 | 10 | 3 | 32 | 2 | 18 | 11 | 32 |
| 52 + 5 | 0 | 5 | 8 | 5 | 5 | 3 | 3 | 8 | 1 | 13 | 2 | 3 | 3 | 13 |
| 52 + 6 | 2 | 5 | 3 | 3 | 5 | 1 | 3 | 5 | 2 | 7 | 5 | 5 | 2 | 8 |
| 52 + 7 | 2 | 4 | 3 | 1 | 3 | 3 | 1 | 4 | 0 | 4 | 2 | 1 | 0 | 4 |
| 52 + 8 | 2 | 1 | 3 | 3 | 3 | 1 | 0 | 3 | 1 | 3 | 1 | 1 | 2 | 3 |
| 52 + 9 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 0 | 1 |
| 52 + 10 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 |
| Sum | 18 | 38 | 82 | 38 | 39 | 40 | 10 | 66 | 17 | 392 | 17 | 110 | 33 | 496 |
| Incidence (%) | 3.63 | 7.66 | 16.53 | 7.66 | 7.86 | 8.06 | 2.02 | 13.31 | 3.43 | 79.03 | 3.43 | 22.18 | 6.65 | |
| Monosomic addition (%) | 0.45 | 0.90 | 3.17 | 0.45 | 1.81 | 2.26 | 0.00 | 5.43 | 0.45 | 82.81 | 0.90 | 1.36 | 0.00 |
Toward development of a complete set of MAALs
We characterized a total of 496 plants using the GISH technique and 160 SSR markers to identify chromosomes of G. australe in G. hirsutum during the isolation of a set of MAALs (Table 2, Fig. 3a–l). Our results indicated that the majority of plants (221) carried one chromosome from G. australe, 115 carried two, and 48 plants carried three alien chromosomes. Among the 221 MAALs identified by SSR markers distributed on the homoeologous chromosomes in the Dt-subgenome of the tetraploids, the vast majority of the plants were MAAL-10Ga (183 plants), followed by MAAL-8Ga (12), MAAL-3Ga (3), MAAL-6Ga (5), MAAL-5Ga (4), MAAL-12Ga (3), MAAL-2Ga and MAAL-11Ga (2 plants each), and MAAL-1Ga, MAAL-4Ga and MAAL-9Ga (1 plant each). There were no MAALs isolated for chromosomes 7Ga and 13Ga but double monosomic addition lines for chromosomes 13Ga with 5Ga, and triple monosomic addition lines for chromosomes 7Ga with 8Ga and 10Ga were identified in G. hirsutum.
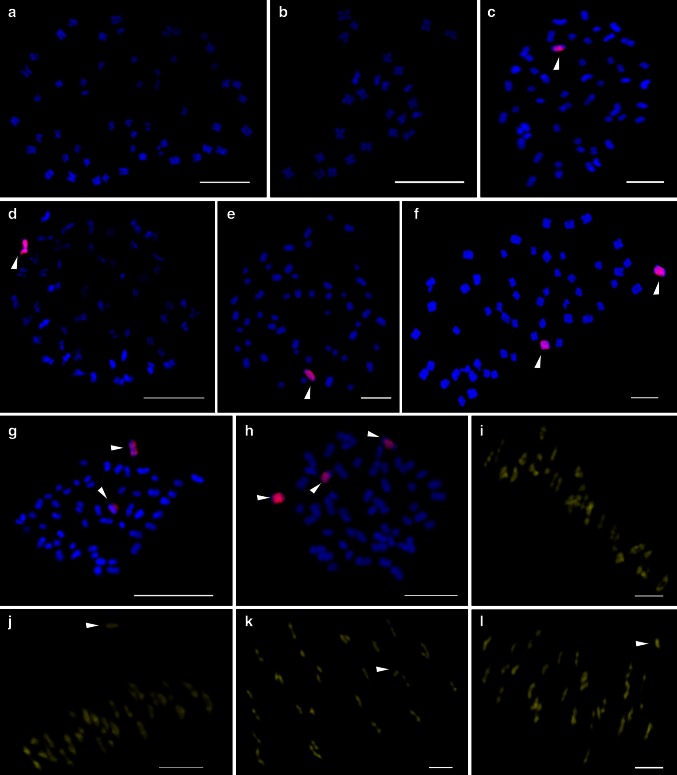
Fig. 3.
a–l Genomic in situ hybridization of the putative alien chromosomes of G. australe in the G. hirsutum background shows the chromosomal constitutions. a Mitotic chromosome spread of the 52 chromosomes of G. hirsutum. b Mitotic chromosome spread of the 26 chromosomes of G. australe. c, d and e Mitotic chromosome spread showing the 52 G. hirsutum (blue) chromosomes and individual chromosomes 3Ga, 6Ga, and 10Ga of G. australe (red, white arrowhead), respectively. f, g and h Mitotic chromosome spread showing the 52 G. hirsutum (blue) chromosomes and two (2Ga, 10Ga), two (10Ga, 13Ga) and three (7Ga, 8Ga, 10Ga) chromosomes of G. australe (red, white arrowhead), respectively. i Meiotic chromosome spread showing 26 bivalents of G. hirsutum. j, k and l Meiotic chromosome spread showing 26 bivalents of G. hirsutum plus 1 univalent of chromosomes 5Ga, 6Ga, and 10Ga from G. australe (white arrowhead), respectively. Scale bar 3 μm
In conclusion, GISH analysis and SSR markers distributed on the homoeologous chromosomes of the Dt-subgenome of tetraploid cotton enabled the isolation of the first complete set of alien addition lines of G. australe chromosomes in G. hirsutum. Eleven of these lines are monosomic additions, while chromosomes 7Ga and 13Ga are in multiple addition lines. The MAALs for 1Ga and 11Ga are the first to be reported for these chromosomes.
Transmission rates of chromosome 10Ga in G. hirsutum through the male and female gametes and in different generations
In this study, interestingly, we found that chromosome 10Ga from G. australe showed a transmission rate of >90 % in G. hirsutum. To determine which gamete transmitted 10Ga, the 10Ga chromosome addition lines were reciprocally crossed with TM-1. The results indicated that the 10Ga chromosome was mainly transmitted through the female gamete, and its transmission rate was as high as 96.5 %, while the transmission rate was only 5.7 % through the male gamete (Table 3). Theoretically, there would be 5.5 % disomic 10Ga addition lines in the self-pollinated progeny of monosomic 10Ga addition lines. In fact, to date, we have been unable to isolate any disomic 10Ga chromosome addition lines from self-pollinating the monosomic 10Ga addition lines.
Table 3.
Transmission rates of alien chromosome 10Ga from G. australe through the female and male gametes in G. hirsutum, respectively
| Cross | No. of plants | Total | |
|---|---|---|---|
| 2n (%) | 2n + 1 (%) | ||
| (2n + 1) × 2n a | 4 (3.5) | 109 (96.5) | 113 |
| 2n a × (2n + 1) | 50 (94.3) | 3 (5.7) | 53 |
a2n denotes the recurrent parent, G. hirsutum acc. TM-1
To elucidate the pattern of transmission of chromosome 10Ga in successive backcross generations, we observed the frequencies and incidences of chromosome 10Ga in three generations from the BC2 to BC4. The results indicated that the frequencies and incidences of chromosome 10Ga decreased slightly from BC2 to BC4, while the frequencies of chromosome 10Ga inheritance were as high as 100 % when the 10Ga addition lines were self-pollinated at each BC generation (Table 4).
Table 4.
Transmission rates of the alien chromosome 10Ga in generations BC2 to BC4
| Lineage | No. of progenies from MAAL-10Ga self-pollinated | Total | |
|---|---|---|---|
| 2n (%) | 2n + 1 (%) | ||
| BC2F1 | 3 (6.4) | 44 (93.6) | 47 |
| BC2F2 | 0 (0.00) | 8 (100.0) | 8 |
| BC3F1 | 10 (12.2) | 72 (87.8) | 82 |
| BC3F2 | 0 (0.00) | 18 (100.0) | 18 |
| BC4F1 | 27 (14.7) | 157 (85.3) | 184 |
| BC4F2 | 0 (0.00) | 30 (100.0) | 30 |
Morphological traits of MAALs
Most of the MAALs exhibited slow growth rates compared to the G. hirsutum accession TM-1. In addition, their morphological traits differed from one another, and they also differed from the normal diploid G. australe in fundamental morphological features such as growth habit, height, shape and length of leaves, and pollen fertility (Tables 5, 6; Figs. 4a–b, 5, 6). In the present experiment, all the MAALs isolated had slow growth rates except MAAL-9Ga and MAAL-4Ga, which were as vigorous as G. hirsutum TM-1.
Table 5.
Morphological characters in eleven MAALs
| Trait | P1 | P2 | Monosomic alien addition line | ||
|---|---|---|---|---|---|
| TM-1 | G. australe | 1G | 2G | 3G | |
| Petal color | Creamy | Mauve | Creamy | Creamy | Creamy |
| Pollen color | Creamy | Gray | Creamy | Creamy | Creamy |
| Petal length (cm) | 4.44 ± 0.14 | 3.46 ± 0.07 | 4.41 ± 0.10 | 3.77 ± 0.13 | 4.68 ± 0.16 |
| Petal width (cm) | 4.15 ± 0.11 | 3.35 ± 0.08 | 4.44 ± 0.15 | 3.90 ± 0.15 | 4.00 ± 0.11 |
| Style length (cm) | 3.50 ± 0.11 | 2.12 ± 0.04 | 2.80 ± 0.05 | 2.66 ± 0.10 | 3.16 ± 0.17 |
| Stigma length (cm) | 1.00 ± 0.07 | 0.56 ± 0.05 | 0.80 ± 0.05 | 0.81 ± 0.04 | 0.87 ± 0.07 |
| Another number | 125.20 ± 2.39 | 101.40 ± 2.41 | 111.40 ± 3.97 | 65.50 ± 3.82 | 105.40 ± 3.65 |
| Pedicel length (cm) | 1.52 ± 0.08 | 0.32 ± 0.04 | 1.54 ± 0.11 | 0.42 ± 0.12 | 1.02 ± 0.08 |
| Calyx teeth length (cm) | 0.24 ± 0.08 | 0.74 ± 0.08 | 0.36 ± 0.15 | 0.26 ± 0.10 | 0.22 ± 0.07 |
| Leaf length (cm) | 12.47 ± 0.70 | 8.40 ± 0.36 | 14.00 ± 0.87 | 13.83 ± 0.79 | 13.93 ± 0.55 |
| Leaf width (cm) | 15.57 ± 0.86 | 7.93 ± 0.93 | 20.37 ± 2.23 | 17.17 ± 0.97 | 13.43 ± 0.81 |
| Petiole length (cm) | 11.30 ± 1.91 | 3.40 ± 0.95 | 13.60 ± 4.01 | 9.63 ± 2.12 | 8.40 ± 1.51 |
| Trait | Monosomic alien addition line | ||||
|---|---|---|---|---|---|
| 4G | 5G | 6G | 8G | 9G | |
| Petal color | Creamy | Creamy | Creamy | Creamy | Creamy |
| Pollen color | Creamy | Creamy | Creamy | Creamy | Creamy |
| Petal length (cm) | 4.14 ± 0.10 | 3.78 ± 0.16 | 5.04 ± 0.15 | 4.21 ± 0.12 | 4.28 ± 0.12 |
| Petal width (cm) | 3.78 ± 0.16 | 3.76 ± 0.16 | 4.78 ± 0.13 | 3.79 ± 0.12 | 3.99 ± 0.11 |
| Style length (cm) | 2.67 ± 0.13 | 2.76 ± 0.18 | 3.62 ± 0.08 | 3.42 ± 0.19 | 3.22 ± 0.20 |
| Stigma length (cm) | 0.82 ± 0.08 | 0.80 ± 0.04 | 1.28 ± 0.08 | 0.96 ± 0.09 | 0.87 ± 0.04 |
| Another number | 125.60 ± 2.41 | 70.40 ± 3.91 | 122.60 ± 5.73 | 114.60 ± 2.70 | 112.00 ± 2.55 |
| Pedicel length (cm) | 0.58 ± 0.08 | 0.56 ± 0.09 | 1.68 ± 0.08 | 1.54 ± 0.05 | 1.00 ± 0.20 |
| Calyx teeth length (cm) | 0.24 ± 0.08 | 0.27 ± 0.12 | 0.22 ± 0.09 | 0.28 ± 0.10 | 0.24 ± 0.10 |
| Leaf length (cm) | 8.07 ± 0.40 | 11.23 ± 0.55 | 8.30 ± 0.26 | 12.10 ± 0.53 | 9.07 ± 0.42 |
| Leaf width (cm) | 9.53 ± 0.56 | 13.00 ± 1.50 | 8.97 ± 0.65 | 13.77 ± 0.51 | 11.20 ± 0.98 |
| Petiole length (cm) | 14.60 ± 0.12 | 8.42 ± 1.72 | 9.67 ± 0.60 | 13.60 ± 0.40 | 13.01 ± 1.33 |
| Trait | Monosomic alien addition line | ||||
|---|---|---|---|---|---|
| 10G | 11G | 12G | |||
| Petal color | Creamy | Creamy | Creamy | ||
| Pollen color | Creamy | Creamy | Light yellow | ||
| Petal length (cm) | 4.78 ± 0.11 | 4.76 ± 0.15 | 4.90 ± 0.18 | ||
| Petal width (cm) | 4.36 ± 0.09 | 4.40 ± 0.10 | 4.04 ± 0.17 | ||
| Style length (cm) | 3.06 ± 0.17 | 3.11 ± 0.14 | 3.00 ± 0.16 | ||
| Stigma length (cm) | 0.86 ± 0.05 | 0.87 ± 0.06 | 0.71 ± 0.09 | ||
| Another number | 121.40 ± 3.13 | 82.00 ± 3.40 | 102.80 ± 1.64 | ||
| Pedicel length (cm) | 1.08 ± 0.08 | 1.11 ± 0.09 | 1.00 ± 0.12 | ||
| Calyx teeth length (cm) | 0.18 ± 0.09 | 0.49 ± 0.10 | 0.26 ± 0.10 | ||
| Leaf length (cm) | 13.83 ± 0.71 | 13.20 ± 0.65 | 9.43 ± 0.75 | ||
| Leaf width (cm) | 15.73 ± 0.81 | 17.10 ± 0.84 | 11.50 ± 0.50 | ||
| Petiole length (cm) | 7.01 ± 2.03 | 6.02 ± 1.64 | 9.42 ± 0.91 | ||
Table 6.
The unique traits of the monosomic alien addition lines
| Alien chromosome added | Unique traits |
|---|---|
| 1Ga | Oblate boll and grooved leaves with irregularly serrate margins |
| 2Ga | Small flowers with few anthers |
| 3Ga | Very high lint percentage and normal fiber length |
| 4Ga | Normal fiber length and normal plant shape |
| 5Ga | Small bracts and flowers. Compact plant type and short fibers ca. 25 mm in length |
| 6Ga | Brown fibers |
| 7Ga | Pink flowers and hard stems |
| 8Ga | Dark green leaf with high concentration of chlorophyll a and chlorophyll b |
| 9Ga | Small bracts and small flowers |
| 10Ga | High rate of alien chromosome transmission |
| 11Ga | Completely sterile |
| 12Ga | Yellow anthers |
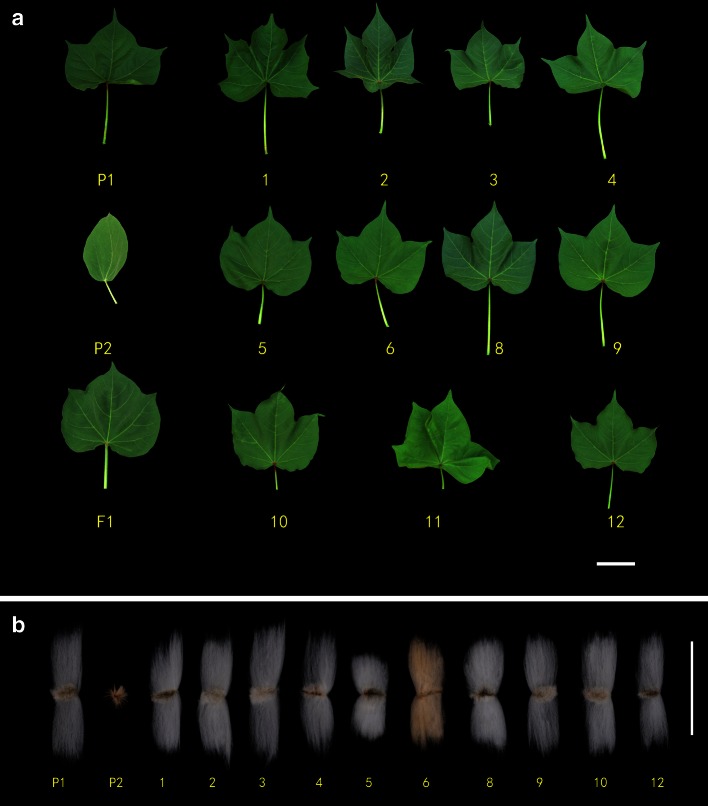
Fig. 4.
a–b Leaf shapes and fiber traits of MAALs of G. australe individual chromosomes in G. hirsutum. P1 G. hirsutum, P2 G. australe, F 1 the hexaploid of G. hirsutum and G. australe. 1–6 and 8–12 are fibers from plants possessing single different individual chromosomes from G. australe, corresponding to 1Ga to 6Ga, and 8Ga to 12Ga (but not 7Ga and 13Ga). The absence of number 11 in B is because there were no bolls set due to the complete sterility of MAAL-11Ga. Scale bar 50 mm
Fig. 5.
a–e Flower traits are shown for MAALs of G. australe individual chromosomes in G. hirsutum. a–d P1 G. hirsutum, P2 G. australe, F 1 the hexaploid of G. hirsutum and G. australe. 1–6 and 8–12 are plants that carry a single different individual chromosome from G. australe, corresponding to 1Ga to 6Ga and 8Ga to 12Ga; 7 is the triple alien addition line of chromosomes 7Ga, 8Ga, and 10Ga. e Except for the absence of 7Ga, 11Ga, and 13Ga, the numbers are the same as in a–d. Scale bar 50 mm
Fig. 6.

Whole plant traits of G. hirsutum acc. TM-1 (left), G. australe (center), and MAAL-11Ga (right)
MAAL-1Ga plant leaves were observed to be grooved with irregularly serrate margins (Fig. 4a), the leaf sizes were larger than those of TM-1, and its bract calyx had more and longer calyx teeth (Fig. 5d, Table 5). The plants grew slowly.
MAAL-2Ga plants had the lowest number of anthers among the 11 MAALs (Tables 5, 6). The flowers with shorter pedicels were significantly smaller than observed in the TM-1 plants. The leaf sizes were slightly larger as well, and the plants grew slowly.
MAAL-3 Ga plants produced normal fiber lengths and had a very high lint percentage due to the extremely low seed index. The flower sizes were similar to those of the TM-1 plants (Table 5).
MAAL-4Ga plants had fewer hairs on the stems and produced round bolls with larger seeds; the seed index was 13.17 g, which was a significant increase of 26.27 % over TM-1 (10.43 g). The flowers and leaves were slightly smaller than that of TM-1 (Table 5).
MAAL-5Ga plants displayed a compact plant type with shorter fibers (Fig. 4b), shorter pedicels and petioles, shorter fruit branches, and smaller flowers (Fig. 5a–e) than those of TM-1. These plants grew slowly. MAAL-6Ga plants were easily distinguishable by its brown-colored fibers and the fact that it had the largest flowers among the 11 MAALs (Figs. 4b, 5a–c). The leaves were smaller than those of TM-1 plants and these plants grew slowly (Table 5). MAAL-7Ga could be identified by the pink flower; even though this chromosome has not been isolated in a single MAAL (Fig. 5a–c), as neither the monosomic alien addition lines that carried 8Ga or 10Ga had a pink flower. Thus, based on the morphological traits, the triple monosomic alien addition line should carry chromosomes 7Ga, 8Ga, and 10Ga simultaneously.
MAAL-8Ga plants were easily distinguishable by its dark green leaves; the chlorophyll a and chlorophyll b contents were as high as 1065.39 and 417.01 mg/g, respectively, significantly higher than the values for TM-1, which were 736.1 and 281.14 mg/g (Table 7). These plants grew slowly.
Table 7.
Photosynthetic pigment contents in G. hirsutum and three MAALs
| Line | Chlorophyll a | Chlorophyll b | Carotenoids |
|---|---|---|---|
| TM-1 | 728.82 ± 8.05 | 288.29 ± 14.57 | 65.79 ± 1.14 |
| MAAL-8Ga (1) | 1,118.35 ± 26.28** | 414.75 ± 22.84** | 101.93 ± 16.33** |
| MAAL-8Ga (2) | 1,109.77 ± 31.15** | 409.09 ± 26.87** | 106.03 ± 19.81** |
| MAAL-8Ga (3) | 1,071.28 ± 6.40** | 402.18 ± 23.39** | 102.35 ± 1.14** |
** Significance at the 1 % level
MAAL-9Ga plants had longer fruit-branch nodes than those of TM-1 plants, and they were pagoda-like in shape. The leaves were smaller than those of TM-1 plants (Table 5).
MAAL-10Ga plants grew slowly and its alien chromosome was transmitted at the high rate of 96.5 %. The flowers were slightly larger than those of the TM-1 plants, and the leaves were similar in size to that of the TM-1 plant (Table 5).
The MAAL-11Ga plant had large light-green leaves with folded lobes, and showed complete male and female sterility—the anthers did not disperse pollen, and no bolls were set when it was used as a maternal parent (Figs. 4a, 6). Chromosome 11Ga seems to carry genes that cause sterility. The flowers were slightly larger than those of the TM-1 plant, while the leaves were larger (Table 5). This plant grew very slowly.
MAAL-12Ga plants were highly distinguishable because of their small, cone-shape bolls (Fig. 5e). The flowers were slightly larger than those of the TM-1 plants, while the leaves were smaller (Table 5). These plants also grew slowly.
Discussion
Challenges in developing hirsutum wild species alien chromosome addition lines
Monosomic alien addition lines (MAALs), which carry an additional chromosome from a related species through interspecific/intergenomic hybridization, have become powerful tools for genetic and genomic research. These MAALs can also be used to improve the process of genetic introgression of desirable genes from donor species into recipient species through the production of chromosome substitution and translocation lines. However, the development of cotton MAALs is an extremely laborious and frustrating process using conventional cytogenetic techniques, because the chromosomes of cotton are numerous (2n = 52) and are too small for reliable identification and characterization in mitotic and meiotic cells (genome size is approximately 2.5 Gb). At present, incomplete sets of MAALs from several wild diploid Gossypium species have been developed in G. hirsutum; however, no complete set of MAALs has been reported, even though many efforts have been made since the early 1980s (Hau 1981; Rooney and Stelly 1991; Mergeai 1992). With the advent of molecular markers and genomic in situ hybridization (GISH) techniques utilized in cotton, this situation has changed drastically. Through the combination of SSR markers and GISH analysis, Ahoton et al. (2003) isolated six of the possible 13 MAALs carrying G. australe chromosomes in a G. hirsutum background. Subsequently, Sarr et al. (2011) also isolated five new MAALs of G. australe in G. hirsutum using the methods of Ahoton et al. (2003). Zhou et al. (2004) also isolated two MAALs of G. somalense chromosomes in G. hirsutum using random amplified polymorphic DNA (RAPD) and classical cytogenetic analysis. In this study, we have developed the first complete set of alien chromosome addition lines in cotton; 11 of these lines are monosomic additions, with chromosomes 7Ga and 13Ga present in multiple addition lines. GISH allowed us to show that one or more alien chromosomes were present, and chromosome-specific markers allowed us to identify the alien chromosomes by genotyping in the cultivated cotton background. Our results indicated that the combination of SSR markers and GISH analysis can facilitate the efficient establishment of monosomic alien addition lines in cotton.
During the isolation of cotton MAALs, we found that most of the alien (G. australe) chromosomes were transmitted through the maternal parent at a very low rate, while one chromosome, 10Ga, was transmitted maternally at an extremely high frequency. Differences in the transmission rates of individual alien chromosomes in monosomic alien addition lines have been reported previously in tomato by Ali et al. (2001). Although there is no clear explanation for the phenomenon, Ali et al. (2001) believed that different genetic backgrounds might influence the transmission of alien chromosomes, so they proposed that the selection of proper backcross populations was important. In our study, alien chromosome transmission biases and different transmission rates were observed, as have been previously reported in cotton (Lopez-Lavalle and Brubaker 2007; Rooney and Stelly 1991) and tomato (Ali et al. 2001). To further facilitate the elucidation of the genetic roles for each alien chromosome in upland cotton in the future, the standard genetic line TM-1 was used as the sole recurrent parent to consecutively backcross to the progeny of G. hirsutum and G. australe, leading to a uniform genetic background in the MAALs. But the genetic background of TM-1 may have differential effects on the transmission of individual chromosomes of G. australe in G. hirsutum during successive backcrossing; for example, no single chromosome addition lines for 7Ga and 13Ga were isolated, because single 7Ga and 13Ga chromosomes were either not transmitted to the progeny or were always transmitted with other alien chromosomes in the TM-1 background. If so, the isolation of monosomic alien addition lines for chromosomes with low transmission rates will necessitate using different parental lines during backcrossing. For isolation of MAALs of 7Ga and 13Ga to complete the set of MAALs of G. australe in G. hirsutum, it will be necessary to test alternative recurrent parents in the future.
Another question is whether homoeologous recombination has occurred between chromosomes of G. australe and G. hirsutum during the development of the MAALs. Theoretically, if homoeologous chromosomes of G. australe and G. hirsutum were to pair at meiosis, recombination could possibly occur. Previous cytological investigations on chromosomal configuration in pollen mother cells at meiosis in a triploid F1 showed that there was a very low rate of chromosome pairing between AADD and G-genome chromosomes (Liang et al. 1997), indicating the distant relationship that exists between chromosomes from the AADD and G genomes. In addition, cytological investigation into the behavior of monosomic alien chromosomes showed that very little or no pairing with AD chromosomes was observed in this study (Fig. 3j–l), implying that there is very little or no homoeologous recombination between them. However, homoeologous recombination in the aneuploid progeny still cannot be completely ruled out. Furthermore, it could be speculated that recombination is more likely to happen between Ah and G chromosomes than between Dh and G chromosomes due to G being phylogenetically closer to Ah than to Dh. These hypotheses are based on the results of GISH (Guan et al. 2008) and molecular phylogenetic evolution analyses for the Gossypium genus by Wendel and Cronn (2003).
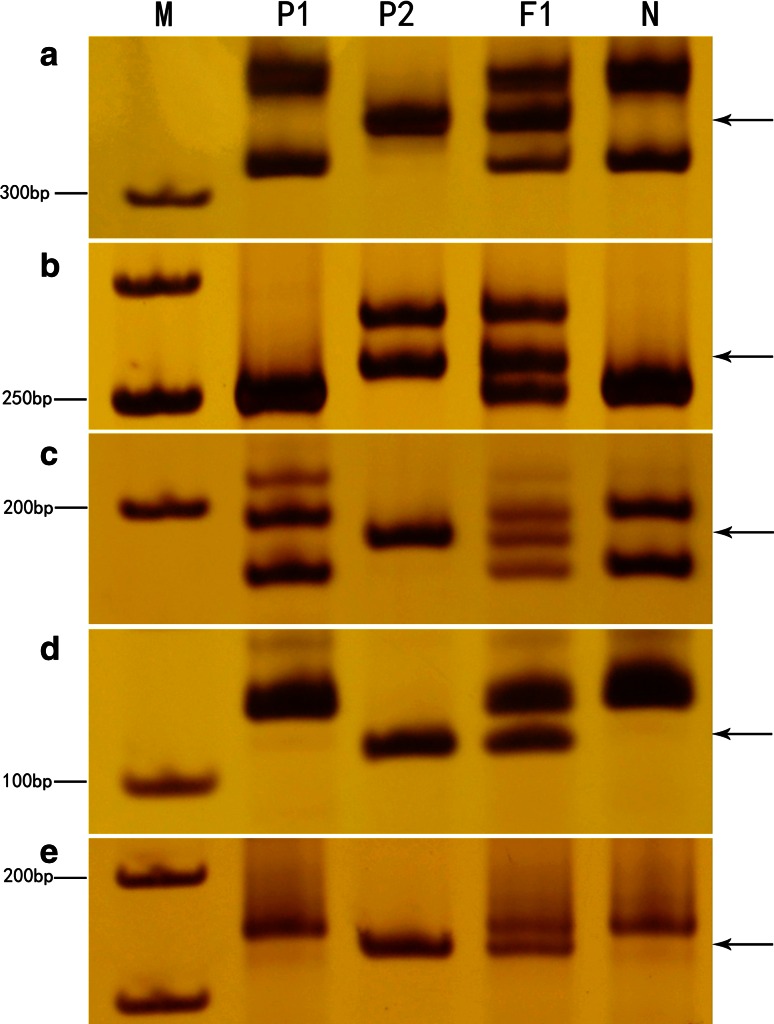
If homoeologous recombination were to occur, it could damage the integrity of the alien chromosome, and such MAALs should be identified. However, it is very difficult with present methods to detect recombination. The GISH technique cannot easily detect recombinant segments in the recipient genome, because cotton chromosomes are very small and there is very little or no repetitive DNA present in the euchromatic regions (Ali et al. 2001). Molecular markers could be used to detect recombinant DNA segments if they were polymorphic. For instance, if a plant was identified by GISH as being a cytological euploid without alien chromosomes, but species-specific markers from the donor parent were present, the occurrence of homoeologous recombination would be confirmed. But no such plant has been found in our study. Thus, it can be concluded that very little, if any, homoeologous recombination occurred during the development of MAALs. Conversely, several MAALs were found to be missing a few species-specific markers (Table 1; Fig. 7, Fig. 1S), which might result from the elimination of some DNA fragments during backcrossing or polyploidization (Fig. 1S). This phenomenon has been reported previously in cotton and other crops (Jiang et al. 2000; Ozkan et al. 2001; Shaked et al. 2001; Skalicka et al. 2005).
Fig. 7.
a–e G. australe-specific DNA fragments detected using SSR markers during backcrossing were deleted in some MAALs in G. hirsutum. a–e Deleted G. australe-specific amplicons were detected by individual chromosome-specific SSR primers for NAU6728 (2Ga), NAU5486 (5Ga), MUSS266 (10Ga), BNL193 (13Ga), and CIR414 (11Ga), respectively. P1 G. hirsutum, P2 G. australe. F 1 the hexaploid of G. hirsutum and G. australe. N indicates MAAL-2Ga (a), MAAL-5Ga (b), MAAL-10Ga (c), MAAL-13Ga (d), and MAAL-11Ga (e). M is the molecular size marker (50 bp ladder). Arrows indicate the deleted G. australe chromosome-specific markers
Strategies for developing cotton cultivars with the glandless-seed and glanded-plant traits
The genus Gossypium is a globally important crop that is used to produce textiles, oil, and protein. Gossypol in cultivated cottonseed, however, is toxic to humans and non-ruminant animals but highly related to insect resistance (Bottger et al. 1964). Due to gossypol mainly existed in the cotton pigment gland, thus, breeding a high-yielding “glandless-seed” and “glanded-plant” cultivar has become an area of interest for researchers. Efforts have been made to eliminate gossypol from cottonseeds while keeping it in foliar vegetative tissues. Since Muramoto first synthesized a fertile hexaploid of (G. hirsutum × G. sturtianum) F1 (Australian wild species, C genome), three Australian wild species [G. bickii, G. australe (G genome) and G. sturtianum] have been used in interspecific breeding through both the hexaploid and tetraploid pathways. In the hexaploid pathway, tetraploid cotton (2n = AADD = 52) was crossed with diploid wild-type species (2n = CC or GG = 26) to produce a triploid F1. Then, the obtained triploid F1 was chromosome doubled and the hexaploid was subsequently developed. Through the hexaploid pathway, breeding efforts for the introgression of glanded-plant and glandless-seed trait were made (Muramoto 1969; Dilday 1986; Altman et al. 1987). He and Sun (1994) studied the hybrid F1 between glandless upland cotton and G. bickii. Liang et al. (1997) developed a new red-flowered germplasm line of cotton selected from a hybrid of Gossypium hirsutum × G. bickii. These have been used in hybrid cotton (Wang and Wang 2008).
He et al. (2000) analyzed the inheritance of the petal color genes introgressed from G. bickii. Using the tetraploid pathway, diploid cotton (2n = AA or DD = 26) was crossed with diploid wild-type species (2n = CC or GG = 26) to produce a diploid F1. The obtained diploid F1 was then chromosome doubled to develop the tetraploid. For example, Mergeai (1992) developed an amphidiploid F1 of G. arboreum × G. sturtianum with glandless-seed and glanded-plant traits, which were sequentially used as bridge material for the transfer of the target trait into upland cotton (Bi et al. 1998; Bi et al. 1998, 1999a, b; Ahoton et al. 2003). Kulkarni et al. (2002) carried out studies on the interspecific hybridization of G. australe and G. herbaceum. Zhang et al. (1994) synthesized allotetraploid cotton with an (AG) complex chromosome set, which has since been used in breeding programs (Zhu et al. 2004). However, the “glandless-seed” and “glanded-plant” cultivar has not been developed at present. The glandless-seed, glanded-plant trait is present only in some Australian wild diploid cotton species with C and G genomes, which are phylogenetically distant from the cultivated upland cotton, G. hirsutum. The formation of gossypol glands in upland cotton is controlled by two main alleles, Gl2 and Gl3 (McMichael 1960). Genetic analysis by Zhu et al. (2004) demonstrated that “glandless-seed” and “glanded-plant” trait from G. bickii, another Australian wild G-genome species, is controlled by a gene located at the Gl 2 locus, which has been temporarily named Gl b2. This gene, Gl b2, is dominant to upland cotton pigment gland alleles Gl 2 and gl 2, but is recessive and epistatic to another glanded gene Gl 3. Based on these results, it is presumed that only the MAAL with Gl b2 Gl 2 Gl 2 gl 3 gl 3 or Gl b2 gl 2 gl 2 gl 3 gl 3 will show the glandless-seed, glanded-plant phenotype. Using normal upland cotton with the dominant gene Gl 3 as the recipient parent, it will be impossible to obtain a MAAL with the glandless-seed, glanded-plant trait, since the genotype of the MAAL is Gl b2 Gl 2 Gl 2 Gl 3 Gl 3. This also could explain why no MAAL with this trait was detected in our study. Thus, to develop a MAAL with glandless-seed, glanded-plant trait, the recipient upland cotton parent used should carry the recessive gl 3 gene. To largely eliminate the role of Gl 3 in conferring gossypol synthesis, the ideal cotton genotype would be Gl b2 Gl b2 Gl 2 Gl 2 without Gl 3 Gl 3. The only way to obtain a “glandless-seed” and “glanded-plant” cultivar with the Gl b2 Gl b2 Gl 2 Gl 2 genotype would be to use radiation to induce chromosomal translocation between the homoeologous chromosomes 12Dt in G. hirsutum and 12Ga in G. australe, since very little or no homoeologous recombination occurs under natural conditions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure 1S The set of putative G. australe chromosome-specific SSR markers that were used to screen for polymorphisms. Markers are based on the backbone map of the Dt-subgenome of tetraploid cotton constructed using the BC1 population of (G. hirsutum × G. barbadense) × G. hirsutum (Guo et al. 2007). Note: Markers in red were eliminated in the corresponding monosomic alien addition lines during backcrossing (DOC 177 kb)
Acknowledgments
This program was financially supported in part by the National Natural Science Foundation of China (30571184 and 31271771) and the Priority Academic Program Development of Jiangsu Higher Education Institutions. We are also grateful to Dr. CL Brubaker (Plant Indusry of CSIRO, Australia) for providing seeds of Gossypium australe and the allohexaploid of G. hirsutum × G. australe.
Conflict of interest
The authors declare that there are no conflicts of interest in the reported research.
Ethical standards
The authors note that this research is performed and reported in accordance with ethical standards of the scientific conduct.
References
- Ahoton L, Lacape JM, Baudoin JP, Mergeai G. Introduction of Australian diploid cotton genetic variation into upland cotton. Crop Sci. 2003;43:1999–2005. doi: 10.2135/cropsci2003.1999. [DOI] [Google Scholar]
- Ali SNH, Ramanna MS, Jacobsen E, Visser RGF. Establishment of a complete series of a monosomic tomato chromosome addition lines in the cultivated potato using RFLP and GISH analyses. Theor Appl Genet. 2001;103:687–695. doi: 10.1007/s001220100652. [DOI] [Google Scholar]
- Altman DW, Stelly DM, Kohel RJ. Introgression of glanded-plant and glandless-seed trait from Gossypium sturtianum Willis into cultivated upland cotton using ovule culture. Crop Sci. 1987;27(5):880–884. doi: 10.2135/cropsci1987.0011183X002700050008x. [DOI] [Google Scholar]
- Bao WK, Leng L. Determination methods for photosynthetic pigment content of bryophyte with special relation of extracting solvents. Chin J Appl Environ Biol. 2005;11:235–237. [Google Scholar]
- Bento M, Gustafson P, Viegas W, Silva M. Genome merger: from sequence rearrangements in triticale to their elimination in wheat-rye addition lines. Theor Appl Genet. 2010;121:489–497. doi: 10.1007/s00122-010-1325-6. [DOI] [PubMed] [Google Scholar]
- Bi IV, Baudoin JP, Mergeai G. Cytogenetics of the ‘gland-less-seed and glanded-plant’ trait from Gossypium sturtianum Willis introgressed into upland cotton (Gossypium hirsutum L.) Plant Breed. 1998;117(3):235–241. doi: 10.1111/j.1439-0523.1998.tb01932.x. [DOI] [Google Scholar]
- Bi IV, Baudoin JP, Mergeai G. Development of high-gossypol cotton plants with low-gossypol seeds using trispecies bridge crosses and in vitro culture of seed embryos. Euphytica. 1999;106(3):243–251. doi: 10.1023/A:1003539924238. [DOI] [Google Scholar]
- Bi IV, Maquet A, Baudoin JP, Jardin PD, Jacquemin JM, Mergeai G. Breeding for “low-gossypol seed and high-gossypol plants” in upland cotton. Analysis of tri-species hybrids and backcross progenies using AFLPs and mapped RFLPs. Theor Appl Genet. 1999;99:1233–1244. doi: 10.1007/s001220051329. [DOI] [Google Scholar]
- Bottger GT, Sheehan ET, Lukefahr MJ. Relation of gossypol content of cotton plants to insect resistance. J Econ Entomol. 1964;57:283–285. [Google Scholar]
- Brubaker CL, Brown AHD. The use of multiple alien chromosome addition aneuploids facilitates genetic linkage mapping of the Gossypium G genome. Genome. 2003;46:774–791. doi: 10.1139/g03-063. [DOI] [PubMed] [Google Scholar]
- Budahn H, Schrader O, Peterka H. Development of a complete set of disomic rape-radish chromosome-addition lines. Euphytica. 2008;162:117–128. doi: 10.1007/s10681-007-9609-x. [DOI] [Google Scholar]
- Chen BY, Simonsen V, Lannrr-Herrera C, Heneen WK. A Brassica campestris-alboglabra addition line and its use for gene mapping, intergenomic gene transfer and generation of trisomics. Theor Appl Genet. 1992;84:592–599. doi: 10.1007/BF00224157. [DOI] [PubMed] [Google Scholar]
- Chen CC, Chen SK, Liu MC, Kao YY. Mapping of DNA markers to arms and sub-arm regions of Nicotiana sylvestris chromosomes using aberrant alien addition lines. Theor Appl Genet. 2002;105:8–15. doi: 10.1007/s00122-001-0829-5. [DOI] [PubMed] [Google Scholar]
- Chen JF, Luo XD, Qian CT, Jahn MM, Staub JE, Zhuang FY, Lou QF, Ren G. Cucumis monosomic alien addition lines: morphological, cytological, and genotypic analyses. Theor Appl Genet. 2004;108:1343–1348. doi: 10.1007/s00122-003-1546-z. [DOI] [PubMed] [Google Scholar]
- Cifuentes M, Benavente E. Wheat-alien metaphase I pairing of individual wheat genomes and D genome chromosomes in interspecific hybrids between Triticum aestivum L. and Aegilops geniculata Roth. Theor Appl Genet. 2009;119:805–813. doi: 10.1007/s00122-009-1090-6. [DOI] [PubMed] [Google Scholar]
- Dilday RH. Development of a cotton plant with glandless seeds and glanded foliage and fruiting forms. Crop Sci. 1986;26:639–641. doi: 10.2135/cropsci1986.0011183X002600030046x. [DOI] [Google Scholar]
- Endrizzi JE, Turcotte EL, Kohel RJ. Genetics, cytology, and evolution of Gossypium. Adv Genet. 1985;23:271–375. doi: 10.1016/S0065-2660(08)60515-5. [DOI] [Google Scholar]
- Fang XH, Gu SH, Xu ZY, Chen F, Guo DD, Zhang HB, Wu NH. Construction of a binary BAC library for an apomictic monosomic addition line of Beta corolliflora in sugar beet and identification of the clones derived from the alien chromosome. Theor Appl Genet. 2004;108:1420–1425. doi: 10.1007/s00122-003-1566-8. [DOI] [PubMed] [Google Scholar]
- Friebe B, Qi LL, Nasuda S, Zhang P, Tuleen NA, Gill BS. Development of a complete set of Triticum aestivum-Aegilops speltoides chromosome addition lines. Theor Appl Genet. 2000;101:51–58. doi: 10.1007/s001220051448. [DOI] [Google Scholar]
- Fryxell PA. A revised taxonomic interpretation of Gossypium L. (Malvaceae) Rheedea. 1992;2:108–165. [Google Scholar]
- Fu S, Lv Z, Qi B, Guo X, Jun Li, Liu B, Han F. Molecular Cytogenetic characterization of wheat-Thinopyrum elongatum addition, substitution and translocation lines with a novel source of resistance to wheat Fusarium head blight. J Genet Genomics. 2012;39:103–110. doi: 10.1016/j.jgg.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Gao D, Guo D, Jung C. Monosomic addition lines of Beta corolliflora Zoss in sugar beet: cytological and molecular-marker analysis. Theor Appl Genet. 2001;103:240–247. doi: 10.1007/s001220100601. [DOI] [Google Scholar]
- Geleta M, Heneen WK, Stoute AI, Muttucumaru N, Scott RJ, King GJ, Kurup S, Bryngelsson T. Assigning Brassica microsatellite markers to the nine C-genome chromosomes using Brassica rapa var. trilocularis–B. oleracea var. alboglabra monosomic alien addition lines. Theor Appl Genet. 2012;125:455–466. doi: 10.1007/s00122-012-1845-3. [DOI] [PubMed] [Google Scholar]
- Gerstel DU, Sarvella PA. Additional observations on chromosomal translocations in cotton hybrids. Evolution. 1956;10:408–414. doi: 10.2307/2407000. [DOI] [Google Scholar]
- Guan B, Wang K, Zhou BL, Guo WZ, Zhang TZ. Establishment of a multi-color genomic in situ hybridization technique to simutaneously discriminated the three interspecific hybrid genomes in Gossypium. J Integr Plant Biol. 2008;50:345–351. doi: 10.1111/j.1744-7909.2007.00623.x. [DOI] [PubMed] [Google Scholar]
- Guo WZ, Wang W, Zhou BL, Zhang TZ. Cross-species transferability of G. arboreum-derived EST-SSRs in the diploid species of Gossypium. Theor Appl Genet. 2006;112:1573–1581. doi: 10.1007/s00122-006-0261-y. [DOI] [PubMed] [Google Scholar]
- Guo WZ, Cai CP, Wang CB, Han ZG, Song XL, Wang K, Niu XW, Wang C, Lu KY, Shi B, Zhang TZ. A microsatellite-based, gene-rich linkage map reveals genome structure, function and evolution in Gossypium. Genetics. 2007;176:527–541. doi: 10.1534/genetics.107.070375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlan JR, de Wart JMJ. Toward rational classification of cultivated plants. Taxon. 1971;20:509–517. doi: 10.2307/1218252. [DOI] [Google Scholar]
- Hau B. Lignées d’addition sur l’espèce Gossypium hirsutum L. I. Utilisation de l’hybridation interspécifique et de la method des lignées d’addition pour l’amélioration du cotonnier. Cot Fib Trop. 1981;36:247–258. [Google Scholar]
- He JX, Sun CW. A scheme for introgression of delayed gland morphogenesis gene from wild Gossypium bickii into cultivated upland cotton (G. hirsutum) Acta Genetica Sinica. 1994;21(1):52–58. [Google Scholar]
- He JX, Jiang RQ, Zhang XX, Li ZS, Zhao GZ. Genetic analysis for introgressive genes of petal color from Gossypium bickii to G. hirsutum. Acta Genetica Sinica. 2000;27(4):344–350. [PubMed] [Google Scholar]
- Ji Y, Chetelat RT. Homoeologous pairing and recombination in Solanum lycopersicoides monosomic addition and substitution lines of tomato. Theor Appl Genet. 2003;106:979–989. doi: 10.1007/s00122-002-1090-2. [DOI] [PubMed] [Google Scholar]
- Jiang CX, Chee PW, Draye X, Morrell PL, Smith CW, Paterson AH. Multilocus interactions restrict gene introgression in interspecific populations of polyploid Gossypium (cotton) Evolution. 2000;54:798–814. doi: 10.1111/j.0014-3820.2000.tb00081.x. [DOI] [PubMed] [Google Scholar]
- Jiang SM, Hu J, Yin WB, Chen YH, Wang RRC, Hu ZM. Cloning of resistance gene analogs located on the alien chromosome in an addition line of wheat-Thinopyrum intermedium. Theor Appl Genet. 2005;111:923–931. doi: 10.1007/s00122-005-0022-3. [DOI] [PubMed] [Google Scholar]
- Kishii M, Yamada T, Sasakuma T, Tsujimoto H. Production of wheat–Leymus racemosus chromosome addition lines. Theor Appl Genet. 2004;109:255–260. doi: 10.1007/s00122-004-1631-y. [DOI] [PubMed] [Google Scholar]
- Kong F, Wang H, Cao A, Qin B, Ji J, Wang S, Wang X. Characterization of T. aestivum-H. californicum chromosome addition lines DA2H and MA5H. J Genet Genomics. 2008;35:673–678. doi: 10.1016/S1673-8527(08)60089-2. [DOI] [PubMed] [Google Scholar]
- Kulkarni VN, Khadi BM, Sangam VS. Pre-breeding efforts for low gossypol seed and high gossypol plant in G. herbaceum L. cotton utilizing G. australe Mueller. Curr Sci. 2002;82(4):434–439. [Google Scholar]
- Kynast RG, Riera-Lizarazu O, Vales MI, Okagaki RJ, Maquieira S, Chen G, Ananiev EV, Odland WE, Russell CD, Stec AO, Livingston SM, Zaia HA, Rines HW, Phillips RL. A complete set of maize individual chromosome additions to the oat genome. Plant Physiol. 2001;125:1216–1227. doi: 10.1104/pp.125.3.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kynast RG, Okagaki RJ, Galatowitsch MW, Granath SR, Jacobs MS, Stec AO, Rines HW, Phillips RL. Dissecting the maize genome by using chromosome addition and radiation hybrid lines. Proc Natl Acad Sci USA. 2004;101:9921–9926. doi: 10.1073/pnas.0403421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang ZL, Jiang RQ, Zhong WN. New red flower germplasm lines of cotton selected from hybrid of Gossypium hirsutum × G. bickii. Sci China (series C) 1997;40:284–292. doi: 10.1007/BF02879089. [DOI] [PubMed] [Google Scholar]
- Lopez-Lavalle LAB, Brubaker CL. Frequency and fidelity of alien chromosome transmission in Gossypium hexaploid bridging population. Genome. 2007;50:479–491. doi: 10.1139/G07-030. [DOI] [PubMed] [Google Scholar]
- McMichael SC. Combined effects of glandless genes gl2 and gl3 on pigment glands in the cotton plant. Agron J. 1960;52:385–386. doi: 10.2134/agronj1960.00021962005200070005x. [DOI] [Google Scholar]
- Mergeai G. New perspectives concerning the methodology to be used for introgression of the glanded plant and glandless seed character in cultivated cotton (Gossypium hirsutum L.) Cot Fib Trop. 1992;47:113–118. [Google Scholar]
- Molnár I, Molnár -Láng M. GISH reveals different levels of meiotic pairing with wheat for individual Aegilops biuncialis chromosomes. Bilo Plant. 2010;54:259–264. doi: 10.1007/s10535-010-0045-8. [DOI] [Google Scholar]
- Multani DS, Khush GS, delos Reyes BG, Brar DS. Alien genes introgression and development of monosomic alien addition lines from Oryza latifolia Desv. to rice Oryza sativa L. . Theor Appl Genet. 2003;107:395–405. doi: 10.1007/s00122-003-1214-3. [DOI] [PubMed] [Google Scholar]
- Muramoto H. Hexaploid cotton: some plant and fiber properties. Crop Sci. 1969;9:27–29. doi: 10.2135/cropsci1969.0011183X000900010009x. [DOI] [Google Scholar]
- Ozkan H, Levy AA, Feldman M. Allopolyploidy induced rapid genome evolution in the wheat (Aegilops–Triticum) group. Plant Cell. 2001;13:1735–1747. doi: 10.1105/tpc.13.8.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Brubaker KL, Wendel JF. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep. 1993;11:122–127. doi: 10.1007/BF02670470. [DOI] [Google Scholar]
- Pertuzé RA, Ji Y, Chetelat RT. Transmission and recombination of homeologous Solanum sitiens chromosomes in tomato. Theor Appl Genet. 2003;107:1391–1401. doi: 10.1007/s00122-003-1384-z. [DOI] [PubMed] [Google Scholar]
- Peterka H, Budahn H, Schrader O, Ahne R, Schutze W. Transfer of resistance against the beet cyst nematode from radish (Raphanus sativus) to rape (Brassica napus) by monosomic chromosome addition. Theor Appl Genet. 2004;109:30–41. doi: 10.1007/s00122-004-1611-2. [DOI] [PubMed] [Google Scholar]
- Reamon-Ramos SM, Wricke G. A full set of monosomie addition lines in Beta vulgaris from Beta webbiana: morphology and isozyme markers. Theor Appl Genet. 1992;84:411–418. doi: 10.1007/BF00229501. [DOI] [PubMed] [Google Scholar]
- Rooney WL, Stelly DM. Identification of four Gossypium monosomic alien addition derivatives from a backcrossing program with G. hirsutum. Crop Sci. 1991;31:337–341. doi: 10.2135/cropsci1991.0011183X003100020024x. [DOI] [Google Scholar]
- Sarr D, Lacape JM, Rodier-Goud M, Jacquemin JM, Benbouza H, Toussaint A, Palm R, Ahoton L, Baudoin JP, Mergeai G. Isolation of five new monosomic alien addition lines of Gossypium australe F. Muell in G. hirsutum L. by SSR and GISH analyses. Plant Breed. 2011;130:60–66. doi: 10.1111/j.1439-0523.2010.01819.x. [DOI] [Google Scholar]
- Sears ER. The aneuploids of common wheat. Univ Mo Res Bull. 1954;572:1–58. [Google Scholar]
- Shaked H, Kashkush K, Ozkan H, Feldman M, Levy AA. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell. 2001;13:1749–1759. doi: 10.1105/tpc.13.8.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J, Panaud O, Vitte C, Mendioro MS, Brar DS. RDA derived Oryza minuta-specific clones to probe genomic conservation across Oryza and introgression into rice (O. sativa L.) Euphytica. 2010;176:269–279. doi: 10.1007/s10681-010-0245-5. [DOI] [Google Scholar]
- Skalicka K, Lim KY, Matyasek R, Matzke M, Leitch AR, Kovarik A. Preferential elimination of repeated DNA sequences from the paternal, Nicotiana tomentosiformis genome donor of a synthetic, allotetraploid tobacco. New Phytol. 2005;166:291–303. doi: 10.1111/j.1469-8137.2004.01297.x. [DOI] [PubMed] [Google Scholar]
- Srinivasan K, Malathi VG, Kirti PB, Prakash S, Chopra VL. Generation and characterisation of monosomic chromosome addition lines of Brassica campestris-B. oxyrrhina. Theor Appl Genet. 1998;97:976–981. doi: 10.1007/s001220050979. [DOI] [Google Scholar]
- Stewart, JM (1995) Potential for crop improvement with exotic germplasm and genetic engineering. In: Constable GA, Forrester NW (eds) Challenging the future: Proceedings of the World Cotton Research Conference-1, Brisbane Australia, February 14–17, pp 313–327, Melbourne, Australia
- Wang LM, Wang JB. The utilization of HB-red flower in hybrid cotton breeding. Cotton Sci. 2008;20:86. [Google Scholar]
- Wang P, Zhang TZ. Genetic dissection of photosynthetic pigment content in cotton interspecific chromosome segment introgression lines. Acta Agronomica Sinica. 2012;38:947–953. doi: 10.3724/SP.J.1006.2012.00947. [DOI] [Google Scholar]
- Wang XE, Chen PD, Liu DJ, Zhang P, Zhou B, Friebe B, Gill BS. Molecular cytogenetic characterization of Roegneria ciliaris chromosome additions in common wheat. Theor Appl Genet. 2001;102:651–657. doi: 10.1007/s001220051693. [DOI] [Google Scholar]
- Wang K, Song XL, Han ZG, Guo WZ, Yu JZ, Sun J, Pan JJ, Kohel RJ, Zhang TZ. Complete assignment of the chromosomes of Gossypium hirsutum L. by translocation and fluorescence in situ hybridization mapping. Theor Appl Genet. 2006;113:73–80. doi: 10.1007/s00122-006-0273-7. [DOI] [PubMed] [Google Scholar]
- Wendel JF, Cronn RC. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. doi: 10.1016/S0065-2113(02)78004-8. [DOI] [Google Scholar]
- Zhang BJ, Li BL, Wan LM, Han YH, Wang RH, Zhu SJ, Wang HM. Synthesis of allotetraploid cotton with [AG] complex chromosome set. Chin Sci Bull. 1994;39(7):644–648. [Google Scholar]
- Zhang J, Wu YT, Guo WZ, Zhang TZ. Fast screening of microsatellite markers in cotton with PAGE/silver staining. Cotton Sci Sin. 2000;12:267–269. [Google Scholar]
- Zhang J, Guo WZ, Zhang TZ. Molecular linkage map of allotetraploid cotton (Gossypium hirsutum L. × Gossypium barbadense L.) with a haploid population. Theor Appl Genet. 2002;105:1166–1174. doi: 10.1007/s00122-002-1100-4. [DOI] [PubMed] [Google Scholar]
- Zhou ZH, Yu P, Liu GH, He JX, Chen JX, Zhang XX. Morphological and molecular characterization of two G. somalense monosomic alien addition lines (MAALs) Chin Sci Bull. 2004;49:558–562. [Google Scholar]
- Zhu SJ, Jiang YR, Reddy N, Ji DF. Breeding, introgression and inheritance of delayed gland morphogenesis trait from Gosspium bickii into upland cotton germplasm. Chin Sci Bull. 2004;49:2470–2476. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1S The set of putative G. australe chromosome-specific SSR markers that were used to screen for polymorphisms. Markers are based on the backbone map of the Dt-subgenome of tetraploid cotton constructed using the BC1 population of (G. hirsutum × G. barbadense) × G. hirsutum (Guo et al. 2007). Note: Markers in red were eliminated in the corresponding monosomic alien addition lines during backcrossing (DOC 177 kb)