Abstract
Multimodality imaging using noncytotoxic triple fusion (TF) reporter genes is an important application for cell-based tracking, drug screening, and therapy. The firefly luciferase (fl), monomeric red fluorescence protein (mrfp), and truncated herpes simplex virus type 1 thymidine kinase SR39 mutant (ttksr39) were fused together to create TF reporter gene constructs with different order. The enzymatic activities of TF protein in vitro and in vivo were determined by luciferase reporter assay, H-FEAU cellular uptake experiment, bioluminescence imaging, and micropositron emission tomography (microPET). The TF construct expressed in H1299 cells possesses luciferase activity and red fluorescence. The tTKSR39 activity is preserved in TF protein and mediates high levels of H-FEAU accumulation and significant cell death from ganciclovir (GCV) prodrug activation. In living animals, the luciferase and tTKSR39 activities of TF protein have also been successfully validated by multimodality imaging systems. The red fluorescence signal is relatively weak for in vivo imaging but may expedite FACS-based selection of TF reporter expressing cells. We have developed an optimized triple fusion reporter construct DsRedm-fl-ttksr39 for more effective and sensitive in vivo animal imaging using fluorescence, bioluminescence, and PET imaging modalities, which may facilitate different fields of biomedical research and applications.
1. Introduction
Multiple molecular imaging techniques have been used not only to monitor complex biological processes both spatially and temporally but also to study the transplanted cell trafficking, long term monitoring of disease (i.e., cancer progression), and therapeutic effectiveness in living animals [1, 2]. These techniques have accelerated the translation of in vitro based drug discovery studies to in vivo imaging-based preclinical and clinical applications [3]. Several noninvasive imaging modalities such as positron emission tomography (PET), single photon emission computed tomography (SPECT), magnetic resonance imaging (MRI), and optical imaging are available for real-time repetitive imaging of reporter gene expression in living subjects [1–4]. Optical imaging is quick and cost-effective and requires no radioisotopes for validating different reporter systems. However, it does not provide optimal tomographic information and quantitative data as compared to 3D radionuclide imaging techniques (i.e., PET, SPECT) [5]. A dual or triple fusion reporter gene construct harboring various fluorescent proteins (i.e., enhanced green fluorescent protein (egfp), red fluorescent protein (rfp), and different luciferase genes (i.e., firefly luciferase (fl), renilla luciferase (rl), and herpes simplex virus type 1 thymidine kinase (tk) gene)) can combine the advantage of fluorescence, bioluminescence, and radionuclide imaging techniques and overcome the shortcomings of each imaging modality [6].
The tk-gfp dual fusion gene allows for microscopic and whole-body fluorescence imaging as well as PET imaging [7]. Another fusion gene, the tksr39-rl (tksr39 is a mutant version of tk [8]), was also validated in living mice bearing transduced tumor xenografts by microPET and bioluminescence imaging (BLI) [9]. A triple fusion (TF) reporter gene, truncated tk-egfp-fl, was constructed to visualize the TF-transduced cells in living animals by optical, fluorescent, and nuclear imaging modalities [10]. However, the carboxyl-terminal (C-terminal) region of tk is involved in the enzymatic catalysis profoundly [11] and fusion of the partner gene such as fl gene to the C-terminal region of tk would lead to a decreased TK activity (note that tk refers to the gene and TK to the protein) [9]. The configuration of tk-egfp-fl TF reporter gene is not favorable due to severe influence on TK folding and enzymatic function from the downstream large bulky EGFP-FLUC fusion protein. Several other TF genes had also been constructed and validated, that is, rl-egfp-ttksr39 [12], rl-mrfp-ttksr39, and fl-mrfp-ttksr39 [3, 13] (mrfp: monomeric red fluorescent protein; ttksr39: truncated tksr39). Because of the intrinsic autofluorescence problem in the living animals, the egfp-harboring TF gene constructs were not chosen for in vivo imaging applications. In contrast, the efficacy of TF gene construct harboring rfp gene in the 293T human embryonic kidney cells or A375M human melanoma cells has been demonstrated in living mice with microPET and optical imaging systems [3]. The fl-mrfp-ttksr39 reporter gene has also been applied in stem cell research. In 2006, Cao et al. developed murine embryonic stem cells (ESCs) stably expressing human ubiquitin-C (UBC) promoter-driven TF fusion gene and demonstrated that kinetics of ESC survival, proliferation, and migration in living mice could be monitored using multimodality imaging systems [14, 15]. Using lentiviral-mediated transduction, the modified myeloproliferative sarcoma virus (mnd) promoter-driven TF gene can be stably expressed in human mesenchymal stem cell (MSC) for in vivo monitoring of the efficacy of stem cell transplantation [16]. It is noteworthy that there are no experimental data using stem cells transduced with an fl-mrfp-ttksr39 gene construct driven by a constitutive cytomegaloviral (CMV) promoter. Several studies have reported epigenetic silencing of CMV promoter after longitudinal passage of stem cells [16, 17]. Thus, we speculated whether the lentiviral-mediated overexpression of fl-mrfp-ttksr39 TF driven by the CMV promoter would induce aberrant characteristics or alter differentiation capacity of stem cells.
For drug screening and validation, a less cytotoxic and highly sensitive reporter gene that faithfully reflects the drug effectiveness and eludes the cytotoxic effect of reporter gene product is required. Furthermore, for cell therapy and trafficking, it is also necessary to adopt a less cytotoxic reporter gene to minimize the cellular stress from the aggregation-prone reporter gene product in cell labeling. However, considering the detection sensitivity and limitation, the reporter gene was usually driven by a strong promoter to increase the reporter gene expression and consequently the aggregation-prone reporter gene products accumulated in the transfected cells and ultimately resulted in cellular stress and cytotoxic effect that hampered the cell growth. Previous reports indicated that the rl-mrfp-ttksr39 or fl-mrfp-wt.ttk (wt.ttk; truncated wild type tk) is the best configuration of TF gene constructs for multimodality imaging techniques, if the rl or fl are chosen for TF [3, 13]. To best preserve the TK activity for radionuclide imaging the ttksr39 should be combined with the rl-mrfp component and the wt.tk should be combined with the resting fl-mrfp component. The enzymatic activity of ttksr39 or wt.tk could be significantly influenced by fusion with different fusion partners. Furthermore, the intracellular localization of TK-harboring fusion proteins changes significantly while different fusion partner is in-frame fused to tk gene or the same fusion partner is in-frame fused to tk at N- or C-terminal. The GFP-TK fusion protein is exclusively and homogenously expressed in the nuclei of the transduced cells, whereas TK-GFP exists as pancellular localization but the majority of proteins are in the nucleus with the massive aggregated form of nuclear bodies [10, 18]. Such intracellular localization of TK-containing fusion protein will result in a reduced enzymatic activity of TK and most likely cause severe cytotoxic effects to cells due to the reporter protein aggregation in the cell nucleus [19, 20]. Because of the advantage of TF reporter construct in the research, we attempted to establish cancer cell clones stably expressing CMV driven fl-DsRedm-ttksr39 TF gene construct. However, our unpublished studies demonstrated that the fast chromophore maturation of TF is similar to primitive tetrameric DsRed protein suggesting that the TF protein in transfected cells might exist in a tetrameric form. The cell clones only transiently expressed the TF fusion protein and the survived cells did not proliferate after G418 selection and failed to establish clonal populations. Thus, we postulated that the fl-DsRedm-ttksr39 protein may be toxic to cells. Since the order of fusion genes in TF determines the mRNA structure that may influence ribosomal accessibility or mRNA stability, we aimed to construct a new TF reporter gene with optimal configuration for multimodality imaging, the p3H (DsRedm-fl-ttksr39), and to compare this novel TF gene with other two TF genes, the p3G (rl-DsRedm-ttksr39) and p3R (fl-DsRedm-ttksr39).
2. Experimental Procedures
2.1. Construction of Triple Fusion Reporter Gene
The rl-DsRedm-ttksr39 (p3G) and fl-DsRedm-ttksr39 (p3R) were constructed as described by Ray et al. [3, 12]. The novel triple fusion reporter plasmid, DsRedm-fl-ttksr39 (p3H), was constructed essentially as described by Ray et al. [3, 12]. Briefly, the monomeric DsRed expression plasmid, pDsRed-Monomer -C1, driven by CMV enhancer/promoter, was purchased from Clontech (BD science, Inc., USA). The pDsRed-Monomer-C1 digested with restriction enzyme of BamH I and Xba I (New England Biolabs, Inc., USA) was purified from 1% agarose gel by PCR/Gel Extraction kit (Geneaid Inc., Taiwan) and used as cloning vector. The truncated tksr39 gene was amplified from the plasmid pttksr39 (generous gifts from Professor FD Chen, TransWorld University, Taiwan) by polymerase chain reaction (PCR) with 5′-end primer ttkUp/BamHI (5′-CAA GAC GGA TCC TCT GGT AAA ATG CCC ACG CTA CTG C-3′), 3′-end primer ttkDn-XbaI (5′-GTA TTC TCT AGA TCA GTT AGC CTC CCC CAT C-3′), and the proof-reading KOD Taq DNA polymerase (Novagen Inc., USA). The ttksr39 PCR products were purified and subjected to restriction enzyme of BamH I and Xba I (New England Biolabs, Inc., USA) digestion and then the purified ttksr39 insert DNAs were ligated with BamH I-XbaI digested pDsRed-Monomer-C1 vector by T4 DNA ligase (New England Biolabs, Inc., USA) generating a DsRedm-ttksr39 dual fusion reporter genetic construct. The DsRedm-ttksr39 plasmid was digested with restriction enzyme of EcoR I and Sal I (New England Biolabs, Inc., USA) and used for a cloning vector. The fl gene from the pGL3 basic plasmid (Promega Corporation, Madison, WI, USA) was amplified by PCR using the same 5′-end primer FLUCUp-EcoRI 5′-AGC ATC GAA TTC TGA GGA CGC CAA AAA CAT AAA G-3′, the 3′end primer FLUCDn-SalI 5′-CTA GTA GTC GAC AGC AAT CTT TCC GCC CTT CT-3′, and the proof-reading KOD Taq DNA polymerase (Novagen Inc., USA). After purification, the fl PCR products were digested with restriction enzyme of EcoR I and Sal I and then were ligated with the EcoR I-Sal I digested DsRedm-ttksr39 cloning vector to create the DsRedm-fl-ttksr39 triple fusion reporter genetic construct. To create the DsRed1-ttksr39 construct, the pDsRed1-C1 plasmid was digested with Age I and Bam HI, and the DsRed1 gene was purified from agarose gel electrophoresis and was used as DNA inserts. The DsRedm-fl-ttksr39 (p3H) plasmid was digested with the same restriction enzymes to remove the DsRedm-fl DNA fragment and was used as a vector for cloning DsRed1 gene. The DNA coding sequences of all constructs were verified by DNA sequencing service (Misson Biotech, Inc., Taiwan) using ABI model 3730 DNA sequencer.
2.2. Cell Culture
The H1299 nonsmall cell lung cancer cells were maintained in RPMI 1640 growth medium supplemented with 10% fetal calf serum at 37°C. Cells were incubated in a humidified incubator at 37°C and 5% CO2 : 95% air.
2.3. Cell Transfection and Luciferase Reporter Assays
Cells were seeded into 12-well plates (Orange Inc., USA) at a concentration sufficient to give 80–90% confluency on the day of transfection, typically 8 × 104 cells/well, and cultured overnight. In all, 1.5 μg trifusion reporter plasmids were cotransfected with 0.5 μg of CMV-rl, in which the rl was driven by CMV promoter, as an internal control for transfection efficiency using Lipofectamine 2000 transfection reagent (Invitrogen, USA) according to the manufacturer's instructions. Briefly, 2 μg of plasmid DNA and 6 μL of Lipofectamine 2000 transfection reagent diluted in serum-free culture medium (Gibco, USA) were used for each transfection. After a 48-hr incubation, cells were analysed for both firefly and renilla luciferase expression using the Dual Luciferase Assay (Promega, USA). All transfections were carried out in triplicate and normalized by the internal control and amounts of protein. Results presented are the means of at least three independent experiments.
2.4. Cellular Uptake Studies
The H1299 cells were cotransfected with reporter gene (p3H, p3R, p3G, CMV-ttksr39, and DsRed1-ttksr39; 1.8 μg of each construct) and CMV-egfp (0.2 μg). The transfected cells (1.0 × 105 cells) were plated in 12-well dishes. Twenty-four hours after 3H-FEAU (1.48 × 103 Bq/mL [0.4 μCi/mL]) was added in each well and incubated at 37°C for 2 h, cells were washed with cold phosphate-buffered saline (PBS) and lysed by protein extraction reagent (CytoBuster, Novagen, USA), and the radioactivity of each well was measured using beta counter (Perkin-Elmer, USA). The same wells were used to determine the total protein. Because the transfection efficiency of different gene constructs in H1299 cell lines may not be identical, the results were normalized by measuring EGFP expression levels. All results were expressed as the net accumulation of the probe in dpm of cells/dpm of medium/EGFP/μg total protein ± standard error (SE).
2.5. GCV-Mediated Growth Inhibition Studies
Twenty-four hours after transfection with CMV-fl, p3H, p3G, p3R, or DsRed1-ttksr39 plasmids, the transfected cells were reseeded in 96-well flat-bottom microtiter cell culture plates and incubated overnight. Cells were then treated with 300 μL of fresh medium in the presence of varying concentrations of ganciclovir (GCV) (Roche, Germany) for five days at 37°C. The MTT [3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl tetrazolium bromide] (Sigma, USA) was used to treat the cells for 2 h at 37°C. To quantify the cell viability, the formazan products solubilized by DMSO (Sigma, USA) were measured by spectrophotometer (Bio-Rad, USA) with 570 nm and 630 nm of wavelength.
The percentage of cell survival was calculated as
| (1) |
where A is the absorbance from the cells incubated with various concentrations of GCV and B is the absorbance from the cells incubated with the medium containing various concentrations of GCV.
2.6. In Vivo MicroPET and Cooled CCD Optical Imaging
The nu/nu nude mice (6~8-week-old, male) were purchased from National Laboratory Animal Center, Taiwan. About 8 × 106 H1299 cells transfected with TF plasmids were subcutaneously injected into the shoulders and legs of the mice. Forty-eight hours posttransfection, each mouse was placed in the black chamber of CCD camera IVIS 50 (Perkin Elmer, USA) equipped with a halogen light source, an excitation filter at 500–550 nm, and an emission filter at 575–650 nm to acquire a whole body imagefor 1 s. For bioluminescence imaging, the mice were injected with D-luciferin (Promega, USA) intraperitoneally for analysis of fl gene expression (n = 4) and injected later with coelenterazine (Promega, USA) through tail vein for analysis of rl gene expression. Subsequently, static images were obtained from the same anesthetized animals intravenously injected with 0.13 MBq (35 μCi) of 18F-FEAU (100 μL) by using microPET R4 (Concord Microsystems, USA) (n = 4). The bioluminescence and fluorescence signals were obtained by a CCD camera thermoelectrically cooled to −70°C. Bioluminescence signals were displayed in pseudocolors and superimposed on the photographic image using Xenogen's Living Image software. Both bioluminescence and fluorescence signals were recorded as maximum photons/second/centimeter2/steradian (photons/ sec/cm2/sr). Regions of interest (ROIs) in microPET image were drawn over the tumor and the ROI counts were converted to percentage of injected dose per gram (%ID/g) using filtered back projection [3].
2.7. Immunofluorescence Staining
H1299 transfected cells were seeded onto poly-(lysine) coated coverslips and fixed with 4% paraformaldehyde. Cells permeabilized with 0.1% of Triton X-100 were blocked in 4% BSA/PBS and subsequently probed with primary antibody anti-HSV tk Ab (a generous gift from Professor Juri Gelovani, MD Anderson Cancer Center, TX, USA) followed by goat anti-mouse IgG antibody conjugated to FITC (Santa Cruz, USA). The 4′,6-diamidino-2-phenylindole (DAPI) was dropped onto the sections to stain the nuclei. Fluorescent signals from DsRed- or HSV1 tk-expressed cells were obtained using confocal microscopy (Leica TCS SP5, Germany).
2.8. Statistical Analysis
The mean and standard error values were calculated for each group and experimental. The 2-tailed Student's t-test and two-way ANOVA were used for statistical analysis using SPSS 15.0 (SPSS, Inc., IL, USA). Statistical significance was defined as P < 0.05.
3. Results
3.1. In Vitro Demonstration of the Efficacy of Triple Fusion Reporter Gene
Plasmid DNA of three different TF vectors (p3G, p3R, and p3H) has been used to transiently transfect the H1299 cells, and pDsRed-Monomer-C1 (BD Science, Clontech, USA) was used for positive control. The transfected cells were first visualized by fluorescence microscopy for monomeric DsRed activity and further assayed for either RLUC or FLUC and tTKSR39 activities. Fluorescence microscopy of transfected cells demonstrated homogenous distribution of positive control protein (pDsRed-Monomer-C1) throughout the nucleus and cytoplasm (Figure 1). However, fluorescence in p3H (DsRedm-fl-ttksr39) transfected cells was predominantly and uniformly distributed in the cytoplasm with slight fluorescence detectable in the cell nucleus. The majority of cells expressing p3G (rl-DsRedm-ttksr39) gene demonstrated tightly packed red fluorescent aggregates mainly in the cytoplasm. Only a small amount of fusion protein appeared in the nucleus. The cells transfected with p3R (fl-DsRedm-ttksr39) gene demonstrated a similar distribution pattern of TF fusion protein as positive control, with less aggregation in the cytoplasm and nucleus as compared with the p3G-transfected cells. Among all gene constructs used in this study, the p3G-transfected cells demonstrated higher red fluorescence intensity, as compared to pDsRed-Monomer-C1 positive control, whereas cells expressing either p3R or p3H gene exhibited relatively lower fluorescence intensity. Curiously, the red fluorescent signal from the p3G and p3R expressing cells could be observed already overnight posttransfection, as compared to pDsRed-Monomer-C1 and p3H transduced cells, of which the fluorescence was not detectable until 36 hrs posttransfection (data not shown). This phenomenon suggests that p3G and p3R TF reporter protein has faster chromophore maturation rate than that of monomeric DsRed protein. The faster maturation rate is reminiscent of the tetrameric DsRed protein trait. To assess the tTKSR39 enzyme activity, H1299 cells were transiently transfected with TF reporter vectors along with positive control (CMV-ttksr39) and negative control (CMV-DsRed). The uptake of 3H-FEAU in p3G-transfected cells was the highest among the three TF reporter vectors (P < 0.05), about 33% of positive control (Figure 2(a)). In fl-harboring TF vectors, the p3H-transfected cells accumulated more 3H-FEAU than the p3R-transfected cells (23% versus 19%), indicating that higher tTKSR39 activity is achievable using the p3H vector (P < 0.05). Also, we constructed a dual reporter gene (DsRed1-ttksr39) as another positive control. In the cells transfected with DsRed1-ttksr39 gene, a 46% decrease in 3H-FEAU uptake was observed, as compared with the positive control (CMV-ttksr39). However, the tTKSR39 activity in the DsRed1-ttksr39 construct was still higher (54%) than all three TF vectors. Further, luciferase enzyme assay was used to compare the FLUC activity achieved by p3H and p3R vectors; the CMV-fl was used as a positive control for reporter gene expression. The FLUC activity was normalized by measuring the activity of cotransfected CMV-rl using dual-luciferase reporter assay system (http://www.promega.com/tbs/). The highest FLUC activity was observed in cells transfected with CMV-fl (Figure 2(b)). The p3H vector retained about 27% of the FLUC activity measureable with positive control and exhibited threefold higher activity than the p3R vector (P < 0.05).
Figure 1.
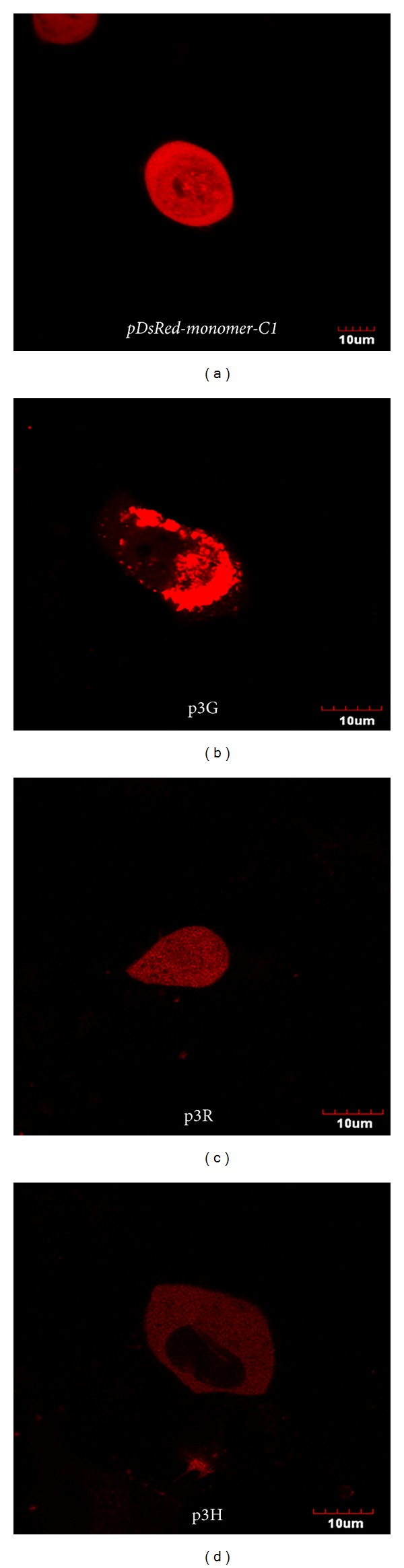
Red fluorescence photomicrographs of H1299 cells expressing pDsRed-Monomer-C1, p3G, p3R, and p3H gene constructs. Bars for the fluorescence micrographs represent 10 μm.
Figure 2.
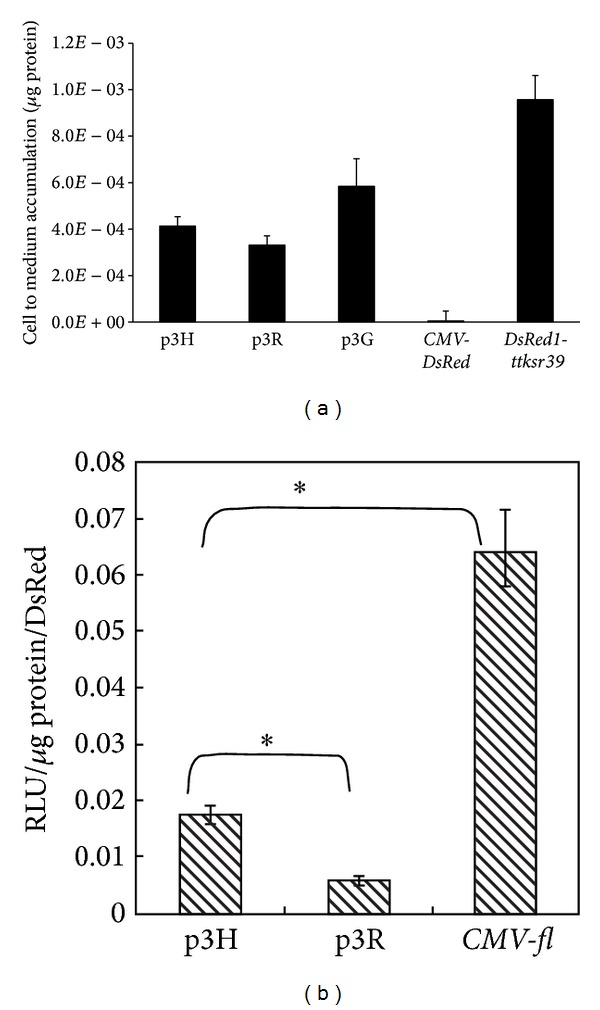
The firefly luciferase and tTKSR39 activity exhibited by H1299 cells transiently transfected with different gene constructs. The H1299 cells were cotransfected with CMV-rl and fl-harboring gene constructs. The tTK activity normalized with CMV-EGFP was expressed as cell to medium accumulation/g protein (a). Values for FLUC activity was normalized with RLUC activity and expressed as relative light units (RLU)/g protein/RLUC activity (b). All the experiments (n = 4) were done in triplicate. *P < 0.05.
3.2. Subcellular Localization of Different Triple Fusion Proteins
Immunofluorescence staining of transfected cells demonstrated distinct differences in subcellular localization of native tTKSR39 and fused tTKSR39 (p3G, p3R, and p3H) proteins. The single tTKSR39 protein exhibited uniform distribution in the nucleus and cytoplasm (Figure 3). Fusion of ttksr39 with p3G, p3R, and p3H genes resulted in predominant cytoplasmic distribution of fusion proteins (Figure 3). This effect was more pronounced in p3H-expressing cells than p3G- and p3R-expressing cells. Moreover, the cells transfected with p3R showed a tendency to form small aggregation in the perinuclear region and the cytoplasm.
Figure 3.
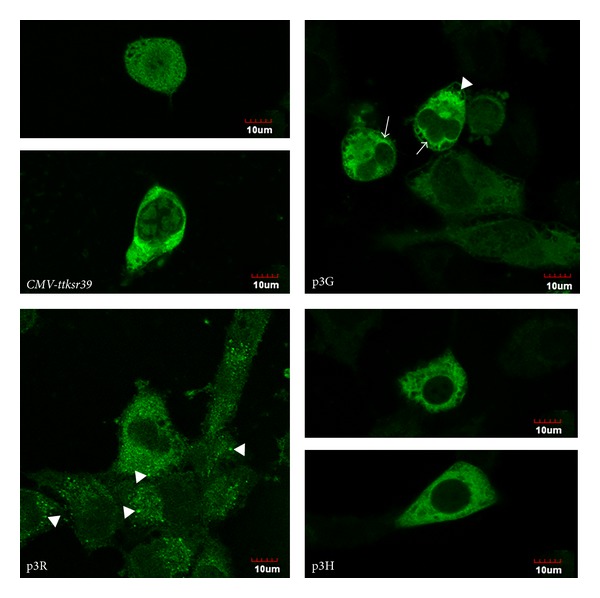
Immunofluorescence staining for TKSR39 protein expressed by cells transfected with CMV-ttksr39, p3G, p3R, or p3H gene constructs. Arrow heads indicate aggregated proteins of p3G and p3R in cytoplasm. Arrows indicate peri-nuclear distribution of p3G proteins. Bars for the fluorescence micrographs represent 10 μm.
3.3. GCV-Mediated Cytotoxicity in Triple Fusion Reporter Gene-Transfected Cells
Efficient tTKSR39 expression in the TF-transfected cells should facilitate the activation of GCV and subsequently enhance cell killing. Cells transfected with either CMV-fl or CMV-ttksr39 were used as negative and positive controls, respectively. The slight decrease in cell viability observed in negative control was due to the nature toxicity of GCV to cells (Figure 4). Consistent with the results of in vitro 3H-FEAU accumulation studies, the positive control showed the highest level of tTKSR39 resulting in significant decrease in cell viability (Figure 4). In contrast, all three TF-transfected cell lines exhibited less pronounced GCV-induced cell death than the positive control cells; the cell death induced by GCV in p3R-transduced cells was the lowest among three TF vectors.
Figure 4.
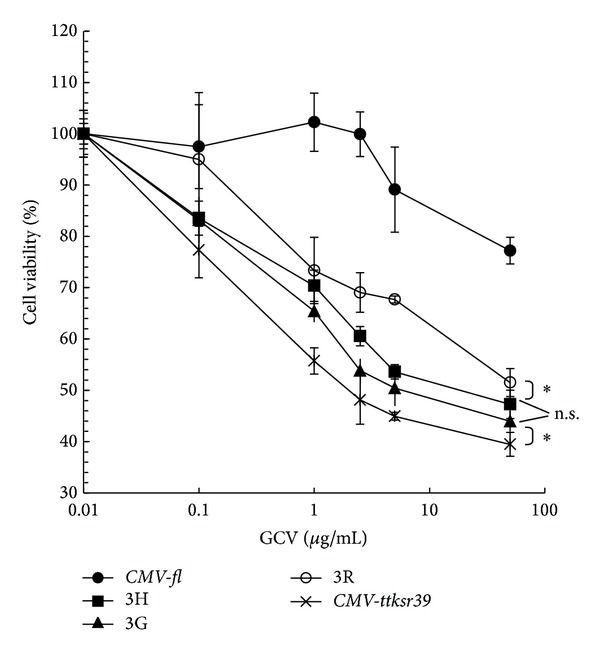
In vitro cytotoxicity effect of GCV in H1299 cells transfected with CMV-fl (●), p3H (■), p3G (▲), p3R (⚪), or CMV-ttksr39 (×) constructs. Cells transfected with these constructs were incubated with various concentrations of GCV for 5 days followed by cell survival estimation using MTT. All the experiments (n = 4) were done in triplicate. *P < 0.05; n.s.: statistically nonsignificant.
3.4. In Vivo Imaging of the Triple Fusion Reporter Gene Expression by Optical and Radionuclide Techniques
Subsequent to in vitro studies, we investigated whether the TF vectors would simultaneously and repeatedly express the three fusion proteins in living subjects and would be detectable using different imaging modalities. The H1299 cells transiently transfected with p3G, p3R, p3H, or DsRed1-ttksr39 vectors were injected subcutaneously into the shoulders and legs of 6~8-week-old nude mice (Figure 5(a)). The red fluorescence was detected in tumors grown from p3G-transfected cells with the signal (80.8 ± 3 × 106 photons/sec/cm2/sr) lower than that observed in positive control tumors established from cells expressing DsRed1-ttksr39 gene (225 ± 37 × 106 photons/sec/cm2/sr) but was not detectable in tumors grown from p3R- or p3H-transfected cells (Figure 5(b)). The expression efficacy of rl reporter gene in tumors established from p3G-transfected cells was demonstrated by bioluminescence imaging (Figure 5(c)). The bioluminescence signal of RLUC (1.65 ± 0.55 × 106 photons/sec/cm2/sr) was observed in tumors established from cells expressing p3G protein. Tumors expressing p3H protein showed 4-fold higher FLUC activity (211 ± 137 × 106 photons/sec/cm2/sr) than tumors expressing p3R protein (50.4 ± 12.44 × 106 photons/sec/cm2/sr) (Figure 5(d)). Because FEAU has a better substrate affinity to tTKsr39 enzyme than acycloguanosine FHBG [21] and the accumulation of FEAU in the gastrointestinal tract is lower than FHBG, we thus selected 18F-FEAU for monitoring of the ttksr39 reporter gene expression in living subjects (Figure 6(a)). Tumors expressing DsRed1-ttksr39 positive control gene showed 1.36-fold higher level of 18F-FEAU uptake (2.66 ± 0.26 %ID/g) than tumors expressing p3G TF gene (1.96 ± 0.38 %ID/g) (Figure 6(b)). A trend of decreasing 18F-FEAU uptake (p3G > p3H > p3R) in all three TF vectors was observed. However, the differences in 18F-FEAU uptake between each tumor did not reach statistical significance.
Figure 5.
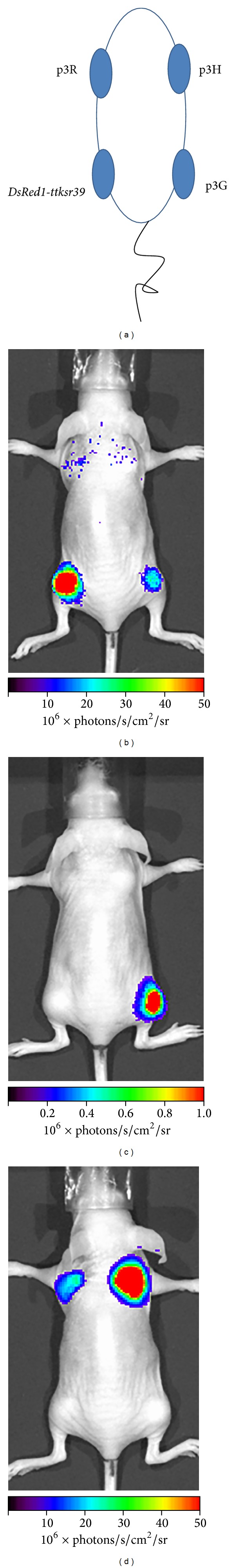
Demonstration of triple fusion gene vector in living subjects by optical imaging. Cells transiently expressing p3R, p3H, p3G, and DsRed1-ttksr39 genes were implanted subcutaneously at four different sites of a nude mouse (a). Red fluorescence signals were detectable only in the regions implanted with DsRed1-ttksr39- and p3G-expressing cells (b). Bioluminescence signals resulted from RLUC were found in p3G-expressing cells (c). Different levels of bioluminescence signals resulted from FLUC were observed in p3H- and p3R-expressing cells (d).
Figure 6.
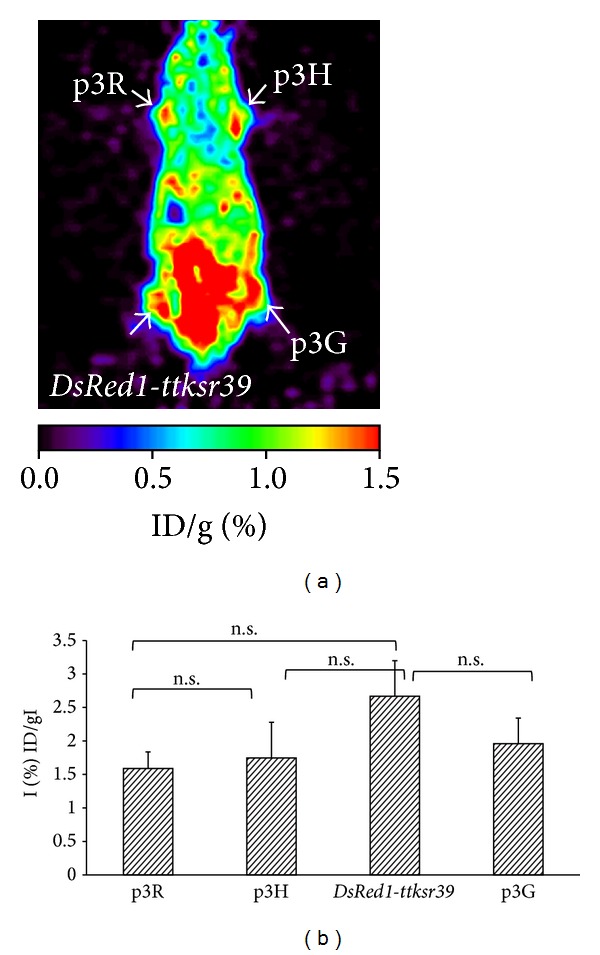
Demonstration of triple fusion gene vector in living subjects by microPET imaging. The same mouse demonstrated previously by optical imaging was then imaged by microPET using 18F-FEAU. Cells expressing the fusion reporter genes and DsRed1-ttk gene showed 18F-FEAU accumulation in the tumor (coronal section) (a). ROIs drawn over the sites of cell implantation of four mice were calculated and expressed as percentage of injected dose per gram of tissue (% ID/g). Each bar represents the mean ± SD (n = 4) (b). n.s.: statistically nonsignificant.
4. Discussion
Genetic fusion reporter systems have greatly facilitated studies on the regulation of gene expression, as well as protein localization and function using multiple imaging modalities. Several triple fusion (TF) reporter genes harboring a bioluminescence reporter genes (i.e., rl or fl), fluorescence reporter genes (i.e., mrfp or egfp), and a radionuclide imaging reporter gene (ttksr39 or wt.tk) have been utilized for in vivo imaging of small animals. Our primary interest was to develop TF reporter gene constructs for research applications in cancer, stem cell, and regenerative medicine. However, we have observed that several types of stem cells lost their proliferation ability and showed morphological abnormalities after transfection with p3R developed by Ray et al. [17]. These problems have been resolved in a novel TF gene construct-p3H by changing the orientation of fl and DsRedm genes in the expression cassette. The C-terminal region of TK is essential for maintaining nucleoside phosphorylation activity of this enzyme [11]. Moreover, the N-terminal tTKSR39 fusion proteins are susceptible to enzymatic cleavage [9]. Therefore, we placed the ttksr39 gene at the carboxyl end of a TF gene construct. The different subcellular distribution pattern of monomeric DsRed or TK in all three TF gene constructs demonstrated that the localization of fusion protein can be influenced not only by the nature of different fusion partner proteins but also by their order in the fusion.
It has been established that chromophore maturation rate and brightness of monomeric DsRed protein are lower than dimeric or tetrameric form [22, 23]. In the current study, fluorescence microscopy demonstrated that chromophore maturation rate in p3G- and p3R-transfected cells (<24 hrs) was faster than that in p3H-transfected cells (>24 hrs), which caused a significant condensation of TF protein in the cytoplasm. One possible explanation of this observation is that the sequence of proteins in p3G and 3R is more prone to formation of DsRed beta-barrel structure, as compared to p3H. Previous studies have also pointed out that due to high levels of reporter gene expression mediated by CMV promoter, the transfected cells may exhibit growth defects [24, 25] and detach from the growth surface [24]. Such cytotoxicity was typical of high levels of RFP expression that causes aggregation of this protein [24, 26]. Significant protein aggregates will lead to the impairment of ubiquitin-proteasome system (UPS) that plays an important role in regulating cell division and apoptosis and may cause cell cycle arrest in G2/M [19, 27]. This mechanism may explain at least in part the failure of our previous efforts to establish stable clones of transduced tumor cells using p3G or p3R TF constructs. By using split synthetic renilla luciferase complementation-based bioluminescence assay, Degrève et al. demonstrated the homodimerization of HSV1-TKSR39 [28]. If the tTKSR39 fusion part in p3G or p3R forms homodimer more easily than p3H, the DsRedm fusion part may also form dimeric or tetrameric form resulting in faster chromophore maturation and obvious protein aggregation. In contrast, in p3H the FLUC positioned between DsRed and tTKSR39 may act as a structural barrier to avoid dimerization or tetramerization mediated by tTKSR39. However, as the result of such positioning of RFP in p3H construct, the brightness of RFP fluorescence is relatively lower than that in p3G and p3R.
Previous crystal structure studies revealed that FLUC has a large N-terminal domain (residues 1–436) and a small C-terminal domain (residues 440–550) [29]. During the enzyme-substrate reaction, the distance between N- and C-terminal needs to be close enough to sandwich the substrate [30]. Previous studies indicated that not only the C-terminal domain of FLUC is indispensable for efficient coupling of adenylation and oxidation steps [21] but also the extreme N-terminal amino acid sequence of FLUC is important in thermal stability and proper conformation of this enzyme [31]. In fact, the N-terminal amino acids from residues 1 to 11 are not directly involved in the active site of FLUC, which is located in the cleft formed by N- and C-terminal domains [21]. The FLUC activity has been shown to be 50-fold higher in cells expressing the wt.tk-fl fusion gene, as compared to cells expressing the fl-wt.tk fusion gene [32]. To test the effect of N-terminal amino acid modification on the FLUC enzyme activity, the GAL4 DNA binding domain was fused with the N terminus of fl, and the results showed that the enzymatic activity of FLUC from GAL4-FLUC fusion protein is as normal as in the nonfused protein [31]. The N terminus of FLUC seems to be more tolerant of fusions than the C terminus. Therefore, we hypothesized that changing the sequence of genes in the fl-mrfp fusion to mrfp-fl may improve the enzymatic activity of FLUC. The results of our studies demonstrated a 3-fold improvement of FLUC enzymatic activity in p3H-expressing cells, as compared to the p3R-expressing cells, which supports our hypothesis. Another probable reason for impaired FLUC activity is that the close proximity of the bulky DsRedtTKSR39 to FLUC may increase the distance between C- and N-terminal domains of FLUC and interfere with the active site formation and/or function [30, 31]. As expected, the functional activity of FLUC was partially preserved in both p3H and p3R TF gene constructs.
The truncated tksr39 we choose for one partner of our TF gene construct lacks the first 45 amino acids and is designed to disrupt its nuclear localization signal (NLS) and a putative cryptic testicular-specific promoter, which causes sterility in transgenic mouse males [28, 33]. TF constructs harboring this truncated protein exhibit pancellular distribution of the tTKSR39 reporter protein, whereas the wild type TK (wt.TK) localizes predominantly to the cell nucleus and results in improved total cellular enzymatic activity [18]. In addition, the TKSR39 exhibits increased sensitivity towards different acycloguanosine analogues when compared with wt.TK [3]. The TKSR39 has been extensively studied as a reporter gene for noninvasive imaging with PET using different radiolabeled acycloguanosine analogues as reporter probes, including [14C]1-(2′-fluoro-2′-deoxy-d-arabinofuranosyl)-5-methyluracil (14C-FMAU), [3H]2′-fluoro-2′-deoxyarabinofuranosyl-5-ethyluracil (3H-FEAU), [14C]2′-fluoro-2′-deoxy-β-d-arabinofuranosyl-5-iodouracil (14C-FIAU), and [3H] penciclovir (3H-PCV) [34]. In the current study, the immunofluorescence images of tTKSR39 protein demonstrated that the p3H-expressing cells have more cytoplasmic localization of TF protein than the p3R-expressing cells indicating that higher tTKSR39 activity is preserved in the p3H construct. Moreover, a significant aggregation of TF protein was observed in the cytoplasm and, especially, in the perinuclear regions of p3R-expressing cells. Such abnormal aggregation of p3R protein had a negative effect on the efficacy of GCV prodrug activating ability of tTKSR39 and resulted in decrease in GCV-induced cytotoxicity, as well as decreased the cellular uptake of 3H-FEAU, as compared to p3G or p3H reporter constructs. Furthermore, in mouse imaging studies we observed the red fluorescence signals in the implanted tumors established from p3G-transfected cells, albeit the RFP gene expression of p3G was significantly lower than the DsRed1-ttksr39 (positive control). This observation is consistent with previous reports demonstrating that DsRed1 gene expressed a tetrameric form that has substantially higher fluorescence than the monomeric form [22, 23]. In addition to the strong red fluorescent signal and relatively high RLUC, the TK activity is also preserved in the aggregation-prone p3G TF protein. This provides evidence that the aggregation of p3G TF protein might not severely inhibit the function of the individual proteins in this TF reporter. The red fluorescence of the p3R- and p3H-expressing cells was weak and easily scattered and attenuated by the surrounding tissues in vivo. The fl gene expression can be successfully monitored in vivo, with 4-fold higher FLUC activity in the p3H-expressing cells than in the p3R-expressing cells indicating that the FLUC activity in TF gene constructs is preserved and is sufficient for in vivo bioluminescence imaging.
In the current work we clearly demonstrated the differences of tTKSR39 activity among three TF gene constructs by in vitro cellular uptake, but the differences in ttksr39 gene expression (1.25- to 1.4-folds) are too small to be discriminated by in vivo PET imaging. The individual differences of mice and the transfection efficiency contributed by different gene constructs may mask the small differences in tTKSR39 activity. Although it seems attractive to perform multiple imaging modalities with p3G TF construct, which retained the most activities of each fusion partners among all three TF constructs, the following criteria are in favor of using fl for BLI instead of rl: (1) in rl, the tissue penetration of light is limited due to emissions peaking at 480 nm; (2) the multidrug resistant MDR1 p-glycoprotein (PgP) which overexpresses in cancer cells [35] can alter the transport of coelenterazine (the RLUC substrate), resulting in a decreased bioluminescence signals; (3) while injected with i.p., the autoluminescence contributes to high background [36]; (4) coelenterazine is more expensive than D-luciferin (FLUC substrate). We also noted that the origin/source of fl reporter gene used by Ray et al. to construct the first and second generation TF gene constructs were different [13]. The nucleotide sequence of a fusion partner gene can influence the structure and stability of TF mRNA and further affect the efficiency of TF protein translation. It may explain why Ray et al. chose the wt.tk as the fusion partner in their second generation TF construct, even though the enzymatic affinity of tTKSR39 mutant to acycloguanisines is higher than that of the wt.TK. In order to eliminate any undesired effect resulting from certain fusion partners of uncertain origin/source, we used commercially available, codon optimized fusion partner genes to construct the p3G, p3R, and p3H. Thus, the different efficacies between p3R and p3H fusion protein demonstrated in this study resulted solely from the orientation of the genes in the reporter construct.
Our results demonstrated a decreased tTKSR39 activity in all fusion constructs. Longer spacer between the two fusion partner genes might improve the function of each protein [9]. Truncation of the first 135bp of the native tksr39 coding sequence shortened the distance between fl and ttksr39 genes. Thus, a longer spacer composed of small flexible amino acids, such as poly-glycine, or the use of the minimally truncated tksr39 gene (only with deletion of N-amino terminal nuclear localization signal) instead of ttksr39 can be considered as another option for the next generation of TF gene. Moreover, human codon optimization could be implemented to alter the NLS and the cryptic testicular promoter regions of the full-length tksr39 for applications in transgenic mice. Such kind of modification can remove the high frequency of CpG island in tksr39 gene and eliminate the CpG methylation associated with transcriptional silencing, resulting in high-yield and long-term gene expression of tk [35]. Several new DsRed variant genes, such as mCherry or mRaspberry, could also be used for the new generation of TF genes, which will benefit from far-red emissions spectra and little overlap with GFP and YFP [37].
In conclusion, we have developed an optimized construct of triple reporter gene for multimodality molecular imaging in vitro and in vivo. The DsRedm-fl-ttksr39 triple reporter gene is less cytotoxic, more effective, and sensitive for in vitro cell imaging as well as in vivo animal imaging using fluorescence, bioluminescence, and PET imaging modalities, which may facilitate different fields of translational biomedical research and applications.
Acknowledgments
This research was supported by the following grants: NSC 102-2314-B-010-038-MY3, NSC 102-2627-M-010-003 (National Science Council, Taiwan), MOHW103-TD-B-111-02 (Department of Health), and V102C-082 (Taipei Veterans General Hospital). The authors thank the technical support from Molecular and Genetic Imaging Core, Taiwan Mouse Clinic (NSC 102-2325-B-001-042), which is funded by the National Research Program for Biopharmaceuticals (NRPB) at the National Science Council (NSC) of Taiwan and Mr. Tsung-Hsun Yu and Ms. Tsuey-Ling Jan for the assistance in preparing the paper.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contribution
Luen Hwu and Ya-Ju Hsieh contributed equally to the study.
References
- 1.Massoud TF, Gambhir SS. Molecular imaging in living subjects: seeing fundamental biological processes in a new light. Genes and Development. 2003;17(5):545–580. doi: 10.1101/gad.1047403. [DOI] [PubMed] [Google Scholar]
- 2.Herschman HR. Molecular imaging: looking at problems, seeing solutions. Science. 2003;302(5645):605–608. doi: 10.1126/science.1090585. [DOI] [PubMed] [Google Scholar]
- 3.Ray P, De A, Min J-J, Tsien RY, Gambhir SS. Imaging tri-fusion multimodality reporter gene expression in living subjects. Cancer Research. 2004;64(4):1323–1330. doi: 10.1158/0008-5472.can-03-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alford R, Ogawa M, Choyke PL, Kobayashi H. Molecular probes for the in vivo imaging of cancer. Molecular BioSystems. 2009;5(11):1279–1291. doi: 10.1039/b911307j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connell-Rodwell CE, Burns SM, Bachmann MH, Contag CH. Bioluminescent indicators for in vivo measurements of gene expression. Trends in Biotechnology. 2002;20(8):S19–S23. [Google Scholar]
- 6.Doubrovin M, Serganova I, Mayer-Kuckuk P, Ponomarev V, Blasberg RG. Multimodality in vivo molecular-genetic imaging. Bioconjugate Chemistry. 2004;15(6):1376–1388. doi: 10.1021/bc0498572. [DOI] [PubMed] [Google Scholar]
- 7.Jacobs A, Dubrovin M, Hewett J, et al. Functional coexpression of HSV-1 thymidine kinase and green fluorescent protein: implications for noninvasive imaging of transgene expression. Neoplasia. 1999;1(2):154–161. doi: 10.1038/sj.neo.7900007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gambhir SS, Bauer E, Black ME, et al. A mutant herpes simplex virus type 1 thymidine kinase reporter gene shows improved sensitivity for imaging reporter gene expression with positron emission tomography. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(6):2785–2790. doi: 10.1073/pnas.97.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ray P, Wu AM, Gambhir SS. Optical bioluminescence and positron emission tomography imaging of a novel fusion reporter gene in tumor xenografts of living mice. Cancer Research. 2003;63(6):1160–1165. [PubMed] [Google Scholar]
- 10.Ponomarev V, Doubrovin M, Serganova I, et al. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. European Journal of Nuclear Medicine and Molecular Imaging. 2004;31(5):740–751. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 11.Saijo M, Suzutani T, Niikura M, Morikawa S, Kurane I. Importance of C-terminus of herpes simplex virus type 1 thymidine kinase for maintaining thymidine kinase and acyclovir-phosphorylation activities. Journal of Medical Virology. 2002;66(3):388–393. doi: 10.1002/jmv.2157. [DOI] [PubMed] [Google Scholar]
- 12.Kim YJ, Dubey P, Ray P, Gambhir SS, Witte ON. Multimodality imaging of lymphocytic migration using lentiviral-based transduction of a tri-fusion reporter gene. Molecular Imaging and Biology. 2004;6(5):331–340. doi: 10.1016/j.mibio.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 13.Ray P, Tsien R, Gambhir SS. Construction and validation of improved triple fusion reporter gene vectors for molecular imaging of living subjects. Cancer Research. 2007;67(7):3085–3093. doi: 10.1158/0008-5472.CAN-06-2402. [DOI] [PubMed] [Google Scholar]
- 14.Cao F, Lin S, Xie X, et al. In vivo visualization of embryonic stem cell survival, proliferation, and migration after cardiac delivery. Circulation. 2006;113(7):1005–1014. doi: 10.1161/CIRCULATIONAHA.105.588954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao F, Drukker M, Lin S, et al. Molecular imaging of embryonic stem cell misbehavior and suicide gene ablation. Cloning and Stem Cells. 2007;9(1):107–117. doi: 10.1089/clo.2006.0E16. [DOI] [PubMed] [Google Scholar]
- 16.Love Z, Wang F, Dennis J, et al. Imaging of mesenchymal stem cell transplant by bioluminescence and PET. Journal of Nuclear Medicine. 2007;48(12):2011–2020. doi: 10.2967/jnumed.107.043166. [DOI] [PubMed] [Google Scholar]
- 17.Kim YH, Lee DS, Kang JH, et al. Reversing the silencing of reporter sodium/iodide symporter transgene for stem cell tracking. Journal of Nuclear Medicine. 2005;46(2):305–311. [PubMed] [Google Scholar]
- 18.Söling A, Simm A, Rainov NG. Intracellular localization of Herpes simplex virus type 1 thymidine kinase fused to different fluorescent proteins depends on choice of fluorescent tag. FEBS Letters. 2002;527(1–3):153–158. doi: 10.1016/s0014-5793(02)03201-5. [DOI] [PubMed] [Google Scholar]
- 19.Massoub TF, Paulmurugan R, Gambhir SS. Molecular imaging of homodimeric protein-protein interactions in living subjects. FASEB Journal. 2004;18(10):1105–1107. doi: 10.1096/fj.03-1128fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 21.Ayabe K, Zako T, Ueda H. The role of firefly luciferase C-terminal domain in efficient coupling of adenylation and oxidative steps. FEBS Letters. 2005;579(20):4389–4394. doi: 10.1016/j.febslet.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nature Biotechnology. 2004;22(12):1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 23.Strongin DE, Bevis B, Khuong N, et al. Structural rearrangements near the chromophore influence the maturation speed and brightness of DsRed variants. Protein Engineering, Design and Selection. 2007;20(11):525–534. doi: 10.1093/protein/gzm046. [DOI] [PubMed] [Google Scholar]
- 24.Strack RL, Strongin DE, Bhattacharyya D, et al. A noncytotoxic DsRed variant for whole-cell labeling. Nature Methods. 2008;5(11):955–957. doi: 10.1038/nmeth.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tao W, Evans B-G, Yao J, et al. Enhanced green fluorescent protein is a nearly ideal long-term expression tracer for hematopoietic stem cells, whereas DsRed-express fluorescent protein is not. Stem Cells. 2007;25(3):670–678. doi: 10.1634/stemcells.2006-0553. [DOI] [PubMed] [Google Scholar]
- 26.Yanushevich YG, Staroverov DB, Savitsky AP, et al. A strategy for the generation of non-aggregating mutants of Anthozoa fluorescent proteins. FEBS Letters. 2002;511(1–3):11–14. doi: 10.1016/s0014-5793(01)03263-x. [DOI] [PubMed] [Google Scholar]
- 27.Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Molecular Cell. 2005;17(3):351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 28.Degreve B, Esnouf R, de Clercq E, Balzarini J. Characterization of multiple nuclear localization signals in herpes simplex virus type 1 thymidine kinase. Biochemical and Biophysical Research Communications. 1999;264(2):338–342. doi: 10.1006/bbrc.1999.1485. [DOI] [PubMed] [Google Scholar]
- 29.Morton RA, Hopkins TA, Seliger HH. The spectroscopic properties of firefly luciferin and related compounds. An approach to product emission. Biochemistry. 1969;8(4):1598–1607. doi: 10.1021/bi00832a041. [DOI] [PubMed] [Google Scholar]
- 30.Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a super-family of adenylate-forming enzymes. Structure. 1996;4(3):287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 31.Sung D, Kang H. The N-terminal amino acid sequences of the firefly luciferase are important for the stability of the enzyme. Photochemistry and Photobiology. 1998;68(5):749–753. doi: 10.1562/0031-8655(1998)068<0749:tntaas>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 32.Söling A, Theiß C, Jungmichel S, Rainov NG. A dual function fusion protein of Herpes simplex virus type 1 thymidine kinase and firefly luciferase for noninvasive in vivo imaging of gene therapy in malignant glioma. Genetic Vaccines and Therapy. 2004;2(1, article 7) doi: 10.1186/1479-0556-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cohen JL, Boyer O, Salomon B, et al. Fertile homozygous transgenic mice expressing a functional truncated herpes simplex thymidine kinase ΔTK gene. Transgenic Research. 1998;7(5):321–330. doi: 10.1023/a:1008893206208. [DOI] [PubMed] [Google Scholar]
- 34.Kang KW, Min J-J, Chen X, Gambhir SS. Comparison of [14C]FMAU, [3H]FEAU, [14C]FIAU, and [3H]PCV for monitoring reporter gene expression of wild type and mutant Herpes simplex virus type 1 thymidine kinase in cell culture. Molecular Imaging and Biology. 2005;7(4):296–303. doi: 10.1007/s11307-005-0010-7. [DOI] [PubMed] [Google Scholar]
- 35.Ponomarev V, Doubrovin M, Serganova I, et al. Cytoplasmically retargeted HSV1-tk/GFP reporter gene mutants for optimization of noninvasive molecular-genetic imaging. Neoplasia. 2003;5(3):245–254. doi: 10.1016/S1476-5586(03)80056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, El-Deiry WS. Bioluminescent molecular imaging of endogenous and exogenous p53-mediated transcription in vitro and in vivo using an HCT116 human colon carcinoma xenograft model. Cancer Biology & Therapy. 2003;2(2):196–202. doi: 10.4161/cbt.2.2.347. [DOI] [PubMed] [Google Scholar]
- 37.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2(12):905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]


