Abstract
Seven phenylalanine derivatives with small ortho substitutions were genetically encoded in Escherichia coli and mammalian cells at an amber codon using a previously reported, rationally designed pyrrolysyl-tRNA synthetase mutant (PylRS(N346A/C348A)) coupled with tRNACUAPyl. Ortho substitutions of the phenylalanine derivatives reported herein include three halides, methyl, methoxy, nitro, and nitrile. These compounds have the potential for use in multiple biochemical and biophysical applications. Specifically, we demonstrated that o-cyano-phenylalanine could be used as a selective sensor to probe the local environment of proteins and applied this to study protein folding/unfolding. For six of these compounds this constitutes the first report of their genetic incorporation in living cells. With these compounds the total number of substrates available for PylRS(N346A/C348A) is increased to nearly 40, which demonstrates that PylRS(N346A/C348A) is able to recognize phenylalanine with a substitution at any side-chain aromatic position as a substrate. To our knowledge, PylRS(N346A/C348A) is the only aminoacyl-tRNA synthetase with such a high substrate promiscuity.
The genetic incorporation of non-canonical amino acids (NCAAs) into recombinant proteins has provided researchers with powerful tools for the study of protein structure and function. The ability to genetically incorporate NCAAs with bioorthogonal functional groups, coupled with click chemistry, allows for the introduction of site-selective protein modifications that have been used for fluorescence labeling1−8and photo-cross-linking,9−13 as well as for the study of posttranslational modifications that are traditionally difficult or impossible to obtain in bacterial expression systems.14−18 Other analytic probes that have been site-selectively introduced using the genetic incorporation approach include infrared, Raman, NMR, and EPR spectroscopic probes and heavy atoms for X-ray crystallographic studies.19−21 Generally, NCAAs are introduced into a protein through the suppression of an in frame nonsense or quadruplet codon by a suppressor tRNA coupled with an evolved aminoacyl-tRNA synthetase (aaRS). One widely used synthetase for this purpose is the tyrosyl-tRNA synthetase (MjTyrRS) derived from Methanocaldococcus jannaschii.22 Indeed, directed evolution of MjTyrRS, coupled with a suppressor tRNACUATyr, for the suppression of the TAG (amber) codon has been highly successful at the incorporation of a number of NCAAs into proteins in Escherichia coli. Despite the success of engineering the MjTyrRS-tRNACUA pair, its weakness lies in that it cannot be used to incorporate NCAAs into eukaryotic organisms owing to the fact that tRNACUATyr contains a recognition element that allows for the aminoacylation of tRNACUA with canonical amino acids by the endogenous aaRSs in eukaryotic organisms.23,24 Alternatives to the MjTyrRS-tRNACUATyr pair include the EcTyrRS-tRNACUA pair, which was derived from E. coli tyrosyl-tRNA synthetase-tRNATyr pair, the EcLeuRS-tRNACUALeu pair derived from E. coli leucyl-tRNA synthetase-tRNALeu pair, and the pyrrolysyl-tRNA synthetase (PylRS)-tRNACUA pair, which naturally exists in certain methanogenic archaea and some bacteria. Both EcTyrRS-tRNACUATyr and EcLeuRS-tRNACUA pairs can be applied in eukaryotic cells but not in bacterial systems.11,25,26 However, tRNACUAPyl is a naturally occurring amber suppressor tRNA that is specifically recognized by PylRS but does not cross-interact with endogenous aaRSs in both bacterial and eukaryotic cells.27−31
To date, the PylRS-tRNACUAPyl pair has been successfully used for the genetic incorporation of NCAAs into recombinant proteins expressed in bacteria and a variety of eukaryotes including mammalian cells and multicellular organisms.32,33 While the ability of the PylRS-tRNACUA pair to be used in both bacterial and eukaryotic cells is advantageous in that it allows for the same NCAA to be used in different model systems, the method still has limitations. The incorporation of a new NCAA typically requires selection of large libraries of aaRS with randomized active site mutations. Schultz and co-workers have developed a powerful method for the directed evolution of aaRSs involving both positive and negative selections based on the ability to suppress a nonsense mutation in the presence of the desired NCAA.34 However, this process still requires multiple rounds of selection for each new substrate. Therefore, a single aaRS with broad substrate promiscuity is ideal to facilitate rapid progress and widespread use of the genetic NCAA incorporation technique. We have previously reported the ability of a single rationally designed PylRS mutant (PylRS(N346A/C348A)), coupled with tRNACUAPyl, to genetically incorporate a wide variety of phenylalanine derivatives with large para and small meta substitutions at amber mutation sites in living cells.35−37 More recent studies indicate that this mutant is also able to recognize more than 10 phenylalanine derivatives with large meta substitutions (Tuley and Liu, unpublished data). PylRS(N346A/C348A) displays high activities toward these NCAAs while remaining relatively orthogonal toward canonical amino acids. Here we reveal that the same enzyme is also able to mediate the genetic incorporation of seven ortho-substituted phenylalanine derivatives, which brings the total substrate availability of this enzyme close to 40 and leads to the conclusion that PylRS(N346A/C348A) is able to recognize phenylalanine derivatives with a substitution at any side-chain aromatic position as a substrate. To our knowledge, PylRS(N346A/C348A) is the only aaRS with such a high substrate promiscuity. We also demonstrate, for the first time, the ability of PylRS(N346A/C348A) to facilitate the genetic incorporation of phenylalanine derivatives in mammalian cells. Aside from o-nitro-phenylalanine,38 which has been genetically encoded in E. coli using an evolved MjTyrRS-tRNACUA pair, this constitutes the first report of the genetic incorporation for all of these ortho-substituted phenylalanine derivatives.39 One of the ortho-substituted phenylalanine derivatives, o-cyano-phenylalanine (oCNF) is a small, environmentally sensitive, fluorescent probe that can be used as a sensor to probe the local environment of proteins, and we applied this to study protein folding/unfolding.
Wang and co-workers previously determined the crystal structure of a PylRS mutant (OmeRS) in complex with O-methyl-tyrosine.40 Figure 1 presents the active site structure of this enzyme–substrate complex superimposed on the wild-type PylRS with pyrrolysyl-AMP bound. The two overlaid structures clearly show that the N346 side chain is adjacent to the ortho position of the O-methyl-tyrosine side-chain phenyl group. Removing the N346 side chain would potentially leave space for adding an additional substitution to this position. Since PylRS(N346A/C348A) is expected to bind to a phenylalanine derivative in a similar fashion as OmeRS, we suspect that PylRS(N346A/C348A) may well recognize an ortho-substituted phenylalanine derivative and, together with tRNACUAPyl, mediate its incorporation at an amber mutation site. We exploited an E. coli cell system harboring plasmids pEVOL-pylT-PylRS(N346A/C348A) and pET-pylT-sfGFPS2TAG to assess the ability of the PylRS(N346A/C348A)-tRNACUA pair to incorporate ortho-substituted phenylalanine derivatives in response to an amber mutation. The plasmid pEVOL-pylT-PylRS(N346A/C348A) contains genes coding for PylRS(N346A/C348A) and tRNACUAPyl, while pET-pylT-sfGFPS2TAG contains genes coding for tRNACUA and superfolder green fluorescent protein (sfGFP) with an amber mutation at its S2 position and a 6 × His tag at the C-terminus.35 Growing cells in GMML medium (a minimal medium supplemented with 1% glycerol and 0.3 mM l-leucine) led to translation termination at the amber mutation site of sfGFP; therefore no full-length sfGFP was expressed. However, supplementing the medium with 2 mM of one of seven ortho-substituted phenylalanine derivatives (1–7, shown in Figure 2A) promoted overexpression of full-length sfGFP (Figure 2B). Incorporation of NCAAs was confirmed via electrospray ionization mass spectrometry (ESI-MS) (Figure 2C). For each of the seven NCAAs, the detected molecular weight agreed well with the theoretical value (Table 1). Except for 6, this is the first report of the genetic incorporation of these NCAAs in living cells, and for all of the NCAAs this is the first report of their incorporation using an aaRS derive from PylRS.
Figure 1.
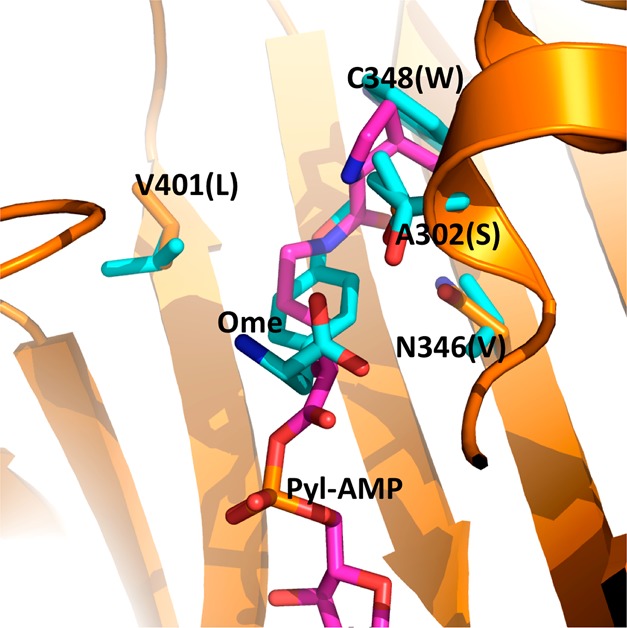
Superimposed structures of the OmeRS complex with O-methyltyrosine and the PylRS complex with pyrrolysyl-adenosyl monophosphate (Pyl-AMP). The structures are based on PDB entries 2Q7H and 3QTC. The PylRS complex with Pyl-AMP is shown in orange for the protein carbon atoms and pink for the Pyl-AMP carbon atoms. Four mutated residues in OmeRS and the p-methoxy-phenylalanine ligand are shown in cyan for the carbon atoms. Letters in parentheses indicate the four mutated residues in OmeRS.
Figure 2.
(A) Structures of 1–7. (B) Site-specific incorporation of 1–7 into sfGFP at its S2 site. N/A indicates no NCAA was available in the medium. ND represents nondetected. (C) Deconvoluted ESI-MS of sfGFP with NCAAs 1–7 incorporated at the S2 position. sfGFP-X in which X is one of 1–7 represents the specific sfGFP variant with X incorporated at its S2 site.
Table 1. ESI-MS Characterization of sfGFP Variants Incorporated with Different NCAAs.
| NCAA | calculated mass (Da) | detected mass (Da)a |
|---|---|---|
| 1 | 27764 | 27761 |
| 2 | 27808 | 27804 |
| 3 | 27855 | 27855 |
| 4 | 27743 | 27741 |
| 5 | 27759 | 27759 |
| 6 | 27774 | 27773 |
| 7 | 27754 | 27751 |
With an error of ±1 Da.
Along with 1–7, two additional amino acids were tested, namely, o-trifluoromethyl-phenylalanine (8) and o-fluoro-phenylalanine (9) (Supplementary Figure 1). In GMML supplemented with 2 mM NCAA 8, we obtained minimal expression of full-length sfGFP. The mass spectra of the purified protein suggested the incorporation of both 8 and phenylalanine at the S2 site (Supplementary Figure 2), indicating the recognition of 8 by PylRS(N346A/C348A) is not significantly better than that of phenylalanine itself. Minimal incorporation of phenylalanine at an amber codon is a typical observation for evolved MjTyrRS-tRNACUATyr and PylRS-tRNACUA pairs.41 In a previous study, we found that while PylRS(N346A/C348A) was able to incorporate m-fluoro-phenylalanine at an amber codon, ESI-MS analysis of the expressed protein indicated that this NCAA was also incorporated at phenylalanine codons.36 Given the size similarity of fluorine and a proton, we attributed the miss-incorporation of m-fluoro-phenylalanine at phenylalanine sites to an inability of the endogenous phenylalanyl-tRNA synthetase to distinguish between the two substrates. Similarly, when testing the incorporation of 9 at the S2 site of sfGFP, we found that the miss-incorporation rate was high. Indeed, major ESI-MS peaks of the purified protein suggest the incorporation of 9 into sfGFP at multiple sites (Supplementary Figure 3). Taken together, these observations suggest that while the PylRS(N346A/C348A)-tRNACUAPyl pair is able to direct the genetic incorporation of a monofluorinated phenylalanine at amber codons, E. coli cells with a more stringent phenylalanyl-tRNA synthetase that excludes monofluorinated phenylalanine as a substrate will need to be engineered for cleaner incorporation.
In one of our previous publications, we showed that PylRS(N346A/C348A) recognizes phenylalanine and mediates its incorporation at an amber mutation site in coordination with tRNACUAPyl.35 However, this background incorporation was suppressed when a NCAA that serves as a better substrate of PylRS(N346A/C348A) was provided. For instance, the PylRS(N346A/C348A)-tRNACUA pair induced significant background phenylalanine incorporation at an amber mutation at F27 of sfGFP in LB medium. However, providing m-trifluoromethyl-phenylalanine obviated this background incorporation, and the expressed sfGFP had quantitative occupancy of m-trifluoromethyl-phenylalanine at F27.36 We used the same system to test the incorporation efficiencies of seven ortho-substituted phenylalanine derivatives (2 mM) in LB. Our data show that, except for 5, all NCAAs led to sfGFP expression significantly better (1.8–3.9-fold) than the background (Supplementary Figure 4). Further, although the sfGFP expression yield in the presence of 5 was comparable to the background, the major peak in the ESI-MS spectrum of purified sfGFP indicated occupancy of 5 at the designated amber mutation site (Supplementary Figure 5), indicating 5 was efficiently incorporated. Presumably, further optimization studies that modify expression conditions, such as increasing the concentration of 5, may lead to a higher incorporation efficiency. These data demonstrate that the practical use of the PylRS(N346A/C348A)-tRNACUAPyl pair in LB for the genetic incorporation of NCAAs is dependent on the NCAA identities and their concentrations.
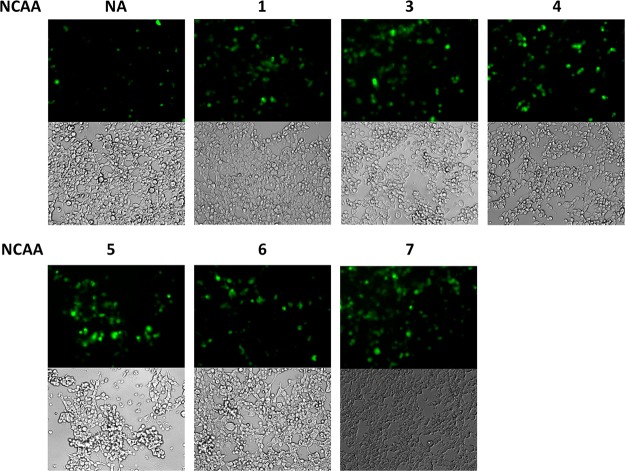
Next we asked whether the PylRS(N346A/C348A)-tRNACUAPyl pair could be used to incorporate NCAAs into proteins in mammalian cells. Given that rich media containing high concentrations of canonical amino acids is required for mammalian cell growth, PylRS(N346A/C348A) must be more reactive toward a NCAA than phenylalanine in the medium for its practical use in mammalian cells. To test the ability of the PylRS(N346A/C348A)-tRNACUA pair to facilitate the selective incorporation of NCAAs into proteins in mammalian cells, HEK293T cells were transfected with plasmids containing genes encoding a C-terminally GFP-fused epidermal growth factor receptor (EGFR-GFP) with an amber mutation at the N128 position as well as PylRS(N346A/C348A) and tRNACUAPyl.42 Cells in cultures supplemented with 5 mM of NCAAs 1 or 3–7 displayed much greater GFP fluorescence intensities compared to cells in the absence of a NCAA, suggesting enhanced suppression of the amber codon in the presence of NCAAs (Figure 3). Supplementing cultures with 5 mM 2 led to cell death. This is likely due to the insolubility of 2 in the growth medium. As expected, a low level of background protein expression was observed in the control culture, which was probably attributable to background phenylalanine incorporation at the amber mutation.
Figure 3.
Expression of EGFP-N128→X in HEK293T cells cotransfected with the plasmid containing genes encoding PylRS(N346A/C348A) and tRNACUAPyl. Cell cultures were supplemented with 5 mM of 1–7 or no NCAA (NA). Supplementing the cultures with 5 mM of compound 2 resulted in nearly 100% cell mortality. The relative ratio of expressed EGFP proteins compared to control based on the fluorescent intensity is 2.6, 4.3, 3.2, 4.1, 2.2, and 4.9 for NCAAs 1, 2, 4, 5, 6, and 7, respectively.
Of the seven ortho-substituted phenylalanine substrates available to PylRS(N346A/C348A), 3 has the potential to be used for protein labeling via the palladium-catalyzed Suzuki–Miyaura cross-coupling reaction,37,43 and 6 can be used to cleave the polypeptide backbone upon irradiation at 365 nm, as a fluorescent distance probe due to its ability to quench the fluorescence of tryptophan, and as an IR probe due to the strong vibrational absorption of the nitro group.38,44,457 could also be used as an IR probe due to the strong vibrational absorption of the nitrile group19 and as a sensor of protein local environment, protein folding/dynamics, and protein–ligand interactions due to the environmentally sensitive fluorescent features of the benzonitrile moiety. Indeed, an isomeric NCAA of oCNF, p-cyano-phenylalanine (pCNF), has been widely used as a reporter of protein local environment to study folding/dynamic analysis and protein–peptide interactions.46,47 Additionally, pCNF can be selectively excited in the presence of both tryptophan and tyrosine and has considerable spectral overlap with both fluorophores; therefore, its ability to form a Förster resonance energy transfer (FRET) with these residues was also recently explored.48,49oCNF could potentially serve as an alternative to pCNF and, because of its naturally occurring orthogonal tRNACUAPyl, could be used in eukaryotic systems. Since the intermediate polarity of the nitrile group is well tolerated in both polar and nonpolar environments and the nitrile group is relatively small, replacing phenylalanine with oCNF will minimally disturb the protein’s structure and folding pattern.49 Our preliminary measurements revealed that 7 has absorption and fluorescence spectra similar to those of pCNF and displays a nearly 10-fold increase in fluorescence when going from a hydrophobic to hydrophilic solvent (Supplementary Figure 6), suggesting that 7 could be used as a probe of protein local environment.
To test whether 7 could be used as a fluorescent probe of protein local environment, two sfGFP proteins were expressed: sfGFP-N135→7 and sfGFP-F27→7 with 7 incorporated at N135 and F27, respectively. N135 is solvent exposed and 7 is expected to be highly fluorescent at this site; however, F27 is buried within the hydrophobic interior of the protein, and therefore 7 incorporated at this position should display reduced fluorescence. Previous measurements indicate that mutations at these two positions do not significantly alter the folding of sfGFP.36 The pEVOL-pylT-PylRS(N346A/C348A) plasmid was cotransformed into E. coli Top10 cells along with pBAD-sfGFP that contained sfGFP with an amber mutation at either the 27th or 135th position. Cultures were grown in a synthetic autoinduction medium50 supplemented with 2 mM NCAA 7 to afford full-length sfGFP-N135→7 and sfGFP-F27→7 in yields of 140 and 220 mg/L, respectively. No protein was detected in the absence of 7 (Supplementary Figure 7). Figure 4 shows the fluorescence spectra of the folded proteins in phosphate buffered saline at pH 7.5 when excited at 240 nm. In the folded state, sfGFP-F27→7 displays very little fluorescence, consistent with the positioning of 7 in the hydrophobic interior of the protein. However, sfGFP-N135→7 displays nearly an 8-fold increase in fluorescence intensity consistent with 7 being solvent exposed in this protein. As a control, we also measured the fluorescence of the wild-type protein under the same conditions. The wild-type protein displayed fluorescence intensity similar to that of sfGFP-F27→7 with a slightly red-shifted spectrum. These results indicate that oCNF can be selectively excited in the presence of both tyrosine and tryptophan, as sfGFP contains one tryptophan residue and nine tyrosine residues, and demonstrate that 7 serves as an effective indicator of local environment within a protein.
Figure 4.
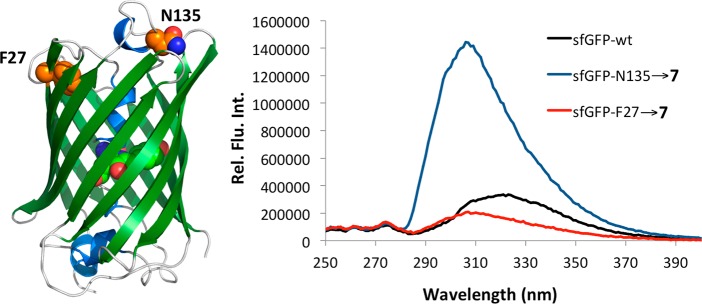
Fluorescence spectra of sfGFP-F27→7 (red line), sfGFP-N135→7 (blue line), and wild-type sfGFP (black line) excited at 240 nm. Proteins used were 20 μM in phosphate buffered saline at pH 7.5. The difference in fluorescence is due to the increased solvent exposure of 7 in sfGFP-N135→7. The structure of sfGFP is presented in the left side of the figure with F27 and N135 labeled.
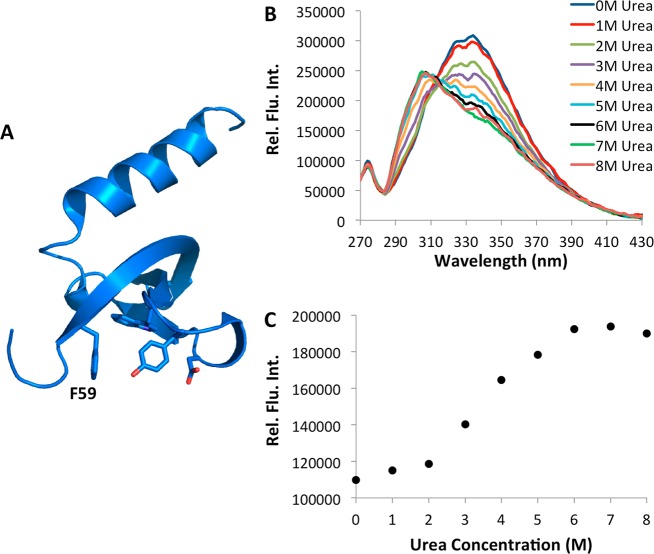
Next we demonstrated the use of 7 for the study of protein folding using the chromodomain of M-phase phosphoprotein 8 (MPP8). MPP8 has been shown to bind specifically to methylated lysine 9 of histone 3 and overexpression of MPP8 is linked to increase tumor metastasis.51 The crystal structure of MPP8 revealed that F59 is located in the hydrophobic cage that forms a binding pocket for the methylated lysine (Figure 5A).52 Therefore, in the folded protein, this residue is not exposed to the solvent and an increase in the fluorescence of 7 would be expected as the protein is denatured. The chromodomain of MPP8, containing an amber mutation at F59, was expressed as an N-terminal fusion protein to sfGFP in E. coli Top10 cells that also expressed the PylRS(N346/C348A)-tRNACUAPyl pair. Fusion to sfGFP allowed for high levels of the soluble MPP8 to be expressed.53 The cells were grown in the synthetic autoinduction medium supplemented with 2 mM NCAA 7 to afford the full length fusion protein (Supplementary Figure 8). Figure 5B displays the fluorescence spectra of MPP8 in the presence of varying concentrations of urea. At low concentrations of the denaturant, a peak corresponding to the fluorescence of 7 was not detected, consistent with this residue being buried in the hydrophobic cage. As the concentration of urea is increased from 0 to 8 molar, an increase in 7 fluorescence at 297 nm was observed accompanied by a decrease in the tryptophan fluorescence of the protein, which is consistent with both 7 and tryptophan becoming solvent exposed. No increase in fluorescence at 297 nm was observed for the wild-type protein (Supplementary Figure 9). Figure 5C shows the two-state unfolding curve for MPP8-F59→7 obtained by exciting the protein at 240 nm and measuring the resulting fluorescence at 297 nm.
Figure 5.
(A) The structure of MPP8 (based on the PDB entry 3QO2). F59 is labeled. (B) Fluorescence spectra of the MPP8-F59→7 at varied concentrations of urea. (C) Fluorescence intensity of MPP8-F59→7 measured at 297 nm as a function of the urea concentration. Proteins used were 2.5 μM in phosphate buffered saline at pH 7.5. The excitation wavelength was 240 nm. A seven point, second order Savitsky–Golay filter was applied to Figure 4B to refine curves.
In conclusion, we have demonstrated an expanded ability of the PylRS(N346A/C348A) mutant to incorporate phenylalanine derivatives with substitutions at the ortho position. The enzyme demonstrates remarkable substrate promiscuity while retaining relative orthogonality toward canonical amino acids, making it a valuable tool for the genetic incorporation of NCAAs. Nearly 40 phenylalanine derivatives have been hitherto genetically encoded at amber codons using the PylRS(N346A/C348A)-tRNACUAPyl pair. In addition, we have demonstrated the ability of PylRS(N346A/C348A) to facilitate the genetic incorporation of NCAAs in mammalian cell lines. Finally, we have demonstrated the ability of oCNF to act as a sensitive fluorescent probe of protein local environment and protein folding. oCNF shows significant differences in fluorescence intensity between hydrophobic and hydrophilic environments, and the nitrile substitution at the ortho position makes it an extremely small and noninvasive substitution in protein studies. We believe this advancement will greatly promote the adoption of the genetic NCAA incorporation approach in a broader biochemistry research field.
Acknowledgments
This work was supported in part by the National Institute of Health (grant 1R01CA161158 to W.R.L.), the National Science Foundation (grant CHEM-1148684), and the Welch Foundation (grant A-1715 to W.R.L.). We thank Y. Rezenom from the Laboratory for Biological Mass Spectrometry at Texas A&M University for characterizing proteins with electrospray ionization mass spectrometry and J. Schlessinger from Yale University for providing us the EGFR-GFP gene.
Supporting Information Available
Plasmid constructions, protein expression and purification, fluorescent analysis, and additional ESI-MS spectra. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Wang L.; Zhang Z.; Brock A.; Schultz P. G. (2003) Addition of the keto functional group to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 100, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiters A.; Schultz P. G. (2005) In vivo incorporation of an alkyne into proteins in Escherichia coli. Bioorg. Med. Chem. Lett. 15, 1521–1524. [DOI] [PubMed] [Google Scholar]
- Summerer D.; Chen S.; Wu N.; Deiters A.; Chin J. W.; Schultz P. G. (2006) A genetically encoded fluorescent amino acid. Proc. Natl. Acad. Sci. U.S.A. 103, 9785–9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.; Xie J.; Schultz P. G. (2006) A genetically encoded fluorescent amino acid. J. Am. Chem. Soc. 128, 8738–8739. [DOI] [PubMed] [Google Scholar]
- Lang K.; Davis L.; Torres-Kolbus J.; Chou C.; Deiters A.; Chin J. W. (2012) Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem. 4, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K.; Davis L.; Wallace S.; Mahesh M.; Cox D. J.; Blackman M. L.; Fox J. M.; Chin J. W. (2012) Genetic Encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels-Alder reactions. J. Am. Chem. Soc. 134, 10317–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass T.; Milles S.; Koehler C.; Schultz C.; Lemke E. A. (2011) Genetically encoded copper-free click chemistry. Angew. Chem., Int. Ed. 50, 3878–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass T.; Milles S.; Koehler C.; Szymanski J.; Mueller R.; Wiessler M.; Schultz C.; Lemke E. A. (2012) Amino acids for Diels-Alder reactions in living cells. Angew. Chem., Int. Ed. 51, 4166–4170. [DOI] [PubMed] [Google Scholar]
- Chin J. W.; Martin A. B.; King D. S.; Wang L.; Schultz P. G. (2002) Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 11020–11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin J. W.; Santoro S. W.; Martin A. B.; King D. S.; Wang L.; Schultz P. G. (2002) Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 124, 9026–9027. [DOI] [PubMed] [Google Scholar]
- Chin J. W.; Cropp T. A.; Anderson J. C.; Mukherji M.; Zhang Z.; Schultz P. G. (2003) An expanded eukaryotic genetic code. Science 301, 964–967. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy M.; Dugan A.; Nwokoye A.; Fung Y. H.; Lancia J. K.; Majmudar C. Y.; Mapp A. K. (2011) Caught in the act: covalent cross-linking captures activator-coactivator interactions in vivo. ACS Chem. Biol. 6, 1321–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Lin S.; Song X.; Liu J.; Fu Y.; Ge X.; Fu X.; Chang Z.; Chen P. R. (2011) A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat. Chem. Biol. 7, 671–677. [DOI] [PubMed] [Google Scholar]
- Liu W. R.; Wang Y. S.; Wan W. (2011) Synthesis of proteins with defined posttranslational modifications using the genetic noncanonical amino acid incorporation approach. Mol. BioSyst. 7, 38–47. [DOI] [PubMed] [Google Scholar]
- Neumann H.; Peak-Chew S. Y.; Chin J. W. (2008) Genetically encoding N(epsilon)-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234. [DOI] [PubMed] [Google Scholar]
- Nguyen D. P.; Garcia Alai M. M.; Kapadnis P. B.; Neumann H.; Chin J. W. (2009) Genetically encoding N(epsilon)-methyl-L-lysine in recombinant histones. J. Am. Chem. Soc. 131, 14194–14195. [DOI] [PubMed] [Google Scholar]
- Virdee S.; Kapadnis P. B.; Elliott T.; Lang K.; Madrzak J.; Nguyen D. P.; Riechmann L.; Chin J. W. (2011) Traceless and site-specific ubiquitination of recombinant proteins. J. Am. Chem. Soc. 133, 10708–10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. S.; Wu B.; Wang Z.; Huang Y.; Wan W.; Russell W. K.; Pai P. J.; Moe Y. N.; Russell D. H.; Liu W. R. (2010) A genetically encoded photocaged N(epsilon)-methyl-L-lysine. Mol. Biosyst. 6, 1557–1560. [DOI] [PubMed] [Google Scholar]
- Schultz K. C.; Supekova L.; Ryu Y.; Xie J.; Perera R.; Schultz P. G. (2006) A genetically encoded infrared probe. J. Am. Chem. Soc. 128, 13984–13985. [DOI] [PubMed] [Google Scholar]
- Xie J.; Wang L.; Wu N.; Brock A.; Spraggon G.; Schultz P. G. (2004) The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination. Nat. Biotechnol. 22, 1297–1301. [DOI] [PubMed] [Google Scholar]
- Wang L.; Schultz P. G. (2004) Expanding the genetic code. Angew. Chem., Int. Ed. 44, 34–66. [DOI] [PubMed] [Google Scholar]
- Wang L.; Brock A.; Herberich B.; Schultz P. G. (2001) Expanding the genetic code of Escherichia coli. Science 292, 498–500. [DOI] [PubMed] [Google Scholar]
- Wang L.; Schultz P. G. (2005) Expanding the genetic code. Angew. Chem., Int. Ed. 44, 34–66. [DOI] [PubMed] [Google Scholar]
- Thibodeaux G. N.; Liang X.; Moncivais K.; Umeda A.; Singer O.; Alfonta L.; Zhang Z. J. (2010) Transforming a pair of orthogonal tRNA-aminoacyl-tRNA synthetase from Archaea to function in mammalian cells. PLoS One 5, e11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.; Brock A.; Chen S.; Chen S.; Schultz P. G. (2007) Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods 4, 239–244. [DOI] [PubMed] [Google Scholar]
- Wu N.; Deiters A.; Cropp T. A.; King D.; Schultz P. G. (2004) A genetically encoded photocaged amino acid. J. Am. Chem. Soc. 126, 14306–14307. [DOI] [PubMed] [Google Scholar]
- Blight S. K.; Larue R. C.; Mahapatra A.; Longstaff D. G.; Chang E.; Zhao G.; Kang P. T.; Green-Church K. B.; Chan M. K.; Krzycki J. A. (2004) Direct charging of tRNA(CUA) with pyrrolysine in vitro and in vivo. Nature 431, 333–335. [DOI] [PubMed] [Google Scholar]
- Polycarpo C.; Ambrogelly A.; Berube A.; Winbush S. M.; McCloskey J. A.; Crain P. F.; Wood J. L.; Soll D. (2004) An aminoacyl-tRNA synthetase that specifically activates pyrrolysine. Proc. Natl. Acad. Sci. U.S.A. 101, 12450–12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan G.; James C. M.; Krzycki J. A. (2002) Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 296, 1459–1462. [DOI] [PubMed] [Google Scholar]
- Mukai T.; Kobayashi T.; Hino N.; Yanagisawa T.; Sakamoto K.; Yokoyama S. (2008) Adding l-lysine derivatives to the genetic code of mammalian cells with engineered pyrrolysyl-tRNA synthetases. Biochem. Biophys. Res. Commun. 371, 818–822. [DOI] [PubMed] [Google Scholar]
- Hancock S. M.; Uprety R.; Deiters A.; Chin J. W. (2010) Expanding the genetic code of yeast for incorporation of diverse unnatural amino acids via a pyrrolysyl-tRNA synthetase/tRNA pair. J. Am. Chem. Soc. 132, 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss S.; Chin J. W. (2011) Expanding the genetic code of an animal. J. Am. Chem. Soc. 14196–14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish A. R.; She X.; Xiang Z.; Coin I.; Shen Z.; Briggs S. P.; Dillin A.; Wang L. (2012) Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl-tRNA synthetase/tRNA pairs. ACS Chem. Biol. 7, 1292–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro S. W.; Wang L.; Herberich B.; King D. S.; Schultz P. G. (2002) An efficient system for the evolution of aminoacyl-tRNA synthetase specificity. Nat. Biotechnol. 20, 1044–1048. [DOI] [PubMed] [Google Scholar]
- Wang Y. S.; Fang X.; Wallace A. L.; Wu B.; Liu W. R. (2012) A rationally designed pyrrolysyl-tRNA synthetase mutant with a broad substrate spectrum. J. Am. Chem. Soc. 134, 2950–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. S.; Fang X.; Chen H. Y.; Wu B.; Wang Z. U.; Hilty C.; Liu W. R. (2013) Genetic incorporation of twelve meta-substituted phenylalanine derivatives using a single pyrrolysyl-tRNA synthetase mutant. ACS Chem. Biol. 8, 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. S.; Russell W. K.; Wang Z.; Wan W.; Dodd L. E.; Pai P. J.; Russell D. H.; Liu W. R. (2011) The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives. Mol. Biosyst. 7, 714–717. [DOI] [PubMed] [Google Scholar]
- Peters F. B.; Brock A.; Wang J.; Schultz P. G. (2009) Photocleavage of the polypeptide backbone by 2-nitrophenylalanine. Chem. Biol. 16, 148–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C. C.; Schultz P. G. (2010) Adding new chemistries to the genetic code. Annu. Rev. Biochem. 413–444. [DOI] [PubMed] [Google Scholar]
- Takimoto J. K.; Dellas N.; Noel J. P.; Wang L. (2011) Stereochemical basis for engineered pyrrolysyl-tRNA synthetase and the efficient in vivo incorporation of structurally divergent non-native amino acids. ACS Chem. Biol. 6, 733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odoi K. A.; Huang Y.; Rezenom Y. H.; Liu W. R. (2013) Nonsense and sense suppression abilitites of original and derivative Methanosarcina mazei pyrrolysyl-tRNA synthetase-tRNAPyl pairs in the Escherichia coli BL21(DE3) cell strain. PLoS One 8, e57035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R. E.; Sorkin A. (1998) Endocytosis of functional epidermal growth factor receptor-green fluorescent protein chimera. J. Biol. Chem. 273, 35000–35007. [DOI] [PubMed] [Google Scholar]
- Chalker J. M.; Wood C. S.; Davis B. G. (2009) A convenient catalyst for aqueous and protein Suzuki-Miyaura cross-coupling. J. Am. Chem. Soc. 131, 16346–16347. [DOI] [PubMed] [Google Scholar]
- Tsao M. L.; Summerer D.; Ryu Y.; Schultz P. G. (2006) The genetic incorporation of a distance probe into proteins in Escherichia coli. J. Am. Chem. Soc. 128, 4572–4573. [DOI] [PubMed] [Google Scholar]
- Smith E. E.; Linderman B. Y.; Luskin A. C.; Brewer S. H. (2011) Probing local environments with the infrared probe: L-4-nitrophenylalanine. J. Phys. Chem. B 115, 2380–2385. [DOI] [PubMed] [Google Scholar]
- Aprilakis K. N.; Taskent H.; Raleigh D. P. (2007) Using the novel fluorescent amino acid p-cyanophenylalanine offers a direct probe of hydrophobic core formation during the folding of the N-terminal domain of ribosomal protein L9 and provides evidence for two-state folding. Biochemistry 46, 12308–12313. [DOI] [PubMed] [Google Scholar]
- Tucker M. J.; Oyola R.; Gai F. (2006) A novel fluorescent probe for protein binding and folding studies: p-cyano-phenylalanine. Biopolymers 83, 571–576. [DOI] [PubMed] [Google Scholar]
- Tucker M. J.; Oyola R.; Gai F. (2005) Conformational distribution of a 14-residue peptide in solution: a fluorescence resonance energy transfer study. J. Phys. Chem. B 109, 4788–4795. [DOI] [PubMed] [Google Scholar]
- Taskent-Sezgin H.; Chung J.; Patsalo V.; Miyake-Stoner S. J.; Miller A. M.; Brewer S. H.; Mehl R. A.; Green D. F.; Raleigh D. P.; Carrico I. (2009) Interpretation of p-cyanophenylalanine fluorescence in proteins in terms of solvent exposure and contribution of side chain quenchers: a combined fluorescence IR and molecular dynamics study. Biochemistry 48, 9040–9046. [DOI] [PubMed] [Google Scholar]
- Hammill J. T.; Miyake-Stoner S.; Hazen J. L.; Jackson J. C.; Mehl R. A. (2007) Preparation of site-specifically labeled fluorinated proteins for 19F-NMR structural characterization. Nat. Protoc. 2, 2601–2607. [DOI] [PubMed] [Google Scholar]
- Kokura K.; Sun L.; Bedford M. T.; Fang J. (2010) Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 29, 3673–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.; Horton J. R.; Bedford M. T.; Zhang X.; Cheng X. (2011) Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding. J. Mol. Biol. 408, 807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.; Wu D.; Lu Z.; Chen W.; Hu X.; Ding Y. (2009) A novel method for high-level production of TEV protease by superfolder GFP tag. J. Biomed. Biotechnol. 2009, 591923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.