Abstract
Cancer metastasis is an important criterion to evaluate tumor malignancy. Matrix metalloproteinases (MMPs) play a crucial role in cancer proliferation and migration by virtue of their proteolytic functions in angiogenesis and extracelluar matrix (ECM) degradation, making them potential targets of anti-metastaic therapeutics. Recently we showed with both in vivo and in vitro experiments that metallofullerenol Gd@C82(OH)22 can effectively inhibit MMP-2 and MMP-9 with high antitumoral efficacy. Furthermore, our in silico study revealed that Gd@C82(OH)22 could indirectly inhibit the proteolysis of MMP-9 via allosteric modulation exclusively at the ligand specificity S1′ loop. Here, we expand our study toward another gelatinase, MMP-2, using molecular dynamics simulations. Despite the high structural similarity with 64.3% sequence identity, their responses to Gd@C82(OH)22 were quite different. Toward MMP-2, Gd@C82(OH)22 could block either the Zn2+-catalylitic site directly or the S1′ loop indirectly. Surface electrostatics uniquely determines the initial adsorption of Gd@C82(OH)22 on MMP-2, and then its further location of the most favorable binding site(s). These findings not only illustrated how the inhibitory mechanism of Gd@C82(OH)22 is distinguished between the two gelatinase MMPs with atomic details, but also shed light on the de novo design of anti-metastatic nanotherapeutics with enhanced target specificity.
Over the past decade, nanomedicine has emerged as an alternative and novel approach for overcoming some of the difficulties experienced by conventional medical approaches in delivering the benefit of small molecule-based medicines1,2,3. There are many applications of nanomedicine, such as drug delivery, diagnostics and therapy. Nanomaterials offer a wide range of applications because of their unmatched flexibility and diversity, endowed by the wide selection of materials including polymers, liposomes, inorganic materials, and carbon allotropes4,5. Another unique property of these nanomaterials, the intrinsically large surface-to-volume ratio, allows multiple biochemical and functional groups or “blocks” to be attached onto their surfaces6, making them effective templates for de novo nanomedicine design to overcome the limitations often exist in conventional drugs, such as low target selectivity and poor release control. Many different platforms have been devised and specified with single or multiple functions aiming at applications for target selectivity, payload optimization, and/or release control. Roughly speaking, nanomedicines can be categorized into diaganostics and therapeutics depending on their general application purposes. Based on their role in pharmacokinetics, therapeutic nanomedicines can be further categorized into nanocarriers for conventional drugs and nanodrugs for direct treatment of the target diseases. There have been considerable efforts in the research of nanocarriers, such as the FDA approved Abraxane and Doxil, but relatively fewer studies on direct nanodrugs (hyperthermia7 and photodynamic therapy3 might be loosely counted into this category). Despite these efforts (and promising achievements), there is still a lack of detailed understanding of molecular interactions between nanomaterials and biomolecules, which is crucial for developing better and smarter nanomedicines, as well as avoiding undesirable side effects8,9,10,11.
Meanwhile, gadolinium ion (Gd3+) has been extensively used as contrast agents for enhanced magnetic resonance imaging (MRI) due to its high proton relaxivity12,13. Recently, fullerene C82 was shown to serve as an effective host to safely secure the toxic Gd3+ with even better contrasting power than conventional agents after water-soluble hydroxylation (i.e., Gd@C82(OH)22)14,15. More importantly, Gd@C82(OH)22 was then found to have strong anti-tumoral efficacy, effectively inhibiting tumor growth and metastasis in human hepatoma and breast cancers xenographted in mice14. Even at an ultra-low dose, Gd@C82(OH)22 shows comparable anti-tumoral efficacy with conventional antineoplastic agents (e.g., cyclophosphamide and cisplatin) without noticeable side effects. Gd@C82(OH)22 also circumvents the acquired resistance to cisplatin by reactivating the impaired endocytosis of human prostate tumor cells16.
Subsequent study showed that Gd@C82(OH)22 simultaneously targets more than 10 angiogenic factors by down-regulating mRNA and protein expression levels, resulting in a dramatic decrease of tumor microvessel density (MVD), thus lowering blood supply to the tumor tissue17. Very recently, we reported that Gd@C82(OH)22 effectively suppresses volume growth of human pancreatic tumor xenografted in mice by >50% compared to the saline control group18. The environmental scanning electron microscope (ESEM) shows clear and smooth surface in the tumor tissue treated with Gd@C82(OH)22, which is again highly contrasted with the saline control one where the typical rough surface is heavily and densely packed with blood vessels. Anti-CD31 antibody stains also showed evidence of fewer MVD and lower CD31 expression in the Gd@C82(OH)22-treated group, indicating restriction of angiogenesis. More specifically, we also identified that Gd@C82(OH)22 effectively inhibits the gelatinases, matrix metalloproteinase-2 (MMP-2) and MMP-9, in both mRNA expression and enzyme activity levels18. Under normal physiological condition, MMPs are tightly regulated, as they govern the proteolysis in various contexts such as in tissue remodeling and wound healing19. Once recruited in the pathological environment such as malignant tumors, however, MMPs are involved in new blood vessel formation and ECM degradation20,21, modifying angiogentic ECM22,23 and cell surface molecules24, as well as mediating the release of ECM-bound pro-angiogenic factors like VEFG and FGF-225,26. Therefore, MMPs, specifically their Zn2+-coordinated active sites, have been designated as a potential target for anti-metastatic therapeutics27. However, similar to kinases28,29,30, the structural similarity in the catalytic sites among all MMPs (at least 28 of them in total) often limits the target selectivity even with favorable binding affinity, resulting in undesired side effects31.
Furthermore, our in silico study with molecular dynamics simulations revealed that Gd@C82(OH)22 could favorably interact with MMP-9 at the ligand specificity S1′ loop, possibly regulating the ligand binding onto the catalytic site remotely18. Interestingly, the S1′ loop is known as the most characteristic region from the catalytic domain in terms of length, sequence similarity, and ligand recognition among the available MMPs19. Thus, our finding, in contrast with conventional MMP inhibitors (MMPi's), proposed an alternative inhibitory pathway that Gd@C82(OH)22 allosterically modulates the native ligand binding on MMP-9 via an indirect but very specific exocite interaction. From a different perspective, this also demonstrates the possibility of applying the carbon-based nanoparticle as a direct nanodrug through the specific interactions with the metastatic protein beyond the general role of nanomedicine as a nanocarrier for conventional drugs32.
In this work, we extend our investigation to the interactions between Gd@C82(OH)22 and the gelatinase MMP-2 using atomistic molecular dynamics simulation. In our previous in vitro studies, activities of both MMP-2 and MMP-9 were inhibited by Gd@C82(OH)22, even though MMP-9 seems to be more dramatically affected (Fig. S1)18. Initially, we presumed that the binding dynamics of Gd@C82(OH)22 would be similar for both gelatinases, since MMP-2 shares a highly conservative catalytic domain structure with MMP-9 (i.e. RMSD = 0.97Å) (Fig. 1). However, our molecular dynamics simulations showed that MMP-2 exhibit quite different binding modes with Gd@C82(OH)22 from MMP-9, thus implying different inhibitory mechanisms between them. The underlying deviations in their molecular binding mechanisms are also illustrated in detail.
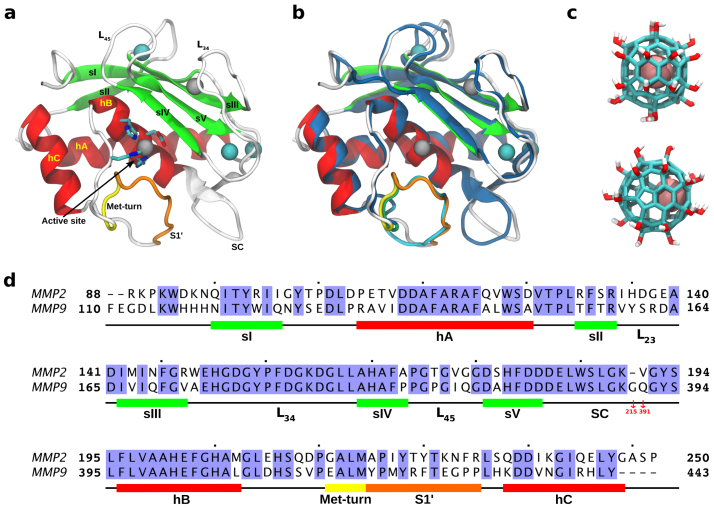
Figure 1. Molecular system (MMP-2 and Gd@C82(OH)22) and sequence alignment between MMP-2 and MMP-9.
(a) Catalytic domain of MMP-2: two Zn2+ and three Ca2+ are depicted in gray and cyan, respectively, and Met-turn and the ligand specificity loop S1′ is highlighted with yellow and orange, respectively. (b) MMP-2 is superimposed on the catalytic domain of MMP-9 marked in blue with RMSD of 0.97 Å. (c) Structure of metallofullerenol Gd@C82(OH)22, where Gd3+ is colored in pink. (d) Sequence alignment between MMP-2 and MMP-9 with 64.3% sequence identity.
Results and discussion
The molecular system was set up with the catalytic domain of MMP-2 (PDB code: IQIB)33 surrounded by multiple nanoparticles of metallofullerenol Gd@C82(OH)22. Due to no prior information on the putative binding modes between MMP-2 and Gd@C82(OH)22, these nanoparticles were initially set aside very far away from the protein MMP-2 with a distance larger than the typical cutoff distance of van der Waals interactions. Following a similar protocol as in our previous study of MMP-918, four Gd@C82(OH)22 were again introduced at the tetrahedral (Td) corners of the simulation water box, with a minimum distance from the MMP-2 protein surface of 25.0 Å. Multiple nanoparticles were used in order to (i) expedite binding site search on MMP-2, (ii) enhance sampling with various initial configurations between MMP-2 and Gd@C82(OH)22, and (iii) simulate the potential clustered binding of multiple Gd@C82(OH)22 nanoparticles on the protein. As a control, we also examined the binding dynamics of the normal fullerenol C82(OH)22 (See details in Methods). The force field parameterization for Gd@C82(OH)22 was adopted from previous studies18,34,35,36, which was developed based on a similar approach widely used in standard force field developments. These parameters are compatible with data obtained from experiments and high level quantum mechanics calculations (see SI text and Fig. S9 of ref. 18).
Overall, both fullerene derivatives Gd@C82(OH)22 and C82(OH)22 frequently interact with various sites of MMP-2 while searching for the most thermodynamically stable sites (see more details below). The nanoparticles were also sometimes observed to cluster together, before or after binding onto MMP-2, as occurred in the previous simulations with MMP-918. This clustering was also observed in the atomic force microscopy (AFM) and synchrotron-radiation small angle X-ray scattering (SR-SAXS) experiments, where Gd@C82(OH)22 can exist as an aggregate with a radius as large as ~22 nm14. This indicates that despite the Gd3+ ion charge and multiple surface hydroxylation, Gd@C82(OH)22 is still largely hydrophobic.
MMP-2 remains intact with Gd@C82(OH)22 interaction
The global structural change of MMP-2 upon interaction with Gd@C82(OH)22 and C82(OH)22 was first investigated here. Figure 2 shows the root mean square deviation (RMSD) and fluctuation (RMSF) of MMP-2 with Gd@C82(OH)22 and C82(OH)22, respectively. MMP-2 appeared quite stable over the simulation time regardless of the interacting fullerene derivatives. The RMSD grew no more than ~3Å in any trajectories, fluctuating between 1Å to 3Å from the referenced X-ray crystal structure, which indicates that the native fold of MMP-2 remains stable, with its secondary structures of 5 β-strands and 3 α-helices all intact, even after frequent contacts with the nanoparticles. The similar interaction of Gd@C82(OH)22 was also found in MMP-9, where RMSD was stable and smaller than 3Å over ~500 ns in multiple trajectories18. Similar to the MMP-9 case, we thus excluded the structural deformation as a potential inhibitory route for Gd@C82(OH)22 on MMP-2. This is in contrast to our previous studies with the single wall carbon nanotubes (SWCNTs) or graphene nanosheets, where protein structures can be significantly distorted/destroyed by the strong hydrophobic and/or π-π stacking interactions between the SWCNT/graphene and hydrophobic core residues of the proteins10,37,38.
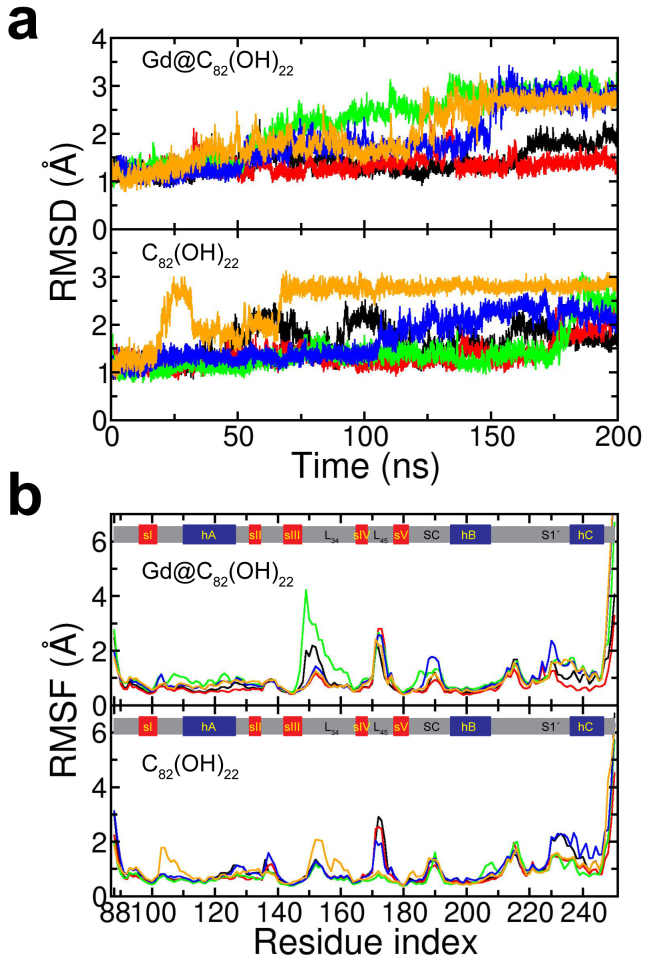
Figure 2. RMSD and RMSF of MMP-2.
(a) RMSD of MMP-2 fluctuates between 1 and 3 Å for both Gd@C82(OH)22 (up) and C82(OH)22 (down), where each trajectories are represented with different colors. (b) RMSF of MMP-2 with Gd@C82(OH)22 (up) and C82(OH)22 (down). Five β-sheets and three α-helices seem stable, while loops from both upper and lower rims are more sensitive to fullerene derivatives.
Even though without any noticeable changes in the overall fold of MMP-2, the RMSF did indicate that these nanoparticles, particularly Gd@C82(OH)22, can sensitively “detect” the surface exposed residues in more sophisticated manner (Fig. 2B). The loop regions seem to be much more “accommodative” to the nanoparticles, in addition to their intrinsic flexibilities. For instance, loops L34 and L45 in the upper rim, as well as loops SC and S1′ in the lower rim (which are involved in the ligand recognition of MMP-2), all display higher flexibility to accommodate the binding of these fullerene derivatives. Both Gd@C82(OH)22 and C82(OH)22 were found to have specific interactions with MMP-2 at certain sites on the surface, such as near the loop L34, where Gd@C82(OH)22 also displayed more selectivity than C82(OH)22 (more below). Interestingly, MMP-9 also showed similar patterns with residues at the loop L34 being more reactive to Gd@C82(OH)22 than C82(OH)22, possibly due to the fact that they share the same fold18. Again, this proposes that Gd@C82(OH)22 inhibition might be through specific bindings on MMP-2 instead of direct structural deformation due to the lack of extreme hydrophobicity as in SWCNT or graphene.
MMP-2 has more diversified interaction with Gd@C82(OH)22 than MMP-9
Next, the nanoparticle binding was characterized in greater detail by residue-specific contact analysis (Fig. 3). The residue contacts were obtained by calculating the fraction of number of contact events over the trajectories. A contact event is counted whenever any heavy-atom pair from a target residue and the nanoparticle is within 5Å. Compared with MMP-9, MMP-2 shows a more diverse interaction with both Gd@C82(OH)22 and C82(OH)22 at various surface sites (cf. Fig. 3 and Fig S2). Overall, 117 and 105 residues of MMP-2 are involved in contacts with Gd@C82(OH)22 and C82(OH)22, respectively, for at least 1% of the entire simulation time. By contrast, only 47 and 79 residues of MMP-9 participated in contact with Gd@C82(OH)22 and C82(OH)22, respectively, for the same time duration.
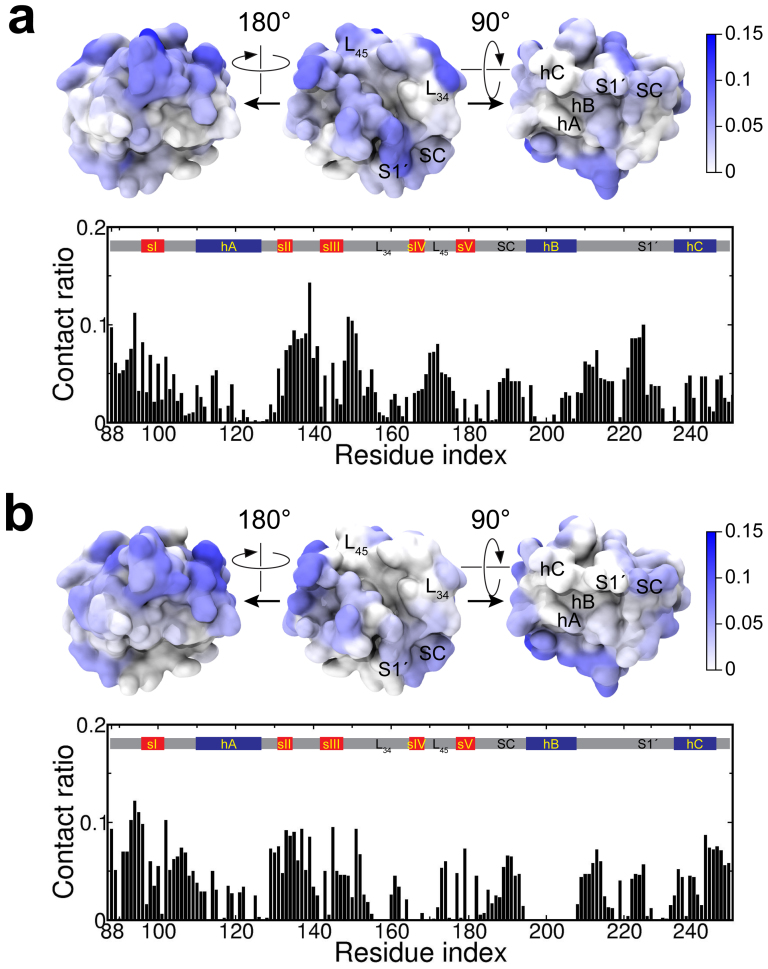
Figure 3. Residue-specific contacts on MMP-2 with Gd@C82(OH)22 and C82(OH)22.
(a) Gd@C82(OH)22 has a favorable contact with MMP-2 specifically at both the ligand specificity S1′ loop and Zn2+-coordinated active site. (b) C82(OH)22 contacts are reduced in both the S1′ and the active site of MMP-2, while interactions with upper rim residues largely remain.
In addition, both Gd@C82(OH)22 and C82(OH)22 interact with much wider range of amino acid types in MMP-2 than in MMP-9. Both fullerene derivatives are effectively interacting with about 17 different amino acid types in MMP-2, whereas only 11 and 14 different types of amino acids were found in MMP-9 for the interactions with Gd@C82(OH)22 and C82(OH)22, respectively (Fig. S3 and Fig. S6 of ref. 18). The effective number Neff of amino acid types were assessed by Neff = exp(S), where S is defined by the sum of –pα ln pα over the contact probabilities pα of 20 amino acids. This clearly indicates that MMP-2 and MMP-9 have different interactions with Gd@C82(OH)22 (and C82(OH)22), and Gd@C82(OH)22 is sensitive enough to distinguish the two gelatinases even with their highly conserved 3-D folds.
Despite the overall similarity in contact number statistics, detailed contact analyses also reveal that there is significant difference between Gd@C82(OH)22 and C82(OH)22 in their characteristic bindings onto MMP-2. C82(OH)22 seems to have its contacts with MMP-2 relatively evenly distributed over the surface, while Gd@C82(OH)22 displays a more concentrated, but multiple, hotspots in binding with MMP-2, manifested in the loop regions L23, L34 and L45 in the upper rim and the ligand specificity S1′ loop regions (Fig. 3a). Meanwhile, Gd@C82(OH)22 is also found to directly contact with the catalytic site of MMP-2, the highly conserved zinc-binding motif 201HExxHxxGxxH211 located between the C-terminus of helix hB and Met-turn. This direct contact, however, is significantly reduced for C82(OH)22, which implies that the Gd encapsulation is crucial for the nanoparticle to have drug efficacy to MMP-2.
This concentrated multi-hotspot binding of Gd@C82(OH)22 on MMP-2 is in great contrast with MMP-9, where a rather dominant single binding mode was observed (Fig. S2b)18. Our previous study showed that Gd@C82(OH)22 mostly contacted with the ligand specificity S1′ loop of MMP-9, and more importantly, the zinc-catalytic site remained intact from direct binding of either Gd@C82(OH)22 or C82(OH)22 even in the extended simulations of up to ~500 ns. This suggested a potential anti-angiogenic mechanism of Gd@C82(OH)22 through MMP-9, where Gd@C82(OH)22 inhibits the proteolytic activity of MMP-9 via allosterically interfering incoming ligands at the ligand specificity S1′ loop, rather than directly blocking the catalytic site. However, the multiple hotspots (binding modes) currently found for MMP-2 indicate that Gd@C82(OH)22 might inhibit MMP-2 via a combined approach with multiple pathways (see more details below).
This contact diversity in MMP-2 may be attributed to the sequence dissimilarity from MMP-9 especially on the surface. Although MMP-2 and MMP-9 share a relatively large portion of amino acids from one-dimensional sequence alignment (i.e., 64.3%), they seem to have quite disproportionate sequence similarity depending on residue locations. Indeed, the buried and exposed sites show 79.0% (of 100 residues) and 38.6% (of 57 residues) sequence similarity, respectively (here a residue is counted as “exposed” if its solvent-accessible surface area is larger than 40% of the total area; otherwise, “buried”) (Fig. 1d). Since the Zn2+-catalylitic domain is largely determined by the buried core with many common residues, it is not surprising to see both gelatinases MMPs share a very similar 3-D scaffold in their catalytic domain (RMSD = 0.97 Å). On the other hand, the ligand specificity between MMP-2 and MMP-9 is mainly regulated by surface residues with relatively lower sequence similarity. This surface dissimilarity influences their specific interactions with Gd@C82(OH)22, resulting in different binding modes and different inhibitory mechanisms.
Interestingly, previous studies have also shown that the binding specificity of Gd@C82(OH)22 is closely related to the surface electrostatics of the target proteins18,34,35. For example, MMP-9 creates a strong negative field around the Zn2+catalytic site, thus presumably facilitating positively charged ligands (Fig. S4). But since the fullerenol cage of Gd@C82(OH)22 is negatively polarized by the Gd3+-ion encapsulation, Gd@C82(OH)22 is likely prohibited from directly contacting the zinc-reactive site. Instead, Gd@C82(OH)22 was driven toward the S1′ loop, which has numerous neutral or polar residues, through specific hydrogen bondings and hydrophobic interaction. On the contrary, MMP-2 forms very different electrostatics, with its active site surrounded by rather neutral and/or positive electrostatic potential (Fig. S4). In addition, the S1′ loop region of MMP-2 seems more positively polarized by K230, R233 and K190 residues, as compared to the mostly neutral S1′ loop in MMP-9. This illustrates how the surface residues can differentiate the binding characteristics for MMP-2 and MMP-9, which in turn influences their native ligand recognition as well as their interaction with nanoparticles. Overall, these electrostatic potential analyses support that MMP-2 may have multiple contacting hotspots with Gd@C82(OH)22, including both the Zn2+ catalytic site and the ligand specificity S1′ loop.
Gd@C82(OH)22 could inhibit MMP-2 in multiple binding pathways
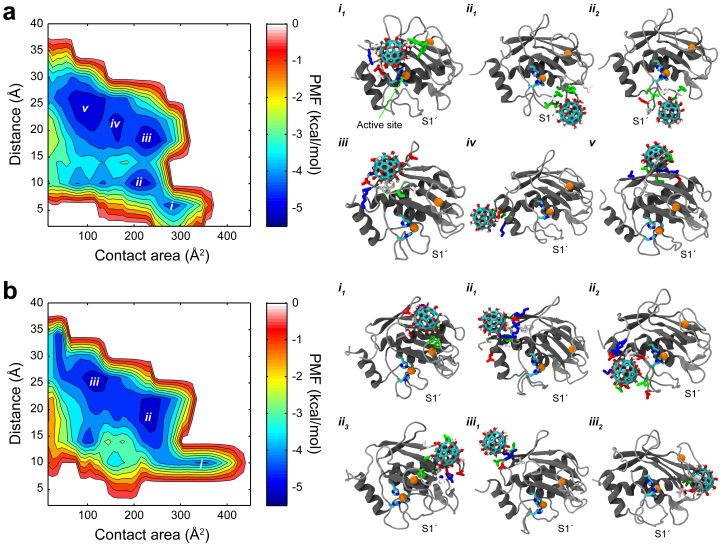
To further illustrate the possible multiple binding modes, we calculated the binding free energy (potential of mean force, PMF) for Gd@C82(OH)22 and C82(OH)22 on MMP-2 (Fig. 4). For consistency, we adopted the same reaction coordinates (Acont, dmin) used in our previous study of MMP-9, where Acont is the contact area between the nanoparticle and MMP-2, and dmin is the minimum distance between the nanoparticle and catalytic Zn2+ ion. The binding free energy is calculated by W(Acont, dmin) = -RT ln p(Acont, dmin), where p(Acont, dmin) is the probability of finding the nanoparticle at position (Acont, dmin). As shown in Fig. 4, the binding free energy surface clearly discriminates multiple binding modes of Gd@C82(OH)22 on MMP-2. We identified two specific binding modes, one near the catalytic Zn2+ ion (i.e., site i in Fig. 4) and one near the S1′ loop (i.e., site ii), and other less-specific binding modes (e.g., iii, iv, and v) relatively distant from the Zn2+ ion, depending on the site selectivity and potential relevance to the inhibitory function to MMP-2. The binding mode i is located at (285 Å2, 6 Å) of the PMF surface with ΔG = −4.34 kcal/mol. This corresponds to the aforementioned direct binding mode onto the putative ligand binding site. The structure (i1) shows that Gd@C82(OH)22 is tightly packed on top of the triad of zinc-coordinating histidines (i.e., H201, H205 and H211) interacting with the nearby residues (e.g., K89, E210, Y155, H166, F168, P90, A169 and L209). As mentioned earlier, this direct blocking is possible partly due to the electrostatically favorable local environment generated by the zinc-reactive center. Here, Gd@C82(OH)22 seems able to approach closely to the Zn2+ ion, which is facilitated by the cation-π interaction39, in addition to the electrostatic attraction between the negatively induced fullerenol cage (i.e., Gd@C82(OH)22[C82(OH)22]3−) and Zn2+ ion40. This mode was absent in MMP-9 even with the highly conservative binding site motif (i.e., MMP-9: HEFGHALGLDH and MMP-2: HEFGHAMGLEH) due to the strong negative electrostatic shielding from nearby acidic residues, such as D113, E111, D182, and D410. Thus, this direct binding represents a straightforward inhibitory pathway of Gd@C82(OH)22 to MMP-2.
Figure 4. Potential of mean force and representative binding modes.
(a) Binding free energy surface of Gd@C82(OH)22 reveals several specific binding modes relevant to inhibition of MMP-2 (i.e., modes i and ii). (b) Binding free energy surface of C82(OH)22 reveals no specific binding modes related with the protein inhibition. In representative binding modes, the Zn2+-coordinated active site is depicted with an orange ball (Zn2+) coordinated with three histidines, and contacting residues to Gd@C82(OH)22 are colored depending on residue polarity and charge: blue, red, green and white for positively charged, negatively charged, polar (non-charged), and non-polar residues, respectively.
Next, Gd@C82(OH)22 was also stabilized near the ligand specificity S1′ loop with ΔG = −4.89 kcal/mol, as indicated as site ii at (205 Å2, 10 Å) on the PMF surface. Our simulations revealed that two representative binding patterns are possible in this energy basin, as shown in structures ii1 and ii2, respectively, which is consistent with those in MMP-9 (cf. Fig. S2b). Gd@C82(OH)22 is capable of binding in-between the S1′ loop with either the SC-loop (i.e., ii1) or the highly conservative Met-turn (i.e., ii2). In both structures, Gd@C82(OH)22 can effectively interact with various types of surface exposed residues. In mode ii1 Gd@C82(OH)22 has frequent contacts with the residues of both SC (e.g., L164, L185, K190, V191, G192, Y193, and S194) and S1′ (e.g., I222, Y223, T227, and Y228) loops, potentially interfering the ligand binding at the pave made of these two loops leading to the Zn2+-catalytic center. Similarly, Gd@C82(OH)22 in mode ii2 has favorable interactions with residues encompassing the Met-turn and S1′ loop (e.g., Q213, D214, P215, G216, A217, A220, P221, I222 and Y223), effectively blocking the tunnel formed by the Met-turn and S1′ loop that also leads to the active site.
On the other hand, the energy minima located at a relatively far distance (>17 Å) from the catalytic zinc ion (i.e., iii, iv, and v) seem less specific in terms of the exact locations, partly due to the reaction coordinates used here (we adopted the same reaction coordinates of MMP-9 for easy comparison, but they might not be ideal). For example, although the energy basin iii (235 Å2, 18 Å) corresponds to a quite stable binding free energy ΔG = −5.08 kcal/mol, it is derived from a mixture of multiple different binding modes which happen to share same values of these two reaction coordinates – contact area and minimum distance from Zn2+ ion – thus, the lower binding energy could be biased by the choice of the reaction coordinates as seen in similar protein folding free energy landscapes41. As exemplified in the three snapshots selected for each energy minimum (i.e., iii, iv, and v), the Gd@C82(OH)22 contacts are highly “diffused” over a large area region (with same minimum distance from Zn2+ ion) in the upper rim of MMP-2. Thus, even with a seemingly strong binding free energy overall, the “diffused” contact areas indicate that Gd@C82(OH)22 makes a non-specific interaction with MMP-2 in this upper rim, as clearly seen in the contact map in Fig. 3 as well. As for the effect of this diffusive large area contact on the native function of MMP-2, it is not immediately clear. But it is possible that the upper rim part of the interaction might also provide some alternative route for allosteric modulation on the enzyme activity42,43. Therefore, in addition to the aforementioned direct binding mode (to the Zn2+-catalytic site) with MMP-2, this diffusive mode is another contrasting point for the binding patterns of Gd@C82(OH)22 on MMP-2 versus MMP-9, where the binding is dominated by the S1′ loop site.
These findings also indicate that Gd@C82(OH)22 can have multiple “drivers” for its specific bindings onto a target protein18,34,35. When Gd@C82(OH)22 is far away from the protein without any contact, the first landing is guided by the long range electrostatic attraction, as shown in many protein and ligand interactions40. Considering the negatively induced fullerenol cage, Gd@C82(OH)22 is likely to be attracted onto the exposed positive residues. For example, we found a cluster of positively charged residues around each detected Gd@C82(OH)22 binding site: R88, K89, and K91 near the binding mode i, and K190, K230, and R233 around the binding mode ii on MMP-2 (Fig. S4). But once it is closer to the protein, the interaction becomes more complicated with multiple forces in play, including cation-π interaction, π-π stacking, hydrophobic interaction, hydrogen bonding, as well as electrostatic interactions39,44. Consequently, Gd@C82(OH)22 can be driven into a thermodynamically more stable site, which may be different from its initial contact point. It is noteworthy that the proposed binding sites (i.e., modes i and ii) are composed of a number of hydrophobic and aromatic residues as well as polar and charged ones. The similar binding reaction pathway was also observed in Gd@C82(OH)22's binding on MMP-9, where Gd@C82(OH)22 was first attracted to MMP-9 via non-specific long range electrostatic interaction with K433 and R440, followed by more specific hydrophobic and hydrogen bonding interactions into the ligand specificity S1′ loop18.
Moreover, we noticed a significant difference between Gd@C82(OH)22 and C82(OH)22 when interacting with MMP-2. While both specific and non-specific binding modes are possible for Gd@C82(OH)22, C82(OH)22 is more likely to interact with MMP-2 via non-specific modes. For example, the binding mode i of C82(OH)22 displayed an even larger contact area than that of Gd@C82(OH)22, but the structure analysis showed that C82(OH)22 was actually located among the three loops (i.e., L23, L34 and L45) in the upper rim with a relatively low binding free energy (i.e., ΔG = −4.19 kcal/mol). More dramatically, the energy basins ii and iii, with relatively stable binding free energies of −4.84 and −5.14 kcal/mol, respectively, were composed of various different binding modes, implying the binding sites are not well specified (i.e. very diffusive) in each energy basin. Compared with Gd@C82(OH)22, the residue contacts are largely reduced near the critical Zn2+-coordinated catalytic site and the ligand specificity S1′ loop, as shown in Fig. 3. Interestingly, C82(OH)22 was also shown to have a non-specific interaction with MMP-9 with significantly reduced inhibitory effect on its proteolytic function (Fig. S2a)18. Thus, our result is consistent with the in vitro experiments that Gd@C82(OH)22 dramatically reduces proteolytic activity of MMP-2 and MMP-9, while the normal fullerenol C82(OH)22 (or C60(OH)20) has much reduced effect on the enzyme activity of the MMPs. (Due to low chemical stability of C82(OH)22, a smaller C60(OH)20 was used as control in our previous experiment. However, we selected C82(OH)22 as control in our simulation to better capture the effect of the metal ion Gd3+ while retaining the molecular size to be comparable with Gd@C82(OH)22. The previous study with both C82(OH)22 and C60(OH)22 as control runs shows similar results18.)
These findings might have shed light on the potential inhibitory mechanism of Gd@C82(OH)22 on MMP-2. Two specific binding modes (i.e., i and ii) were identified for Gd@C82(OH)22's inhibition of MMP-2: (i) one for direct blocking of the active site of MMP-2, and (ii) the other for exocite binding at the ligand specificity S1′ loop. In the former, Gd@C82(OH)22 directly blocks the catalytic site of MMP-2, thus the ligand binding is effectively prohibited or seriously interrupted, resulting in a decrease in the enzyme activity. In the latter, Gd@C82(OH)22 inhibits MMP-2 by allosteric modulation at the S1′ loop. Although it is indirect, the exocite binding can enhance the selectivity of Gd@C82(OH)22 considering the importance of the S1′ loop in the ligand recognition. Despite the dual inhibitory pathways, the overall inhibition capability of Gd@C82(OH)22 toward MMP-2 can be lower than that toward MMP-9, as indicated in our previous in vitro assays (Fig. S1)18. In the MMP-9 case, Gd@C82(OH)22 predominantly binds to the S1′ loop, resulting in a very strong and exclusive binding, while in the MMP-2 case, multiple contact hotspots with many non-specific ones (e.g., mode iii, iv and v in Fig. 4a) co-exist, which might spread the strength thin on each one.
In MMP inhibitor (MMPi) designs, the zinc-active site has been intensively targeted owing to its obvious role in proteolysis. Small organic molecules are often designed as potential MMPi's with high binding affinities, but almost all of them fail in ultimate FDA approval due to the lack of target selectivity and thus resulting in serious side effects31. The lack of target selectivity in these MMPi's is mainly caused by the highly homologous binding site structures among the MMPs19,45. Meanwhile, allosteric modulation was discussed as an alternative route to obtain a better target selectivity27. For example, prymidine dicarboxamide derivatives were shown to selectively inhibit MMP-13 by binding to the ligand specificity S1′ loop with no direct effect on the active site46. The ligand specificity S1′ loop is found to be a region with the least similarity in terms of length and/or sequence among the available MMPs, thus implying a potential target site. Our current simulations show that Gd@C82(OH)22 can potentially inhibit MMP-2 allosterically by interfering an incoming ligand at the ligand specificity S1′ loop, in addition to its direct binding to the zinc catalytic site.
Taken together, the allosteric modulation through the ligand specificity S1′ loop could be a useful and specific inhibitory route for metallofullerenol Gd@C82(OH)22 on both MMP-2 and MMP-9. By acting on the S1′ loop, Gd@C82(OH)22 is shown to effectively interfere the native ligand from the putative binding. While this was found previously as the most probable pathway for MMP-9, our current simulations reveal that MMP-2 can be inhibited by Gd@C82(OH)22 through both direct binding at the Zn2+-catalytic site and indirect binding at the S1′ loop site.
Conclusion
In this study, we used molecular dynamics simulations to study the interaction between the metallofullerenol Gd@C82(OH)22 and matrix metalloproteinases MMP-2 with atomic detail. Although Gd@C82(OH)22 was originally developed as a high contrasting agent for MRI, it has shown eminent efficacy for various cancers including hepatoma, breast cancer, and pancreatic cancer. Among the possible anti-tumor mechanisms, Gd@C82(OH)22 was found to inhibit the proteolytic activity of MMP-2 and MMP-9, which is critical in down-regulating the angiogenesis and tumor extra cellular matrix degradation, and hence restricting tumor proliferation and metastasis. Our results show that Gd@C82(OH)22 can have specific interactions with MMP-2, which are closely related to the proteolytic function of MMP-2. Similar to MMP-9, Gd@C82(OH)22 also acts on the ligand specificity S1′ loop, which implies that Gd@C82(OH)22 can induce allosteric modulation on MMP-2 proteolysis by residing in-between the S1′ loop with either the SC loop or the Met-turn. In addition, Gd@C82(OH)22 can also interact with MMP-2 directly at the zinc-reactive site, which was not found in its binding with MMP-9. Thus, Gd@C82(OH)22 achieves its inhibition to MMP-2 via dual binding modes, with both direct and indirect bindings. Detailed analyses show that the local electrostatic environment differentiates the Gd@C82(OH)22 binding between MMP-2 and MMP-9. The residue constituents on each protein surface are quite different, which results in a favorable electrostatic environment for Gd@C82(OH)22 at the Zn2+-coordinated reaction site of MMP-2, but not MMP-9, even though both share a close fold with a fairly high overall sequence similarity.
Various types of nanomedicines have been actively developed in recent years in order to complement and/or overcome the deficits of traditional small molecule based drugs. Among these efforts, a considerable amount of researches have been devoted to the development of novel nanocarriers for conventional drug delivery. Our current approach focuses on another important route of using nanoparticles directly as nanodrugs for terminal illness such as cancer. This study provides new insights into the de novo design of nanodrugs in addition to a better understanding of the various inhibitory mechanisms.
Methods
In our simulation, we used a 160-residue catalytic domain of human MMP-2 determined from X-ray crystallography33. Four Gd@C82(OH)22 were initially located at the tetrahedral corners of the cubic simulation box at least 25 Å away from MMP-2, since we had no prior information on the putative binding modes of Gd@C82(OH)22 on MMP-2. Multiple copies of metallofullerenols were employed in order to not only facilitate the binding-site searching on MMP-2, but also understand the clustered behavior of the nanomolecules in the aqueous media. The composite system was then immersed in a cubic 90 Å × 90 Å × 90 Å water box, and neutralized with counter ions. Then, the system was ionized with 100 mM NaCl, resulting in the total system of ~69,000 atoms with 22,000 explicit water molecules. Similarly, we configured a separate system for C82(OH)22 as the control run. The CHARMM22 (c32b1 parameter set) force field47 and TIP3P water model48 were used for the protein and water molecules, respectively. The nonbonding parameters for Gd@C82(OH)22 and C82(OH)22 were separately prepared by the DFT-level quantum mechanical calculations, and examined according to the protocol for CHARMM force field development18. The non-bonding interactions were treated with a typical cutoff of 12 Å, while the long-range electrostatic interaction were enumerated with the particle-mesh Ewald method49. Before production runs, the system was prepared with energy minimization for 20,000 steps, followed by 250 ps of equilibration with a 0.5 fs time step. For each system, five trajectories were independently generated for at least 200 ns with a 2 fs time step, where the initial structure was differently configured for each run by randomly rotating MMP-2 in order to avoid statistical bias as much as possible. All simulations were carried out at 1 atm and 310 K with the NAMD2 software massively parallelized for IBM Blue Gene machine50.
Author Contributions
R.Z. designed and supervised the research. S.G.K. and T.H. carried out the simulations. S.G.K., R.Z., R.A-S., D.W., B.W. and T.H. analyzed the data. S.G.K. and R.Z. prepared the manuscript. All authors discussed the results and commented on the manuscript.
Supplementary Material
Supplementary Information
References
- Wagner V., Dullaart A., Bock A. K. & Zweck A. The emerging nanomedicine landscape. Nat. Biotechnol. 24, 1211–1217 (2006). [DOI] [PubMed] [Google Scholar]
- Schroeder A. et al. Treating metastatic cancer with nanotechnology. Nat. Rev. Cancer 12, 39–50 (2012). [DOI] [PubMed] [Google Scholar]
- Doane T. L. & Burda C. The unique role of nanoparticles in nanomedicine: imaging, drug delivery and therapy. Chem. Soc. Rev. 41, 2885–2911 (2012). [DOI] [PubMed] [Google Scholar]
- Petros R. A. & DeSimone J. M. Strategies in the design of nanoparticles for therapeutic applications. Nat. Rev. Drug. Discov. 9, 615–627 (2010). [DOI] [PubMed] [Google Scholar]
- Euliss L. E., DuPont J. A., Gratton S. & DeSimone J. Imparting size, shape, and composition control of materials for nanomedicine. Chem. Soc. Rev. 35, 1095–1104 (2006). [DOI] [PubMed] [Google Scholar]
- Mout R., Moyano D. F., Rana S. & Rotello V. M. Surface functionalization of nanoparticles for nanomedicine. Chem. Soc. Rev. 41, 2539–2544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo D., Lee J. H., Shin T. H. & Cheon J. Theranostic magnetic nanoparticles. Acc. Chem. Res. 44, 863–874 (2011). [DOI] [PubMed] [Google Scholar]
- Nel A. E. et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat. Mater. 8, 543–557 (2009). [DOI] [PubMed] [Google Scholar]
- Lammers T., Aime S., Hennink W. E., Storm G. & Kiessling F. Theranostic Nanomedicine. Acc. Chem. Res. 44, 1029–1038 (2011). [DOI] [PubMed] [Google Scholar]
- Ge C. et al. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 108, 16968–16973 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y. et al. Destructive extraction of phospholipids from Escherichia coli membranes by graphene nanosheets. Nat. Nanotechnol. 8, 594–601 (2013). [DOI] [PubMed] [Google Scholar]
- Weinmann H. J., Brasch R. C., Press W. R. & Wesbey G. E. Characteristics of gadolinium-DTPA complex: a potential NMR contrast agent. AM. J. Roentgenol. 142, 619–624 (1984). [DOI] [PubMed] [Google Scholar]
- Brasch R. C., Weinmann H. J. & Wesbey G. E. Contrast-enhanced NMR imaging: animal studies using gadolinium-DTPA complex. AM. J. Roentgenol. 142, 625–630 (1984). [DOI] [PubMed] [Google Scholar]
- Chen C. et al. Multihydroxylated [Gd@C82(OH)22]n nanoparticles: antineoplastic activity of high efficiency and low toxicity. Nano Lett. 5, 2050–2057 (2005). [DOI] [PubMed] [Google Scholar]
- Kato H. et al. Lanthanoid endohedral metallofullerenols for MRI contrast agents. J. Am. Chem. Soc. 125, 4391–4397 (2003). [DOI] [PubMed] [Google Scholar]
- Liang X. J. et al. Metallofullerene nanoparticles circumvent tumor resistance to cisplatin by reactivating endocytosis. Proc. Natl. Acad. Sci. U. S. A. 107, 7449–7454 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H. et al. Potent angiogenesis inhibition by the particulate form of fullerene derivatives. ACS Nano 4, 2773–2783 (2010). [DOI] [PubMed] [Google Scholar]
- Kang S. G. et al. Molecular mechanism of pancreatic tumor metastasis inhibition by Gd@C82(OH)22 and its implication for de novo design of nanomedicine Proc. Natl. Acad. Sci. U. S. A. 109, 15431–15436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallant C., Marrero A. & Gomis-Ruth F. X. Matrix metalloproteinases: fold and function of their catalytic domains. Biochim. Biophys. Acta. 1803, 20–28 (2010). [DOI] [PubMed] [Google Scholar]
- Deryugina E. I. & Quigley J. P. Pleiotropic roles of matrix metalloproteinases in tumor angiogenesis: contrasting, overlapping and compensatory functions. Biochim. Biophys. Acta. 1803, 103–120 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson U. W. & Dabrosin C. Estradiol and tamoxifen regulate endostatin generation via matrix metalloproteinase activity in breast cancer in vivo. Cancer Res. 66, 4789–4794 (2006). [DOI] [PubMed] [Google Scholar]
- Mott J. D. & Werb Z. Regulation of matrix biology by matrix metalloproteinases. Curr. Opin. Cell. Biol. 16, 558–564 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer 3, 422–433 (2003). [DOI] [PubMed] [Google Scholar]
- Heissig B. et al. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell 109, 625–637 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodele S., Blavier L., Yoon J. M. & DeClerck Y. A. Modifying the soil to affect the seed: role of stromal-derived matrix metalloproteinases in cancer progression. Cancer Metast. Rev. 25, 35–43 (2006). [DOI] [PubMed] [Google Scholar]
- Presta M. et al. Fibroblast growth factor/fibroblast growth factor receptor system in angiogenesis. Cytokine Growth Factor Rev. 16, 159–178 (2005). [DOI] [PubMed] [Google Scholar]
- Jacobsen J. A., Major Jourden J. L., Miller M. T. & Cohen S. M. To bind zinc or not to bind zinc: an examination of innovative approaches to improved metalloproteinase inhibition. Biochim. Biophys. Acta. 1803, 72–94 (2010). [DOI] [PubMed] [Google Scholar]
- Fang Z., Grutter C. & Rauh D. Strategies for the selective regulation of kinases with allosteric modulators: exploiting exclusive structural features. ACS Chem. Biol. 8, 58–70 (2013). [DOI] [PubMed] [Google Scholar]
- Sawa M. Strategies for the design of selective protein kinase inhibitors. Mini-Rev. Med. Chem. 8, 1291–1297 (2008). [DOI] [PubMed] [Google Scholar]
- Cherry M. & Williams D. H. Recent kinase and kinase inhibitor X-ray structures: mechanisms of inhibition and selectivity insights. Curr. Med. Chem. 11, 663–673 (2004). [DOI] [PubMed] [Google Scholar]
- Overall C. M. & Lopez-Otin C. Strategies for MMP inhibition in cancer: innovations for the post-trial era. Nat. Rev. Cancer 2, 657–672 (2002). [DOI] [PubMed] [Google Scholar]
- Yang Z., Kang S. G. & Zhou R. Nanomedicine: de novo design of nanodrugs. Nanoscale 6, 663–677 (2013). [DOI] [PubMed] [Google Scholar]
- Dhanaraj V. et al. X-ray structure of gelatinase A catalytic domain complexed with a hydroxamate inhibitor. Croat. Chem. Acta 72, 575–591 (1999). [Google Scholar]
- Kang S. G., Huynh T. & Zhou R. Non-destructive inhibition of metallofullerenol Gd@C82(OH)22) on WW domain: implication on signal transduction pathway. Sci. Rep. 2, 957 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. G., Huynh T. & Zhou R. Metallofullerenol Gd@C82(OH)22 distracts the proline-rich-motif from putative binding on the SH3 domain. Nanoscale 5, 2703–2712 (2013). [DOI] [PubMed] [Google Scholar]
- Yin X. et al. Impacts of fullerene derivatives on regulating the structure and assembly of collagen molecules. Nanoscale 5, 7341–7348 (2013). [DOI] [PubMed] [Google Scholar]
- Zuo G., Huang Q., Wei G., Zhou R. & Fang H. Plugging into proteins: poisoning protein function by a hydrophobic nanoparticle. ACS Nano 4, 7508–7514 (2010). [DOI] [PubMed] [Google Scholar]
- Zuo G., Zhou X., Huang Q., Fang H. P. & Zhou R. H. Adsorption of Villin Headpiece onto Graphene, Carbon Nanotube, and C60: Effect of Contacting Surface Curvatures on Binding Affinity. J. Phys. Chem. C 115, 23323–23328 (2011). [Google Scholar]
- Woolf T. B., Grossfield A. & Pearson J. G. Indoles at interfaces: Calculations of electrostatic effects with density functional and molecular dynamics methods. Int. J. Quantum Chem. 75, 197–206 (1999). [Google Scholar]
- Wade R. C., Gabdoulline R. R., Ludemann S. K. & Lounnas V. Electrostatic steering and ionic tethering in enzyme-ligand binding: insights from simulations. Proc. Natl. Acad. Sci. U. S. A. 95, 5942–5949 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R. Trp-cage: folding free energy landscape in explicit water. Proc. Natl. Acad. Sci. U. S. A. 100, 13280–13285 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Chen Z., Wang Y., Bonewald L. & Steffensen B. Inhibition of MMP-2 gelatinolysis by targeting exodomain-substrate interactions. Biochem. J. 406, 147–155 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens E. et al. A monoclonal antibody inhibits gelatinase B/MMP-9 by selective binding to part of the catalytic domain and not to the fibronectin or zinc binding domains. Biochim. Biophys. Acta. 1770, 178–186 (2007). [DOI] [PubMed] [Google Scholar]
- Yang Z., Wang Z., Tian X., Xiu P. & Zhou R. Amino acid analogues bind to carbon nanotube via pi-pi interactions: comparison of molecular mechanical and quantum mechanical calculations. J. Chem. Phys. 136, 025103 (2012). [DOI] [PubMed] [Google Scholar]
- Overall C. M. & Kleifeld O. Towards third generation matrix metalloproteinase inhibitors for cancer therapy. Br. J. Cancer 94, 941–946 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel C. K. et al. Structural basis for the highly selective inhibition of MMP-13. Chem. Biol. 12, 181–189 (2005). [DOI] [PubMed] [Google Scholar]
- MacKerell A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998). [DOI] [PubMed] [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- Darden T., York D. & Pedersen L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- Kumar S. et al. Scalable molecular dynamics with NAMD on the IBM Blue Gene/L system. IBM J. Res. Develop. 52, 177–188 (2008). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information