Abstract
The Endosomal Sorting Complex Required for Transport (ESCRT) machinery plays an evolutionarily conserved role in cytokinetic abscission, the final step of cell division where daughter cells are physically separated. Here, we show that Charged Multivesicular Body (MVB) Protein 4C (CHMP4C), a human ESCRT-III subunit, is involved in abscission timing. This function correlated with its differential spatiotemporal distribution during late stages of cytokinesis. Accordingly, CHMP4C functioned in the Aurora B-dependent abscission checkpoint to prevent both premature resolution of intercellular chromosome bridges and accumulation of DNA damage. CHMP4C engaged the Chromosomal Passenger Complex (CPC) via interaction with Borealin suggesting a model whereby CHMP4C inhibits abscission upon phosphorylation by Aurora B. Thus, the ESCRT machinery may protect against genetic damage by coordinating midbody resolution with the abscission checkpoint.
Keywords: Cell Biology, Cell Division, ESCRT, cytokinesis, CPC, abscission
The final separation of daughter cells during cytokinesis is the ancestral function of the Endosomal Sorting Complex Required for Transport (ESCRT) machinery (1-5) which also acts to resolve equivalent membrane scission events in Multivesicular Body (MVB) formation (6, 7) and Human Immunodeficiency Virus-1 (HIV-1) budding (8, 9). Midbody recruitment of ESCRT-III, the filament-forming scission machinery, is an essential event in cytokinesis that is thought to provide constrictive force during abscission (2, 10-12). An Aurora B-dependent abscission checkpoint (NoCut) is thought to retard abscission to prevent damage of lagging chromosomes that are trapped in the midbody (14-16) and may function more generally as an abscission timer (14). However, mechanisms that modulate abscission timing remain poorly understood and the involvement of the core abscission machinery in this process is unclear.
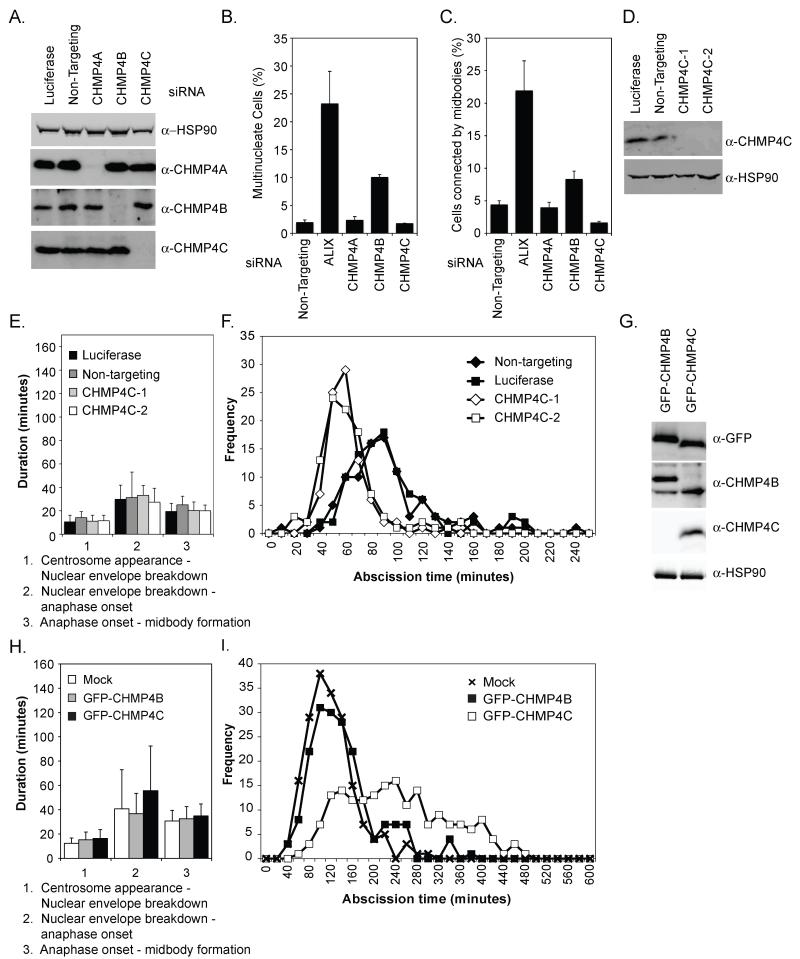
Here, we investigated the function of CHMP4A, -B and –C, human homologs of the yeast ESCRT-III subunit Snf7p. For this purpose, specific siRNAs and antibodies against each of the CHMP4s were developed (17) (Fig. S1). We then analysed ESCRT-dependent endosomal downregulation of Class I Major Histocompatibility Complex (MHC-I) molecules in HeLa cells stably expressing the K3 ubiquitin ligase from Kaposi’s sarcoma-associated herpes virus (KSHV) (18). Similarly to depletion of Tumour Susceptibility Gene 101 (TSG101), depletion of CHMP4B prevented MHC-I degradation, whereas depletion of CHMP4A or CHMP4C had little effect on this process (Fig. S2A and S2B). As expected (13), depletion of CHMP4A or CHMP4C did not inhibit HIV-1 release and only depletion of CHMP4B inhibited this ESCRT-dependent process (Fig. S3). Furthermore, CHMP4A and CHMP4C were dispensable for completion of cytokinesis, whereas CHMP4B was again the sole paralog required (Fig. 1A and 1B). However, in asynchronous cultures of CHMP4C-depleted HeLa cells, fewer cells were connected by midbodies (Fig. 1C), leading us to question whether midbodies were resolved faster in cells lacking CHMP4C.
Figure 1. CHMP4C negatively regulates cytokinesis.
A. Resolved HeLa cell lysates were examined by blotting with a α-CHMP4A, α-CHMP4B, α-CHMP4C or α-Heat Shock Protein 90 kDa (HSP90). B, C. siRNA-transfected HeLa cells were fixed and stained with α-Tubulin. Multinucleate cells (B, n=3±SD) or cells connected by midbodies (C, n=7±SD) were scored visually. D. Resolved lysates from siRNA-transfected HeLa mCh-Tub cells were examined by western blotting with α-CHMP4C or α-HSP90. E-F. Asynchronous cultures of HeLa mCh-Tub cells were transfected with the indicated siRNA and imaged live and mitotic durations quantified. Abscission time (Luciferase: 93±38 minutes, n = 96; Non-targeting: 94±36 minutes, n = 94; CHMP4C-1: 59±17 minutes, n = 88; CHMP4C-2: 61±25 minutes, n = 100) was calculated across 4 independent experiments. G. Resolved cell lysates from HeLa cells stably expressing mCh-Tubulin and either GFP-CHMP4B or GFP-CHMP4C were examined by blotting with α-HSP90, α-GFP, α-CHMP4B or α-CHMP4C. H, I. Cells from G were imaged live and mitotic durations quantified. The more intense imaging (17) resulted in general abscission delays to 116±45 and 137±61 minutes for control or GFP-CHMP4B expressing cells whilst GFP-CHMP4C expressing cells took 240±103 minutes to complete abscission. Data comprises 185 cells per condition from 3 independent experiments.
We then imaged live HeLa cells stably expressing mCherry-Tubulin (HeLa mCh-Tub) (19) to examine mitotic dynamics. CHMP4C-depleted cells (Fig. 1D) showed normal duration of early mitotic phases and centrosome amplification (Fig. 1E and S4A). We next monitored tubulin-disassembly at the midbody as a marker that correlates strongly with abscission (16). Cells treated with control siRNAs resolved their midbodies with similar kinetics and depletion of CHMP4C reduced abscission time by approximately 30 minutes (Fig. 1F, S4A, Movie 1-4). Abscission was also faster in cells co-depleted of CHMP4C and Spastin, an ATPase involved in destabilization of midbody microtubules (20, 21), suggesting that CHMP4C and Spastin regulate distinct stages of abscission (Fig S4B). We next used cell lines stably expressing mCh-tubulin and comparable levels of either Green Fluorescent Protein (GFP)-tagged CHMP4B or CHMP4C (22) (Fig. 1G) for simultaneous imaging of abscission and midbody recruitment of ESCRT-III. Early phases of mitosis and abscission (Fig. 1H and 1I) were similar in control and GFP-CHMP4B-expressing cells. However, expression of GFP-CHMP4C resulted in an abscission delay that could be explained by increased levels of intracellular CHMP4C (Fig. 1I, S5A-D, Movie 5 and 6). As expected (12), GFP-CHMP4B localized transiently to the midbody arms immediately (21±7 minutes) prior to abscission whereas GFP-CHMP4C localized earlier to the midbody, arriving 176±19 minutes prior to abscission (Fig 2A-D). During its recruitment, GFP-CHMP4C localized initially to the midbody arms, before being directed to the central region (Flemming body) (Fig. 2A-D, S5A-D, Movie 5 and 6). Thus, CHMP4B and CHMP4C exhibit differential spatiotemporal distribution during late cytokinesis.
Figure 2. Differential spatio-temporal recruitment of CHMP4 paralogs during cytokinesis.
A-D. GFP-fluorescence intensities of midbody-localised GFP-CHMP4B (A, C n=14±S.D.) or GFP-CHMP4C (B, D n=9±S.D.) during abscission. Abscission indicated by arrow, time in minutes, selected frames presented (C, D). Initial recruitment of GFP-CHMP4C to midbody arms marked (arrowhead). E. ClustalW alignment of the C-terminal regions of CHMP4A, CHMP4B and CHMP4C, S210 indicated by arrow. F. HeLa cells transfected with plasmids encoding the indicated HA-CHMP4 constructs were fixed and stained with α-Tubulin and α-HA. Multinucleate cells were scored (n=5±S.D.). G, H. HeLa mCh-Tub cells stably expressing HA-CHMP4CR, HA-CHMP4CRδINS or HA-CHMP4CR S210A were treated with CHMP4C siRNA, fixed, stained with α-HA and HA-CHMP4C location was scored (n=3±S.D.) Bar is 10 μm.
We sought differences between CHMP4 paralogs that could explain their differential behavior. Alignment of the regulatory region at the C-termini of CHMP4s revealed a CHMP4C-specific insertion (INS) at residues 201-217 (Fig. 2E). This insertion is expanded in mammals (Fig. S6) and contains numerous Serine (S) and Threonine residues. Transiently overexpressed CHMP4 chimeras containing CHMP4C’s C-terminus were more potent inhibitors of cell division (Fig. 2F) and grafting INS into the corresponding region of CHMP4B produced a chimera that inhibited cell division (Fig. 2F), suggesting that INS may be a platform for phosphorylation that inhibits abscission. We next determined the spatial distribution of CHMP4C during abscission by analysing Hemagglutinin (HA)-tagged CHMP4C expressed stably at near-endogenous levels. In early midbodies HA-CHMP4C localized to the midbody arms, whereas in late midbodies, as observed for GFP-CHMP4C in living cells (Fig. 2B), HA-CHMP4C localised to the Flemming-body in an INS-dependent manner (Fig. 2G and 2H). In contrast, CHMP4A and CHMP4B were only observed on midbody arms (Fig. S5E). We speculated that phosphorylation of residues within INS may have directed CHMP4C’s localization and mapped the determinant of Flemming-body localization in late-cyokinesis to S210, a residue conforming to the consensus sequence for Aurora B ([R/K](1-3)-X-[S/T] (23) (Fig. 2G, 2H and S5F).
We next wondered if CHMP4C participated in the Aurora B-dependent NoCut abscission checkpoint. This checkpoint can be activated by partial depletion of Nucleoporin 153 kDa (NUP153) and is evidenced by an accumulation of cells unable to complete abscission and arrested at the midbody stage (24). NoCut activation was prevented by co-depletion of CHMP4C with NUP153 (Fig. 3A and 3B), despite phosphorylated Aurora B persisting at midbodies (Fig. 3C). Chromosomes trapped at the midbody may provide an alternative NoCut activation route. We found that HA-CHMP4C preferentially localized to intercellular chromatin bridges illuminated with Yellow Fluorescent Protein (YFP)-tagged Lamin Associated Protein 2β (LAP2β) (Fig. 3D, S7A and S7B). Moreover, HA-CHMP4C co-localized with activated Aurora B at these chromatin bridges (Fig. 3E). We examined intercellular DNA-bridge resolution in CHMP4C-depleted cells, observing that YFP-LAP2β-positive bridge formation and cellular viability was normal (Fig. S7C and S7D) and found, similarly to Aurora B-inhibited cells (16), faster resolution of YFP-LAP2β-positive chromatin bridges (Fig. 3F, S7E-G, Movies 7-9). Consequently, stable depletion of CHMP4C resulted in increased levels of Histone H2AX phosphorylation (Fig. 3G, S7H), suggesting that deregulation of the abscission checkpoint in these cells results in the accumulation of genetic damage (25).
Figure 3. CHMP4C regulates the abscission checkpoint.
A-C. Cell lysates from siRNA-transfected HeLa cells were examined by blotting with α-NUP153, α-CHMP4C, α-HA and α-HSP90 (A). Alternatively, cells were fixed and stained with α-tubulin (B) or α-tubulin and α-pT232 Aurora B (C) Bar is 10 μm. Multinucleate and midbody-connected cells were scored visually (A, n=6±S.D.). D, E. HeLa cells stably expressing YFP-LAP2β, were transfected with plasmids encoding the indicated HA-CHMP4 constructs. Cells were fixed, stained with α-Tubulin and α-HA (D), or α-pT232 Aurora B and α-HA (E) Bar is 10 μm. F. HeLa cells stably expressing YFP-LAP2β were transfected with the indicated siRNA, imaged live and the duration of LAP2β-bridge resolution (Luciferase: 576±454 minutes, n = 116; CHMP4C-1: 321±308 minutes, n = 112; CHMP4C-2: 291±278 minutes, n = 103) was quantified across 6 independent experiments G. Cell lysates from clonal shRNA-transduced HeLa cells were examined by blotting with α-γH2AX, α-CHMP4C or α-HSP90.
Given the essential role of CHMP4C in the regulation of abscission timing and its participation in the Aurora B-dependent abscission checkpoint, we searched for links between components of the CPC and ESCRT-III by yeast two-hybrid. We found interactions between Borealin and CHMP2A, CHMP4B, CHMP4C and CHMP6 (Fig. 4A and 4B), and mapped these interactions to the C-terminus of Borealin (Fig S8A), a region that recruits adaptor proteins to the CPC. CHMP4C was the strongest interactor with Borealin and we could co-precipitate CHMP4C with Aurora B (Fig. S8B), confirming that the ESCRT-machinery is able to engage the CPC. Co-localisation of HA-CHMP4C and members of the CPC was observed in early, but not late, midbodies (Fig. 4C, S8C and S8D). Accordingly, a λ-phosphatase-sensitive, mobility-shifted form of HA-CHMP4C was detected in mitotic lysates and was enriched upon a phospho-affinity resin (Fig. 4D and S8E). Similarly to Centrosomal protein of 55 kDa (CEP55) (26, 27), CHMP4C phosphorylation occurred at mitotic onset and reverted in a phosphatase-dependent manner as mitosis progressed (Fig. 4E). An Aurora B inhibitor reduced CHMP4C phosphorylation (Fig. 4F) and the epitope detected by α-CHMP4C, which recognizes INS, was masked in the mobility-shifted fraction and revealed upon λ-phosphatase treatment (Fig. 4D), suggesting that residues within CHMP4C’s insertion were phosphorylated during mitosis. Finally, Aurora B could specifically phosphorylate CHMP4C on S210 within INS, the residue required for Flemming Body localisation (Fig. 4G, 4H, S8F and S8G).
Figure 4. Aurora B-dependent phosphorylation of CHMP4C S210 activates the NoCut abscission checkpoint.
A. β-galactosidase assay from yeast co-transformed with the indicated VP16 and GAL4-fused constructs (n=3±S.D.). B. Cell lysates and glutathione-bound fractions from 293T cells transfected with the indicated fusion proteins were examined by western blotting with α-HA. C. HeLa mCh-Tub cells stably expressing HA-CHMP4CR were fixed and stained with α-HA and α-Aurora B. D. Asynchronous and mitotic lysates of HeLa mCh-Tub cells stably expressing HA-CHMP4CR were immunoprecipitated with α–HA and treated as indicated and examined by blotting with α–HA, α-CHMP4C, α-CHMP4B, α–CEP55 and α–HSP90. E. F. Asynchronous or mitotically arrested HeLa mCh-Tub cells stably expressing HA-CHMP4CR were either released into media containing DMSO or the phosphatase inhbitor Okadaic Acid (OA) for the indicated times (E) or were treated overnight during the nocodazole arrest with inhibitors of MEK (U0126), PI 3-kinase (LY294002) or Aurora B (ZM447439) (F). Cell lysates were examined by blotting with α–HA and α–HSP90. G. H. Proteins were immunoprecipitated from 293T cells with α–HA and subjected to an in-vitro kinase assay with recombinant Aurora B. Incorporated 32P was visualized by phosphorimaging, blotting with α–HA allowed detection of immunoprecipitates. I-K. Asynchronous cultures of HeLa mCh-Tub cells stably expressing HA, HA-CHMP4CR, HA-CHMP4CR δINS, or HA-CHMP4CR S210A were transfected with the indicated siRNA. Resolved cell lysates were examined by blotting with α-CHMP4C, α-HA and α-HSP90 (I). Alternatively, cells were imaged live (J, K) and abscission time (Luciferase, 104±35 minutes, n = 244, CHMP4c siRNA, 71±37 minutes, n = 260; CHMP4c siRNA and HA-CHMP4CR, 118±52 minutes, n = 269; CHMP4C siRNA and HA-CHMP4CR δINS, 81±40 minutes, n = 264; CHMP4C siRNA and HA-CHMP4CR S210A, 91±38 minutes, n = 268) quantified across 7 independent experiments. L, HeLa cells stably expressing YFP-LAP2β and either HA or HA-CHMP4CR, HA-CHMP4CR δINS, or HA-CHMP4CR S210A were treated with the indicated siRNA, imaged live and the timing of YFP-LAP2β bridge resolution (Luciferase, 628 ± 382 minutes, n = 41; CHMP4c siRNA, 413 ± 292 minutes, n = 41; CHMP4c siRNA and HA-CHMP4CR, 698 ± 332 minutes, n = 41; CHMP4C siRNA and HA-CHMP4CR δINS, 402 ± 259 minutes, n = 36; CHMP4C siRNA and HA-CHMP4CR S210A, 421 ± 295 minutes, n = 40) quantified from 2 independent experiments.
To investigate the role of Aurora B-phosphorylation of CHMP4C on abscission timing, we employed HeLa mCh-Tub cell lines stably expressing HA-tagged, siRNA-resistant versions of CHMP4C (HA-CHMP4CR, HA-CHMP4CR δINS or HA-CHMP4CR S210A) expressed at similar, near-endogenous levels (Fig. 4I). Interaction of these mutants with known CHMP4C-binding proteins (Fig. S8H) was maintained and early mitotic phases were completed normally in cells depleted of endogenous CHMP4C and reliant upon these proteins (Fig S8I). Importantly, HA-CHMP4CR rescued the faster abscission induced by CHMP4C depletion (Fig. 4J) whereas cells reliant upon HA-CHMP4CR δINS and HA-CHMP4CR S210A could not (Fig. 4K). We propose that Aurora B-dependent phosphorylation of S210 allows CHMP4C localization to the Flemming body and acts as a brake upon the late stages of cytokinesis. Furthermore, cells stably expressing YFP-LAP2β and HA-CHMP4CR S210A were unable to delay abscission in response to intracellular chromatin bridges, despite the presence of these proteins at the chromatin bridge (Fig S9A), indicating deregulation of the NoCut checkpoint in these cells (Fig 4L).
Here we found that CHMP4C acts as an essential regulator of the Aurora B-mediated abscission checkpoint. In the absence of CHMP4C, cells complete abscission faster. We suggest that Aurora B-dependent phosphorylation of CHMP4C upon S210 directs its Flemming-body localization and delays abscission through activation of NoCut, possibly by preventing assembly of a productive abscission complex. A phospho-mimetic mutation (S210D) had no apparent effect on known CHMP4C interactions (Fig S9B) suggesting phospho-regulation of abscission timing may involve as-yet unknown CHMP4C-binding partners. That the S210 consensus site is conserved only in mammals suggests that this mechanism might have emerged late in evolution as a safety belt in addition to the interactions of the CPC with CHMP2A and CHMP6. CHMP4C depletion circumvents the NoCut abscission checkpoint, allows faster resolution of chromatin-bridges and induces the accumulation of phosphorylated H2AX. These observations are consistent with a role of CHMP4C in protection against DNA-damage accumulation In this context, as well as being Charged MVB Proteins (28), CHMPs were originally reported as Chromatin-Modifying Proteins (29) that could associate with condensed chromatin, suggesting that the physical interaction of lagging chromosomes and ESCRT-III at the midbody may trigger activation of NoCut.
Supplementary Material
Acknowledgements
We thank P. Bieniasz for kind gifts of GFP-CHMP4-expressing cells. JM-S was funded by the Medical Research Council (G0802777), the Lister Institute for Preventative Medicine and the EMBO Young Investigators Program. JM-S and M.A. were funded by Wellcome Trust grant (WT093056MA). J.G.C. was funded by Wellcome Trust Value in People award (092429/Z/10/Z). We acknowledge the NIHR Comprehensive Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London for an equipment grant.
Footnotes
SOM: Supporting Information for this manuscript contains information on materials and methods, 9 supplemental figures (S1-S9), 9 supplemental movies (Movie S1-Movie S9) and supplemental references 30-32.
References and Notes
- 1.Carlton JG, Martin-Serrano J. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 2007;316:1908–1912. doi: 10.1126/science.1143422. [DOI] [PubMed] [Google Scholar]
- 2.Morita E, et al. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samson RY, Obita T, Freund SM, Williams RL, Bell SD. A role for the ESCRT system in cell division in archaea. Science. 2008;322:1710–1713. doi: 10.1126/science.1165322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caballe A, Martin-Serrano J. ESCRT machinery and cytokinesis: the road to daughter cell separation. Traffic. 2011;12:1318–1326. doi: 10.1111/j.1600-0854.2011.01244.x. [DOI] [PubMed] [Google Scholar]
- 6.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 7.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev. Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 8.Weiss ER, Göttlinger H. The role of cellular factors in promoting HIV budding. J. Mol. Biol. 2011;410:525–533. doi: 10.1016/j.jmb.2011.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin-Serrano J, Neil SJD. Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 2011;9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- 10.Carlton JG, Agromayor M, Martin-Serrano J. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10541–10546. doi: 10.1073/pnas.0802008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guizetti J, et al. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 12.Elia N, Sougrat R, Spurlin TA, Hurley JH, Lippincott-Schwartz J. Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc. Natl. Acad. Sci. U.S.A. 2011;108:4846–4851. doi: 10.1073/pnas.1102714108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morita E, et al. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norden C, et al. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- 15.Mendoza M, et al. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat. Cell Biol. 2009;11:477–483. doi: 10.1038/ncb1855. [DOI] [PubMed] [Google Scholar]
- 16.Steigemann P, et al. Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell. 2009;136:473–484. doi: 10.1016/j.cell.2008.12.020. [DOI] [PubMed] [Google Scholar]
- 17.Information on materials and methods is available on Science Online.
- 18.Hewitt EW, et al. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. EMBO J. 2002;21:2418–2429. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agromayor M, et al. Essential role of hIST1 in cytokinesis. Mol. Biol. Cell. 2009;20:1374–1387. doi: 10.1091/mbc.E08-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Connell JW, Lindon C, Luzio JP, Reid E. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 2009;10:42–56. doi: 10.1111/j.1600-0854.2008.00847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang D, et al. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 2008;15:1278–1286. doi: 10.1038/nsmb.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meraldi P, Honda R, Nigg EA. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr. Opin. Genet. Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 24.Mackay DR, Makise M, Ullman KS. Defects in nuclear pore assembly lead to activation of an Aurora B-mediated abscission checkpoint. J. Cell Biol. 2010;191:923–931. doi: 10.1083/jcb.201007124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: the histone guardian of the genome. DNA Repair (Amst.) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 26.Bastos RN, Barr FA. Plk1 negatively regulates Cep55 recruitment to the midbody to ensure orderly abscission. J. Cell Biol. 2010;191:751–760. doi: 10.1083/jcb.201008108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fabbro M, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev. Cell. 2005;9:477–488. doi: 10.1016/j.devcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Howard TL, Stauffer DR, Degnin CR, Hollenberg SM. CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J. Cell. Sci. 2001;114:2395–2404. doi: 10.1242/jcs.114.13.2395. [DOI] [PubMed] [Google Scholar]
- 29.Stauffer DR, Howard TL, Nyun T, Hollenberg SM. CHMP1 is a novel nuclear matrix protein affecting chromatin structure and cell-cycle progression. J. Cell. Sci. 2001;114:2383–2393. doi: 10.1242/jcs.114.13.2383. [DOI] [PubMed] [Google Scholar]
- 30.Katoh K, et al. The ALG-2-interacting Protein Alix Associates with CHMP4b, a Human Homologue of Yeast Snf7 That Is Involved in Multivesicular Body Sorting. J. Biol. Chem. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- 31.Garrus JE, et al. Tsg101 and the Vacuolar Protein Sorting Pathway Are Essential for HIV-1 Budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 32.Mackay DR, Elgort SW, Ullman KS. The Nucleoporin Nup153 Has Separable Roles in Both Early Mitotic Progression and the Resolution of Mitosis. Mol. Biol. Cell. 2009;20:1652–1660. doi: 10.1091/mbc.E08-08-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.