Abstract
A β-(1,3),(1,6)-D-glucan produced by A. pullulans (AP-PG) is known to be an immune stimulating agent. In this study, we demonstrate that the stimulation with AP-PG effectively induces the interferon (IFN) stimulated genes (ISGs) in macrophage-like cell lines. The ISGs, Mx1, ISG15, and viperin mRNAs were significantly increased in RAW264.7 cells after stimulation with AP-PG. The stimulation with AP-PG transiently induced IFN-β mRNA. However, the expression of viperin mRNA was also increased after stimulation with AP-PG even when new protein synthesis was completely blocked by treatment with cycloheximide. Further, in IFN-α receptor knockdown RAW264.7 cells, AP-PG stimulation more effectively induced viperin mRNA compared with that of IFN-α stimulation. The phosphorylation of Ser 727 in STAT1 involved in the enhancement of STAT1 activation was immediately increased after stimulation with AP-PG. In addition, viperin mRNA expression induced after stimulation with IFN-α was significantly increased by combined stimulation with AP-PG. These results suggest that stimulation with AP-PG effectively induces the ISGs through the induction of IFN and the enhancement of STAT1-mediated transcriptional activation.
Black yeast, Aureobasidium pullulans extracellularly produces a β-(1,3),(1,6)-D-glucan (β-glucan) at certain growth conditions1,2. The A. pullulans-produced β-glucan (AP-PG) consists of a β-1,3-D-glycosidic linked main chain and β-1,6-D-glycosidic linked side chains, and has an immune modulating activity3. A. pullulans-cultured fluid containing AP-PG as a main compound is thought to induce beneficial effects on health, and is marketed as a food supplement.
Interferons (IFNs) play pivotal roles in the elimination of viral infections through the activation of host anti-viral responses. The replication of the influenza A virus is strongly affected by production of IFNs4. Because of the significance of IFNs, several influenza A virus proteins are involved in the inhibition of the signaling pathway for production of IFNs5,6,7, and dysfunction of this inhibitory activity is closely related to the effective replication and hence pathogenicity of the influenza A virus. The stimulation with IFNs activates transcription of IFN stimulated genes (ISGs) and exhibits antivirus activity. Several ISGs effective to prevent influenza A virus replication have been reported. One of these is Mx1 etc. Mx1 (myxovirus resistance 1), the mouse homologue of human MxA gene, was the first ISG identified as effective for the inhibition of influenza A virus replication8,9,10. In addition, viperin (virus inhibitory protein, endoplasmic reticulum-associated, IFN-inducible; also known as RSAD2)11, ISG15 (IFN stimulated gene, 15 kDa)12, and IFITM3 (IFN-induced transmembrane protein 3)13 have been independently identified as important host-cellular molecules induced after stimulation with IFNs for the inhibition of influenza A virus replication.
For the activation of innate immunity against virus infections including influenza A virus infection, IRF3 (IFN regulatory factor 3) plays a pivotal role through the transcriptional activation of type I IFN mRNAs14. Activation of IRF3 is regulated by IKKε (inhibitor of κB kinase ε) and TBK (TNF receptor-associated factor family member-associated NF-κB activator binding kinase) which are signaling molecules located downstream of the pattern recognition receptors, TLR (Toll-like receptor) and RLR (RIG-I like receptor)15,16. Stimulation of these pattern recognition receptors with extracellular pathogen-derived molecules, such as double stranded RNA, 5′-triphosphate RNA, and lipopolysaccharide, activates IKKε- and TBK-mediated phosphorylation of IRF3 at Ser 39717.
A transcriptional factor, STAT1 (signal transduction and activator of transcription) is known to be essential in the induction of ISGs in response to stimulation with IFNs18,19. Activation of STAT1 is regulated by phosphorylation at Tyr 701 mediated by JAK (Janus kinase), the downstream kinase of the IFN-α and IFN-γ receptors. The phosphorylation of Tyr 701 in STAT1 is critical for the primary activation of STAT1, and is required for dimerization and nuclear translocation of STAT120. In addition, phosphorylation of STAT1 at Ser 727 is also important for activation of STAT1-mediated transcription21.
Previously, we have demonstrated that oral administration of AP-PG containing A. pulluans-cultured fluid is effective to protect from influenza A virus infections in mice, and that stimulation with AP-PG inhibits influenza A virus replication in a murine macrophage-like cell line, RAW264.7 cells22. In this report, we focus on this in vitro anti-virus effect of AP-PG, and demonstrate that stimulation with AP-PG effectively induces ISGs through induction of IFNs and enhancement of the IFN-mediated transcriptional activation of ISGs.
Results
Stimulation with AP-PG induces ISGs responsible for eliminating influenza A virus infection in RAW264.7 cells
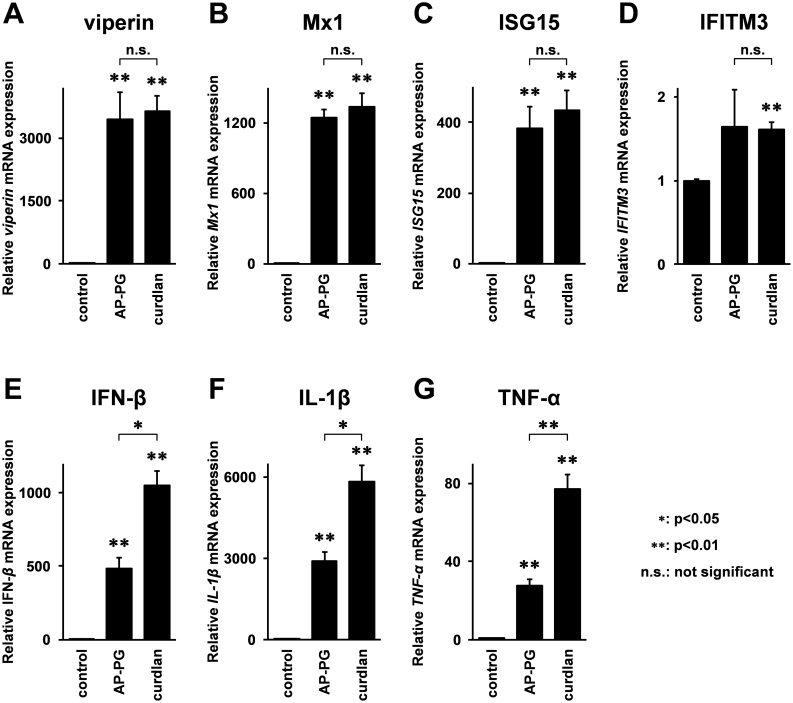
Previously, we have reported that stimulation with AP-PG effectively induces RIG-I (retinoic acid-inducible gene-I) and MDA5 (Melanoma differentiation associated gene-5) mRNAs in RAW264.7 cells22. Since both RIG-I and MDA5 are known to be ISGs23,24, this suggests the possibility that other ISGs are also induced after stimulation with AP-PG. To investigate this, mRNA expressions of ISGs involved in the inhibition of influenza A virus replication after the stimulation with AP-PG were monitored by real-time RT-PCR analysis. As shown in Figure 1A–C, virperin, Mx1, and ISG15 mRNAs were significantly increased after the stimulation with AP-PG. The increment in IFITM3 mRNA expression after stimulation with AP-PG was not statistically significant (Figure 1D).
Figure 1. Stimulation with AP-PG induces ISGs in RAW264.7 cells.
(A–G) RAW264.7 cells were stimulated with 100 μg/ml of AP-PG for 6 hours, and then the total RNAs isolated from the cells were subjected to real-time RT-PCR analysis using specific primer sets. Data represent relative expression values compared with the control after normalization to GAPDH mRNA expression. Error bars indicate standard deviations which were calculated by three independent experiments.
Next, to compare this ISGs induction activity of AP-PG with a structurally distinct β-glucan consisting of β-(1,3)-linked glucose as the main chains, curdlan which is a bacterial β-glucan produced by A. faecalis was used, and the mRNA expressions of these ISGs were monitored after stimulation with curdlan. As shown in Figure 1A–D, the results indicate that viperin, Mx1, and ISG15 mRNAs were also significantly increased after stimulation with curdlan in RAW264.7 cells with efficiencies similar to AP-PG. In addition to these ISGs, the expressions of IFN-β, IFN-γ, IL-1β, and TNF-α mRNAs were monitored using real-time RT-PCR. The results show that the expressions of IFN-β, TNF-α, and IL-1β mRNAs were more strongly induced after stimulation with curdlan than with AP-PG. (Figure 1E–G). Here, the induction of IFN-γ mRNA was quite weak, weaker than the basal expression of IFN-β mRNA (data not shown).
Stimulation with AP-PG also induces ISGs in human macrophage-like cells
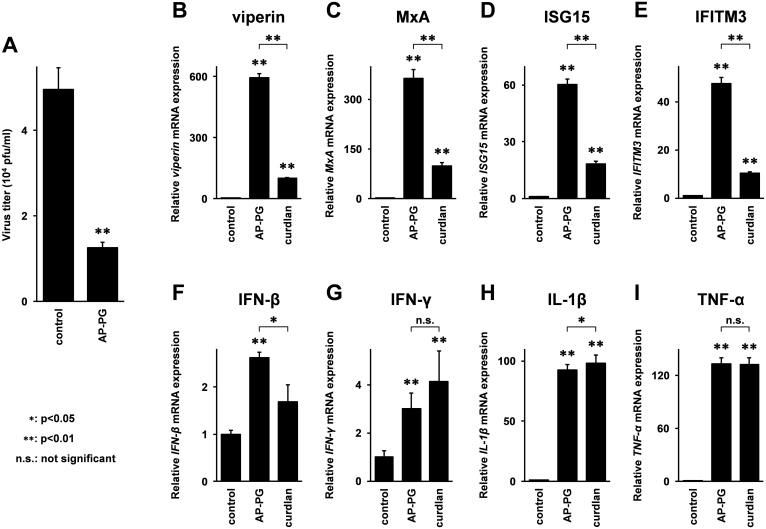
Before the analysis of the ISG expressions, the anti-virus effect of pretreatment with AP-PG in macrophage-differentiated THP-1 cells was confirmed. The results showed that similar to the RAW264.7 cells22, replication of influenza A virus in macrophage-differentiated THP-1 cells was also significantly inhibited by pretreatment with AP-PG (Figure 2A).
Figure 2. Stimulation with AP-PG increases expression of ISGs mRNAs in macrophage-differentiated THP-1 cells.
(A) Macrophage-differentiated THP-1 cells were stimulated with 100 μg/ml of AP-PG for 24 hours, and then the cells were infected with the PR8 strain of influenza A virus (MOI = 0.1). After an additional 48 hour incubation period, the virus titer in the medium was quantified by plaque assay. (B–I) Macrophage-differentiated THP-1 cells were stimulated with 100 μg/ml of AP-PG for 6 hours, and subsequently the total RNAs isolated from the cells were subjected to real-time RT-PCR analysis using specific primer sets. Data represent relative expression values compared with the control after normalization with GAPDH mRNA expression. Error bars indicate standard deviations which were calculated by three independent experiments.
Next, the expressions of these ISG mRNAs after stimulation with AP-PG and curdlan in macrophage differentiated THP-1 cells were investigated. The results showed that also in macrophage-differentiated THP-1 cells, expressions of viperin, MxA, and ISG15 mRNAs were significantly increased after stimulation with AP-PG (Figure 2B–D). Further, different from the results in RAW264.7 cells, the expression of IFITM3 mRNA was significantly increased after stimulation with AP-PG (Figure 2E). These ISG mRNA expressions were also significantly increased after stimulation with curdlan (Figure 2B–E). However, the induction activity of these ISG mRNAs by curdlan was significantly weaker than that of AP-PG. The induction levels of IFN-β mRNA after stimulation with AP-PG were slightly stronger than those of curdlan (Figure 2F), the induction activities of INF-γ, IL-1β, and TNF-α mRNAs by curdlan were very similar to those of AP-PG (Figure 2G–I). These results suggest that stimulation with AP-PG more effectively induces these ISGs than stimulation with curdlan in macrophage-differentiated THP-1 cells.
Stimulation with AP-PG induces ISGs independent of production of type I IFNs
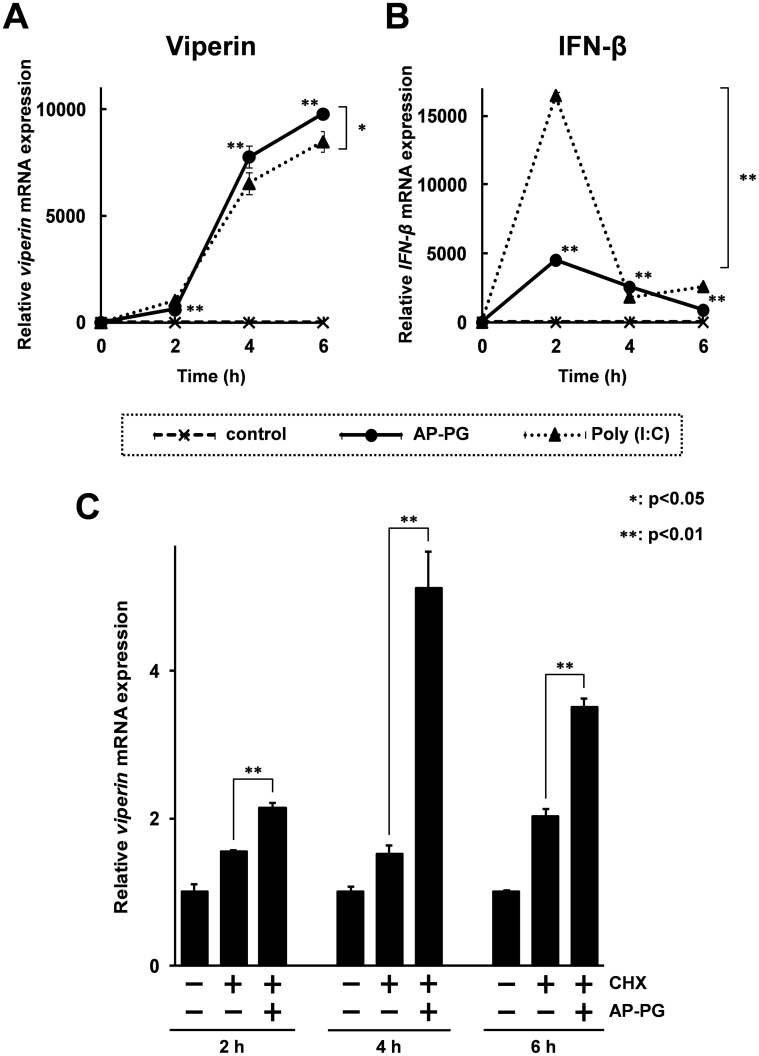
To investigate whether the induction of ISGs is a primary response to the stimulation with AP-PG, a time-course analysis was performed. As shown in Figure 3A, the results show that viperin mRNA expression was significantly induced after stimulation with AP-PG after only 2 hours (an about 600 fold increment was recorded). However, the viperin mRNA expression was more strongly increased 4 hours after stimulation with AP-PG than at 2 hours after the stimulation, and reached an about 9,800 fold increase over that of the initial time point. The expression of IFN-β mRNA was immediately but transiently increased after stimulation with AP-PG, and peaked at 2 hours post-stimulation (Figure 3B).
Figure 3. Stimulation with AP-PG also induces viperin mRNA after treatment with cycloheximide.
(A), (B) RAW264.7 cells were stimulated for the indicated times with 100 μg/ml of AP-PG or poly (I:C), and then the total RNAs isolated from the cells were subjected to real-time RT-PCR analysis using specific primer sets. The data represent relative expression values compared with the mRNA expression at the initial time point after normalization with the GAPDH mRNA expression. (C) RAW264.7 cells were treated with 5 mM cycloheximide (CHX) for 30 min. Subsequently, the cells were stimulated with or without 100 μg/ml of AP-PG. After the indicated incubation periods, the cells were harvested, and the viperin mRNA expression was analyzed by real-time RT-PCR. The data are represented as relative expression values compared with the mRNA expression in untreated cells after the normalization with GAPDH mRNA expression. Error bars indicate standard deviations calculated by three independent experiments.
To compare the induction activities of viperin and IFN-β mRNAs of AP-PG with other immune stimulating agents, expressions of these mRNAs after stimulation with poly (I:C) were investigated. A synthetic double-stranded RNA, poly (I:C), is a well known immune stimulator for the induction of the type I IFN response25. As shown in Figure 3B, IFN-β mRNA was more strongly increased after stimulation with poly (I:C) than with that of AP-PG. Different from the IFN-β mRNA expression, induction levels of viperin mRNA after stimulation with AP-PG were comparable to those with poly (I:C) (Figure 3A).
Next, the induction of viperin mRNA after stimulation with AP-PG was monitored under the condition when new protein synthesis was inhibited, using cycloheximide. The RAW264.7 cells were pretreated with 5 mM cycloheximide for 30 minutes, and subsequently stimulated with AP-PG. It has been reported that new protein synthesis is almost completely inhibited under this condition26. As shown in Figure 3C, the results showed that viperin mRNA was significantly increased after stimulation with AP-PG, and peaked at 4 hours after the start of stimulation. However, the increment of viperin mRNA expression in this condition is clearly weaker than without the cycloheximide treatment. These results suggest that stimulation with AP-PG is able to induce ISGs without IFN production, and further that the IFN production in response to stimulation with AP-PG is required for enhancement of ISG inductions.
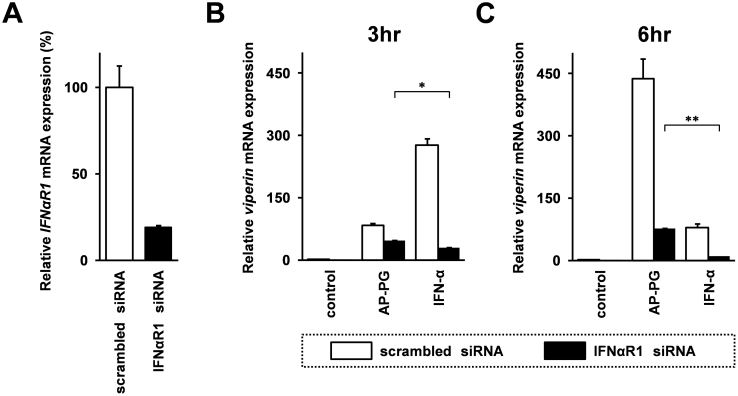
Finally, effects of AP-PG stimulation on induction of viperin mRNA in type I IFN receptor knockdown cells using siRNA were investigated. As shown in Figure 4A, the siRNA targeting IFN-α receptor 1 (IFNαR1) mRNA used in this study was effectively silenced IFNαR1 mRNA expression, and the gene silencing efficiency was calculated at 81.3%. In control scrambled siRNA transfected cells, the viperin mRNA induction after stimulated with AP-PG was weaker than that of IFN-α stimulation at 3 hours post-stimulation (Figure 4B). After 6 hours post-stimulation, viperin mRNA expression after stimulation with AP-PG was higher than that of stimulation with IFN-α in control scrambled siRNA transfected cells (Figure 4C). On the other hand, viperin mRNA expression in IFNαR1 knockdown cells was significantly higher in AP-PG-stimulated cells than that of IFN-α-stimulated cells at both time points. The results suggest that other signaling pathway independent from type I IFN receptor-mediated signaling pathway is involved in the induction of viperin mRNA after stimulation with AP-PG.
Figure 4. Effect of knockdown of TypeI IFN receptor 1 on viperin mRNA induced by AP-PG stimulation.
(A) Gene silencing efficiency of siRNA targeting IFN-α receptor 1 (IFNαR1) mRNA. The IFNαR1 mRNA specific siRNA or control scrambled siRNA were transfected into RAW264.7 cells. After 72 hrs, the cells were harvested, and IFNαR1 mRNA expression was analyzed by real-time RT-PCR. Data represent relative expression of IFNαR1 mRNA compared with control scrambled siRNA-transfected cells after the normalization with GAPDH mRNA expression. (B), (C) RAW264.7 cells were transfected with siRNA targeting IFNαR1 mRNA of control scrambled siRNA. After 72 hrs, the cells were stimulated with 100 μg/ml AP-PG or 10,000 U/ml IFN-α for 3 hrs (B) or 6 hrs (C), and then viperin mRNA expression was monitored by real-time RT-PCR. Data represent relative expression values compared with the control after normalization with GAPDH mRNA expression. Error bars indicate standard deviations calculated by three independent experiments. Single asterisk (*) and double asterisk (**) indicate statistically significant differences (p < 0.05 and p < 0.01 respectively) between stimulations with IFN-α and AP-PG.
Stimulation with AP-PG activates STAT-1 independently of the activation of the IFN receptor-mediated signaling pathway
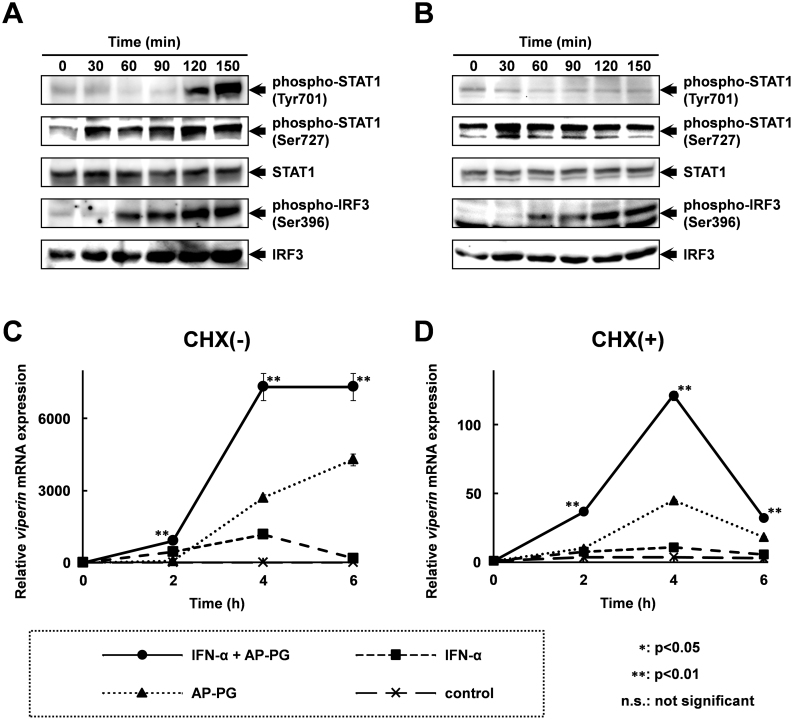
To evaluate the molecular mechanism involved in the mRNA induction of these ISGs after the stimulation with AP-PG, phosphorylations of STAT1 and IRF3 were monitored by immunoblotting using specific antibodies. The results show that the phosphorylation of STAT1 at Ser 727 was increased 30 min after stimulation with AP-PG (Figure 5A). Phosphorylation of STAT1 at Tyr 701 in response to stimulation with AP-PG was slower than that of the phosphorylation at Ser 727, and was strongly increased from 120 min after stimulation with AP-PG. The phosphorylation of IRF3 at Ser 396, which is involved in transcriptional activation of type I IFN genes, was increased 60 min after the stimulation with AP-PG, and peaked at 120 min. Phosphorylation of Tyr 701 in STAT1 is known to be dependent on JAK, a kinase which is activated in response to stimulation with type I IFNs20. Therefore, these results suggest that phosphorylation of Ser 727 in STAT1 and Ser 396 in IRF3 are primary responses to stimulation with AP-PG, and that the phosphorylation of STAT1 at Tyr 701 can be considered to be increased in response to the IFNs induced after AP-PG stimulation. To confirm this, phosphorylations of STAT1 and IRF3 after stimulation of AP-PG under the condition of which new protein synthesis was blocked by high concentration of cycloheximide were investigated. The results show that although the basal level phosphorylation of STAT1 at Ser 727 was increased, Ser 727 phosphorylation of STAT1 was immediately increased after stimulation with AP-PG (Figure 5B). In addition, phosphorylation of Ser 396 in IRF3 was increased in the cycloheximide treated cells as similar to that in untreated cells. However, phosphorylation of Tyr 701 in STAT1 was not increased in the presence of cycloheximide. These findings indicate that production of type I IFNs followed by activation of IFN-mediated signaling pathway was thought to be almost completely inhibited after the high dose treatment with cycloheximide. Further, these results strongly suggest that stimulation with AP-PG is able to increase phosphorylation of Ser 727 in STAT1 independently from production of type I IFNs. The increment of phosphorylation of Ser 727 in STAT1 after stimulation with AP-PG was more rapidly decreased in the cycloheximide treated cells than that in untreated cells. This may suggest that prolonged activation of STAT1 through the phosphorylation of Ser 727 requires protein synthesis, such as production of type I IFNs and other cytokines.
Figure 5. Phosphorylation of STAT1 at Ser 727 is immediately increased after stimulation with AP-PG.
(A), (B) RAW264.7 cells were pretreated with (A) or without (B) 5 mM cycloheximide (CHX) for 30 min. After the treatment, the cells were stimulated with 100 μg/ml of AP-PG. The cells were harvested at the indicated time points, and the whole cell lysates prepared from the cells were analyzed by immunoblotting using specific antibodies. Full length blots are presented in supplementary information (Supplementary Figure S1 and S2). (C), (D) RAW264.7 cells were pretreated with (D) or without (C) 5 mM cycloheximide (CHX) for 30 min, and then the cells were stimulated with 10,000 U/ml of IFN-α together with or without 100 μg/ml of AP-PG. After the indicated incubation periods, the cells were harvested, and the expression of viperin mRNA was analysed by real-time RT-PCR using a specific primer set. The data represent relative expression amounts compared with that at the initial time point after normalization with GAPDH mRNA expression. Double asterisk (**) show statistically significant differences (p < 0.01) compared to independent stimulations with IFN-α or AP-PG.
To investigate the effect of the increase in STAT1 phosphorylation at Ser 727 after stimulation with AP-PG on the IFN-mediated transcriptional activation of ISGs, RAW264.7 cells were stimulated with IFN-α together with AP-PG, and the expression of viperin mRNA was monitored. As shown in Figure 5C, the combined stimulation with AP-PG and IFN-αinduced viperin mRNA more strongly than the independent (separate) stimulations. Further, the viperin mRNA expression after stimulation with IFN-α was also significantly increased by the combined stimulation with AP-PG also when the RAW264.7 cells were pretreated with cycloheximide (Figure 5D). These results suggest that stimulation with AP-PG is involved in the induction of IFNs as well as it is effective in increasing the IFN-induced expression of ISGs.
Discussion
This study demonstrates that stimulation with AP-PG effectively induces ISGs involved in the elimination of influenza A virus infections in RAW264.7 cells and macrophage-differentiated THP-1 cells. Previously we have demonstrated that pretreatment with AP-PG effectively inhibits the replication of the influenza A virus in RAW264.7 cells22. In addition to RAW264.7 cells, this study shows that pretreatment with AP-PG is also effective to inhibit influenza A virus replication in macrophage-differentiated THP-1 cells. The study demonstrates the anti-virus activity of the ISGs examined in this study. This would suggest that induction of these ISG mRNAs may be considered to be involved in the in vitro anti-virus effects of pretreatment with AP-PG with these macrophage-like cell lines.
The results of the immunoblotting analysis indicate that stimulation with AP-PG induces phosphorylation of STAT1 at Ser 727, an effect that is important for the enhancement of STAT1 activation (Figure 5A and 5B). The results also suggest that AP-PG stimulation is not directly involved in phosphorylation of STAT1 at Tyr 701 which is responsible to primary activation of STAT1. Therefore, stimulation with AP-PG is thought to be involved in enhancement of the transcriptional activity of the STAT1 activated by the IFN-mediated signaling pathway. This line of reasoning is supported by the results of the combined stimulation with IFN-α and AP-PG. As shown in Figure 5C and 5D, the IFN-α induced viperin mRNA expression is significantly increased when combined with AP-PG stimulation. The signaling molecules involved in the phosphorylation of STAT1 at Ser 727 after stimulation with AP-PG have not been identified. It has been reported that phosphorylation of Ser 727 in STAT1 is upregulated by the activation of the MAPK pathway21. In addition, the MAPK pathway is known to be activated after stimulation with β-glucan27. Therefore, the MAPK pathway may possibly be involved in the Ser 727 phosphorylation of STAT1 induced after stimulation with AP-PG. The results of real-time RT-PCR analysis show that basal level expression of viperin mRNA was increased after treatment with cycloheximide (Figure 3C). In addition, the basal level phosphorylation of STAT1 at Ser 727 in the cycloheximide was increased in the cells treated with cycloheximide in comparison with that in untreated cells (Figure 5A and 5B). It has been reported that treatment with cycloheximide causes sustained activation of MAPK28. The increment of STAT1 phosphorylation at Ser 727 after treatment with cycloheximide is thought to be involved in this sustained activation of MAPK, and this may affect to the basal level expression of viperin mRNA.
In macrophage-differentiated THP-1 cells, stimulation with AP-PG more effectively induces the ISG mRNAs than stimulation with curdlan, while TNF-α and IL-1β mRNAs are effectively induced after stimulation with curdlan similar to the effect of AP-PG stimulation (Figure 2). Further, as shown in Figure 1, the induction of activity with curdlan stimulation on IL-1β, IFN-β, and TNF-α mRNAs is stronger than with stimulation by AP-PG, however the level of activity induced in ISGs is very similar to that with AP-PG. Macrophages are important to activate the immune system in response to stimulation of β-glucans consisting of β-(1,3)-linked main chains, and CR3 (complement receptor 3) and dectin-1 (dendritic cell-associated C-type lectin-1) are known to be major receptors for recognition of these β-glucans in macrophages29. It has been reported that AP-PG does not exhibit binding activity to dectin-130, while curdlan is known to be an agonist for dectin-1 and is recognized by dectin-131,32. This difference in dectin-1 binding affinities between curdlan and AP-PG may be related in the differences in the induction efficiency of ISGs.
In this study, we propose a molecular mechanism for the in vitro anti-influenza A virus activity arising from pretreatment with AP-PG in macrophage-like cell lines. We have previously demonstrated that oral administration of A. pullulans-cultured fluid containing AP-PG as a main compound also exhibits antiviral activity against influenza A virus in mice. However, it is not clear whether this molecular mechanism works with in vivo anti-influenza A virus effects. A previous report demonstrated that murine alveolar macrophages are activated by oral administration of a β-glucan produced by the fungus Sclerotinia sclerotiorum33. Therefore, it is possible that similar to the β-glucan produced by S. sclerotiorum, oral administration of AP-PG may affect to the immune activities of alveolar macrophages. Further investigation is required to understand the molecular mechanisms of the protective activity arising from oral administration of AP-PG against influenza A virus.
Methods
Cell culture
A murine macrophage-like cell line, RAW264.7 cells (ATCC TIB-71)34, and a human monocyte-derived cell line, THP-1 cells (ATCC TIB-202)35, were cultured in RPMI 1640 medium (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 mg/ml streptomycin (Sigma-Aldrich), and were grown at 37°C in 5% CO2 in a humidified incubator. For differentiation of THP-1 cells into macrophages, the THP-1 cells were stimulated with 100 nM phorbol-12-myristate-13-acetate (PMA) for 72 hours. Then the cells were grown in fresh medium without PMA for 24 hours, and used for the assay.
Preparation of purified β-glucan
The method for purification of AP-PG from A. pullulans-cultured fluid is described elsewhere22. The purity of AP-PG was estimated to be more than 99.9% using high performance liquid chromatography (HPLC). Purified curdlan derived from Alcaligenes faecalis used in this study was obtained from commercially available products (Sigma-Aldrich).
Virus infection and plaque assay
Propagation of A/Puerto Rico/8/1934 (PR8; H1N1) strains of influenza A virus, and quantification of the virus titer were performed as previously described22.
Real-time reverse transcription polymerase chain reaction (RT-PCR)
The method for the extraction of total RNA from the cultured cells followed by real-time PCR analysis is described elsewhere22. The real-time RT-PCR reaction was performed using CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). To determine relative expression of the mRNAs, the real-time RT-PCR data was analyzed by delta delat-Ct method using the CFX manager software Version 1.6 (Bio-Rad Laboratories). Sequences of specific primer sets used in this study are listed in Table 1.
Table 1. List of primer sequences used for RT-PCR analysis in this study.
| Target Gene | Sequence (5′ → 3′) | Reference | |
|---|---|---|---|
| Mouse | |||
| Viperin | Sense | TGCTGGCTGAGAATAGCATTAGG | |
| Antisense | GCTGAGTGCTGTTCCCATCT | ||
| Mx1 | Sense | TCCCTGGGAAGGAGTGAAGCCAG | |
| Antisense | CTTGCCACTGGGGTCAGGCAC | ||
| ISG15 | Sense | TGACGCAGACTGTAGACACG | 36 |
| Antisense | TGGGGCTTTAGGCCATACTC | ||
| IFITM3 | Sense | CTGAGAAACCGAAACTGCCG | |
| Antisense | GTTCATGGTGCGGAGCAAAG | ||
| IFN-β | Sense | GCCTCGTGCTGTCGGACC | |
| Antisense | TGTCGTTGCTTGGTTCTCCTTG | ||
| IFN-γ | Sense | GTCTCTTCTTGGATATCTGGAGGAACT | |
| Antisense | GTAGTAATCAGGTGTGATTCAATGACGC | ||
| IL-1β | Sense | GCCTCGTGCTGTCGGACC | |
| Antisense | TGTCGTTGCTTGGTTCTCCTTG | ||
| TNF-α | Sense | GACCCTCACACTCAGATCATCTTCT | 37 |
| Antisense | CCACTTGGTGGTTTGCTACGA | ||
| Human | |||
| Viperin | Sense | CGTGAGCATCGTGAGCAATG | |
| Antisense | GCTGTCACAGGAGATAGCGA | ||
| Mx1 | Sense | CCAGCTGCTGCATCCCACCC | |
| Antisense | AGGGGCGCACCTTCTCCTCA | ||
| ISG15 | Sense | TGGCGGGCAACGAATT | |
| Antisense | GGGTGATCTGCGCCTTCA | ||
| IFITM3 | Sense | TCCCACGTACTCCAACTTCCA | 38 |
| Antisense | AGCACCAGAAACACGTGCACT | ||
| IFN-β | Sense | CTCCTGTTGTGCTTCTCCACT | |
| Antisense | GGCAGTATTCAAGCCTCCCA | ||
| IFN-γ | Sense | CTTTAAAGATGACCAGAGCATCCA | 39 |
| Antisense | ATCTCGTTTCTTTTTGTTGCTATTGA | ||
| IL-1β | Sense | GCAGCCATGGCAGAAGTACCTGA | |
| Antisense | CCAGAGGGCAGAGGTCCAGGTC | ||
| TNF-α | Sense | CCCAGGGACCTCTCTCTAATC | |
| Antisense | ATGGCTACAGGCTTGTCACT |
siRNA and transfection
The siRNA targeting IFNαR1 mRNA (Sigma-Aldrich) and control scrambled siRNA (Ambion, Austin, TX, USA) were purchased from commercially available products. Transfection of siRNA into RAW264.7 cells was performed using LipofectAMINE RNAimax transfection reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) in accordance with manufacturer's instructions.
Immunoblotting
Rabbit monoclonal antibodies for IRF3, phosphor-IRF3 (Ser 396), STAT1 and phosphor-STAT1 (Tyr701 and Ser727) were purchased from commercially available products (Cell Signaling Technology, Beverly, MA, USA). The cells were lysed with RIPA buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate) supplemented with protease inhibitor (Complete Mini; Roche) and phosphatase inhibitor cocktail (Sigma-Aldrich). After the cell debris was removed by centrifugation, the extracts (20 μg/lane) were subjected to immunoblotting analysis.
Statistical analysis
To check for significant differences between the indicated pairs of data, a two-tailed unpaired Student's t-test was performed in this study.
Author Contributions
Conceived and designed the experiments: D.M., K.K., T.M., H.K. and A.I. Performed the experiments: D.M., K.K., S.A. and A.I. Analyzed the data: D.M., K.K. and A.I. Contributed reagents/materials/analysis tools: H.U., M.O., T.M. and H.K. Wrote the paper: D.M., H.K. and A.I.
Supplementary Material
Supplementary information
Footnotes
There is potential competing interest. This study was funded by by Aureo Co., Ltd., Kimitsu, Japan, and Aureo-Science Co., Ltd., Sapporo, Japan. However, the funders had no role in the study design, data collection, or analysis, decision to publish, or preparation of the manuscript. No additional external funding was received for this study. D.M., K.K., S.A. and A.I. are employees of Aureo-Science Co., Ltd., and K.K., H.M., M.O. and A.I. are employees of Aureo Co., Ltd. The AP-PG-containing A. pullulans-cultured fluid and its derivatives are marketed by Aureo Co., Ltd., and by Aureo-Science Co., Ltd. There are no other patents, products in development, or marketed products to declare.
References
- Hamada N. et al. Ascorbic acid stimulation of production of a highly branched, beta-1,3-glucan by Aureobasidium pullulans K-1--oxalic acid, a metabolite of ascorbic acid as the stimulating substance. Biosci Biotechnol Biochem 64, 1801–6 (2000). [DOI] [PubMed] [Google Scholar]
- Moriya N. et al. Improved beta-glucan yield using an Aureobasidium pullulans M-2 mutant strain in a 200-L pilot scale fermentor targeting industrial mass production. Biotechnology and Bioprocess Engineering 18, 1083–1089 (2013). [Google Scholar]
- Tada R. et al. Induction of IFN-gamma by a highly branched 1,3-beta-d-glucan from Aureobasidium pullulans in mouse-derived splenocytes via dectin-1-independent pathways. Biochem Biophys Res Commun 404, 1105–10 (2011). [DOI] [PubMed] [Google Scholar]
- Hayden F. G., Schlepushkin A. N. & Pushkarskaya N. L. Combined interferon-alpha 2, rimantadine hydrochloride, and ribavirin inhibition of influenza virus replication in vitro. Antimicrob Agents Chemother 25, 53–7 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mibayashi M. et al. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol 81, 514–24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai A. et al. Influenza A virus polymerase inhibits type I interferon induction by binding to interferon beta promoter stimulator 1. J Biol Chem 285, 32064–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga Z. T., Grant A., Manicassamy B. & Palese P. Influenza virus protein PB1-F2 inhibits the induction of type I interferon by binding to MAVS and decreasing mitochondrial membrane potential. J Virol 86, 8359–66 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenmann J., Lane C. A. & Hobson D. The Resistance of A2g Mice to Myxoviruses. J Immunol 90, 942–51 (1963). [PubMed] [Google Scholar]
- Haller O., Arnheiter H., Lindenmann J. & Gresser I. Host gene influences sensitivity to interferon action selectively for influenza virus. Nature 283, 660–2 (1980). [DOI] [PubMed] [Google Scholar]
- Pavlovic J., Zurcher T., Haller O. & Staeheli P. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human MxA protein. J Virol 64, 3370–5 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Hinson E. R. & Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2, 96–105 (2007). [DOI] [PubMed] [Google Scholar]
- Lenschow D. J. et al. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A 104, 1371–6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeley E. M. et al. IFITM3 inhibits influenza A virus infection by preventing cytosolic entry. PLoS Pathog 7, e1002337 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem 282, 15325–9 (2007). [DOI] [PubMed] [Google Scholar]
- Fitzgerald K. A. et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4, 491–6 (2003). [DOI] [PubMed] [Google Scholar]
- Kawai T. & Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci 1143, 1–20 (2008). [DOI] [PubMed] [Google Scholar]
- Servant M. J. et al. Identification of the minimal phosphoacceptor site required for in vivo activation of interferon regulatory factor 3 in response to virus and double-stranded RNA. J Biol Chem 278, 9441–7 (2003). [DOI] [PubMed] [Google Scholar]
- Meraz M. A. et al. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell 84, 431–42 (1996). [DOI] [PubMed] [Google Scholar]
- Durbin J. E., Hackenmiller R., Simon M. C. & Levy D. E. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell 84, 443–50 (1996). [DOI] [PubMed] [Google Scholar]
- Ihle J. N. et al. Signaling by the cytokine receptor superfamily: JAKs and STATs. Trends Biochem Sci 19, 222–7 (1994). [DOI] [PubMed] [Google Scholar]
- Wen Z., Zhong Z. & Darnell J. E. Jr Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell 82, 241–50 (1995). [DOI] [PubMed] [Google Scholar]
- Muramatsu D. et al. beta-Glucan derived from Aureobasidium pullulans is effective for the prevention of influenza in mice. PLoS One 7, e41399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S. N. & Sen G. C. Novel functions of proteins encoded by viral stress-inducible genes. Pharmacol Ther 103, 245–59 (2004). [DOI] [PubMed] [Google Scholar]
- Kang D. C. et al. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci U S A 99, 637–42 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou L., Holt A. C., Medzhitov R. & Flavell R. A. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732–8 (2001). [DOI] [PubMed] [Google Scholar]
- Vabulas R. M. & Hartl F. U. Protein synthesis upon acute nutrient restriction relies on proteasome function. Science 310, 1960–3 (2005). [DOI] [PubMed] [Google Scholar]
- Lee S. Y. et al. Mitogen activated protein kinases are prime signalling enzymes in nitric oxide production induced by soluble beta-glucan from Sparassis crispa. Arch Pharm Res 33, 1753–60 (2010). [DOI] [PubMed] [Google Scholar]
- Lin W. W. & Hsu Y. W. Cycloheximide-induced cPLA(2) activation is via the MKP-1 down-regulation and ERK activation. Cell Signal 12, 457–61 (2000). [DOI] [PubMed] [Google Scholar]
- Chan G. C., Chan W. K. & Sze D. M. The effects of beta-glucan on human immune and cancer cells. J Hematol Oncol 2, 25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada R. et al. Induction of IFN-gamma by a highly branched 1,3-beta-d-glucan from Aureobasidium pullulans in mouse-derived splenocytes via dectin-1-independent pathways. Biochem Biophys Res Commun 404, 1105–10 (2011). [DOI] [PubMed] [Google Scholar]
- Brown G. D. & Gordon S. Immune recognition. A new receptor for beta-glucans. Nature 413, 36–7 (2001). [DOI] [PubMed] [Google Scholar]
- Kankkunen P. et al. (1,3)-beta-glucans activate both dectin-1 and NLRP3 inflammasome in human macrophages. J Immunol 184, 6335–42 (2010). [DOI] [PubMed] [Google Scholar]
- Sakurai T. et al. Enhancement of murine alveolar macrophage functions by orally administered beta-glucan. Int J Immunopharmacol 14, 821–30 (1992). [DOI] [PubMed] [Google Scholar]
- Ralph P. & Nakoinz I. Antibody-dependent killing of erythrocyte and tumor targets by macrophage-related cell lines: enhancement by PPD and LPS. J Immunol 119, 950–54 (1977). [PubMed] [Google Scholar]
- Tsuchiya S. et al. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int J Cancer 26, 171–6 (1980). [DOI] [PubMed] [Google Scholar]
- Nacionales D. C. et al. Type I interferon production by tertiary lymphoid tissue developing in response to 2,6,10,14-tetramethyl-pentadecane (pristane). Am J Pathol 168, 1227–40 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth I. et al. Osmoprotective transcription factor NFAT5/TonEBP modulates nuclear factor-kappaB activity. Mol Biol Cell 21, 3459–74 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J. et al. Gene-expression profiling in Chinese patients with colon cancer by coupling experimental and bioinformatic genomewide gene-expression analyses: identification and validation of IFITM3 as a biomarker of early colon carcinogenesis. Cancer 113, 266–75 (2008). [DOI] [PubMed] [Google Scholar]
- Wilson N. J. et al. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol 8, 950–7 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information