Summary
pAMPK and pmTOR favorably predicted outcome in early NSCLC. The differences were small. Phosphoprotein lability makes routine clinical use and validation difficult. Protein IHC is unlikely to be clinically useful and numerous efforts to create predictive models to select resected patients for therapy have been unsuccessful.
In this issue of Clinical Cancer Research Gold et al attempted to define prognostic signatures to predict outcomes in early stage lung cancer patients who undergo surgical resection but are at risk for recurrence following the surgery (1). This study not only took on this difficult challenge but also attempted to develop a signature based on analysis of protein and/or activated protein expression by immunohistochemistry, targeting proteins identified through gene array data. The authors report that they were able to find several proteins that were significantly associated with either progression free or overall survival some of which overlapped in both categories. Point out the difficulties in creating predictive and prognostic risk models. Among these were pmTOR and pAMPK. These findings will add to evidence of their role in NSCLC and could lead to an explanation of why high mTOR expression was associated with a favorable outcome. However, the magnitude of the differences in progression-free and overall survival was small. As acknowledged by the authors, there are many technical difficulties to the routine assessment of activated (phosphorylated) proteins by immunohistochemistry. Furthermore, the study was complicated by a heterogeneous patient population, which could theoretically result in the impact of this approach being understated. Thus, the authors appropriately concluded that their findings were unlikely to be adopted clinically. Their findings also highlight the importance of the distinction between prognostic and predictive factors and the difficulty of validating proposed signatures for either prognosis or prediction of optimal therapy. The proposed commercial development of gene signatures highlights the importance of the issues.
Since the reports from Duke Univ. that gene signatures could predict outcomes in surgically resected NSCLC patients, there have been numerous reports that failed to validate their findings and numerous other reports that reported prognostic signatures with a different panel of genes (2–12). Still other studies have reported that DNA methylation profiles and microRNA signatures may also have prognostic relevance (13,14). So why do we not have a clinically useful prognostic signature in lung cancer such as the mammoprint® or Oncotype DX® signatures used in breast cancer? The lack of annotated tissues for validation and the lack of prospective studies in early stage NSCLC are part of the reason. Interestingly, there is little overlap in the genes and proteins that best predicted outcomes and the results of validation studies invariably showed lesser degrees of prognostic distinction (2–12). The studies illustrate many of the difficulties encountered in attempting such studies, including inadequate cases with tissue available for study, variations in assay methodology, variations in histology and clinical features, and the use of differing adjuvant therapies. Despite these difficulties, there are several prognostic signatures that have been validated not only in the original series but also in some follow-up series demonstrating relatively large differences in the outcomes based on the signatures. In addition, the Squamous Lung Cancer Consortium supported by the NCI’s Specialized Program for Evaluation of Cancer Signatures is re-evaluating many published gene signatures under standardized circumstances both in terms of tissue processing, RNA extraction, histology and clinical features both in a large prospective test set and defined validation set.
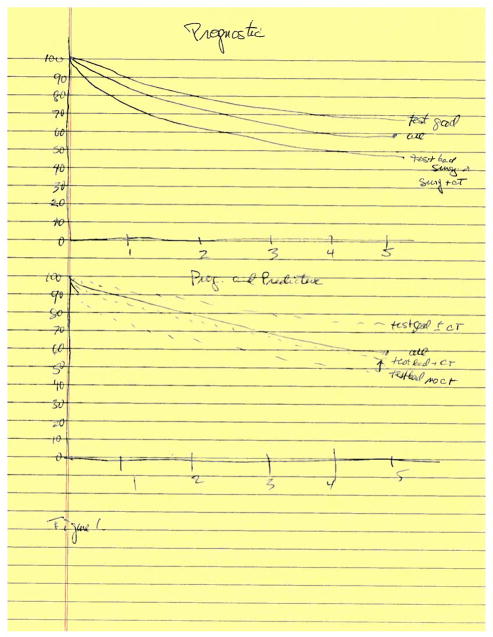
The major question is what is the clinical relevance of these differences. Are any of these prognostic signatures predictive of benefit from adjuvant chemotherapy? Would the knowledge that a resected patient has a high or a low chance of relapse associate with benefit (or lack there of) from chemotherapy and thus affect the subsequent management? If this knowledge would affect subsequent follow-up or therapy, then the utility of such an approach is clear. Figure 1 illustrates the differences between a signature that is prognostic only and one that is prognostic and predictive. Proof that one of the prognostic signatures is predictive would require a prospective randomized trial with randomization to adjuvant or no adjuvant chemotherapy.
Figure 1.
1A, A prognostic signature can separate patients into those with a superior survival “good signature” and those with an inferior survival “poor signature” compared to the entire group as illustrated. The signature may divide the patients into 3 or 4 groups if this is clinically desirable compared to a division into 2 groups. The signature performance may differ by histology and stage and generally the differences are less prognostic in validation sets. 1B. A signature that is prognostic and predictive requires a prospective randomized trial in which those with both a “good” and a “poor” signature would be separately randomized to receive or not receive adjuvant chemotherapy following surgical resection. Survival would be superior in those randomized to the chemotherapy.
Current therapeutic guidelines recommend that patients with completely resected Stage I NSCLCs receive no adjuvant therapy and that patients with stage II and IIIA NSCLC receive adjuvant chemotherapy (15,16) Unfortunately, there is no convincing evidence that current gene signatures which predict a poor outcome in stage I would also indicate that such patients would have their poor outcome reversed by adjuvant chemotherapy. And there is no convincing evidence that stage II or III patients with a good signature would fail to benefit from adjuvant chemotherapy. Moreover, none of the signatures developed to date were designed to predict the best therapy that might be given in the adjuvant setting. Thus, there is a need for separating prognostic- and predictive signatures, even if overlap may occur.
The design issues of these trials can be overcome but it is unclear whether such trials are feasible with our current clinical trial infrastructure and accrual rates. The NCI supported Alliance attempted such a trial but it was closed for lack of accrual. We must hope that we can develop less toxic therapeutic choices and biomarker/signatures that better predict outcomes from these therapies as well as associate with prognosis. Currently, there are no prospective trials that are likely to identify signatures that are prognostic or predictive although it is possible that the Alchemist trials evaluating specific molecular changes with specific molecular therapies could serve some of this functionality. Obstacles include the potential need to evaluate multiple prognostic approaches from one source of tissue, controlling for adjuvant therapy selected with appropriate planned subgroup analysis, controlling for the prognostic information inherently seen in well-defined driver alterations and the need to evaluate single-analyte approaches as well as complex ‘signatures’. The difficulties seen in this and other studies highlight the need for additional worldwide early stage lung cancer clinical trials!
Acknowledgments
Supported in part by grants P50 CA 058187 (SPORE) andU01CA157715 (SPECS) from the National Cancer Institute
Footnotes
Potential Conflicts:
Paul A. Bunn, Jr, MD Dr. Bunn has received consulting honoraria from Myriad Genetics and Life Technologies and grant support from the National Institutes of Health
Fred R. Hirsch, MD, PhD Dr. Hirsch has received consulting honoraria from Life Technologies, Myriad Genetics and research funding (through University of Colorado) from Ventana-Roche and grant support from The US National Cancer Institute/NIH.
Dara Aisner, MD Dr. Aisner has received consulting honoraria from Boehringher Ingelheim and honoraria from Abbott Molecular and Illumina
References
- 1.Gold KA, Kim ES, Liu DD, Yuan P, Behrens C, Solis L, et al. Prediction of survival in resected non-small cell lung cancer using a protein-expression based risk model: implications for personalized chemoprevention and therapy. Clin Can Res. 2014 doi: 10.1158/1078-0432.CCR-13-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Potti A, Mukherjee S, Petersen R, Dressman H, Bild A, Koontz J, et al. Retraction: A genomic strategy to refine prognosis in early-stage non-small cell lung cancer. N Engl J Med. 2011;364:1176. doi: 10.1056/NEJMc1101915. [DOI] [PubMed] [Google Scholar]; N Engl J Med. 2006;355:570–80. [Google Scholar]
- 3.Wistuba I, Behrens C, Lombardi F, Wagner S, Fujimoto J, Raso MG, et al. Validation of a proliferation-based expression signature as prognostic marker in early stage lung adenocarcinoma. Clin Cancer Res. 2013;19:6261–67. doi: 10.1158/1078-0432.CCR-13-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandoval J, Mendez-Gonzalez J, Nadal E, Chen G, Carmona F, Sayols S, et al. A prognostic DNA methylation signature for stage I non-small cell lung cancer. J Clin Oncol. 2013;31:4140–47. doi: 10.1200/JCO.2012.48.5516. [DOI] [PubMed] [Google Scholar]
- 5.Shedden K, Taylor JMG, Enkemann SA, Tsao M, Yeatman T, Gerald S, et al. Gene expression based survival prediction in lung adenocarcinoma, blinded validation study. Nat Medicine. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadara H, Behrens C, Yuan P, Solis L, Liu D, Gu X, et al. A five gene and corresponding protein signature for stage I lung adenocarcinoma prognosis. Clin Can Res. 2011;17:1490–501. doi: 10.1158/1078-0432.CCR-10-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo NL, Wan YW, Tosun K, Msiska Z, Flynn DC, Remick SC, et al. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin Can Res. 2008;14:8212–20. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du SP, Sykes J, Pintilie M, Zhu CQ, Strumpf D, Liu N, et al. Validation of histology independent prognostic gene signature for early stage non-small cell lung cancer including stage IA patients. J Thorac Oncol. 2013 doi: 10.1097/JTO.0000000000000042. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 9.Zhu CQ, Ding K, Stumpf D, Weir BA, Meyerson M, Pennell N, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small cell lung cancer. J Clin Oncol. 2010;28:4417–24. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boutros PC, Lau SK, Pintilie M, Liu N, Shepherd F, Der S, et al. Prognostic gene signatures for non-small cell lung cancer. Proc Natl Acad Sci. 2009;106:2824–28. doi: 10.1073/pnas.0809444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kratz JR, He J, Van Den Eeden SK, Zhu ZH, Gao W, Pham PT, et al. A practical molecular assay to predict survival in resected non-squamous, non-small cell lung cancer: Development and international validation studies. Lancet. 2012;379:823–832. doi: 10.1016/S0140-6736(11)61941-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kratz JR, Van Den Eeden SK, He J, Jablons D, Mann M. A prognostic assay to identify patients at high risk of mortality despite small node-negative lung tumors. JAMA. 2012;308:1629–31. doi: 10.1001/jama.2012.13551. [DOI] [PubMed] [Google Scholar]
- 13.Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, et al. Serum micro-RNA signatures identified in a genome-wide serum micro-RNA expression profiling predict survival in non-small cell lung cancer. J Clin Oncol. 2010;28:1721–26. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 14.Sandoval J, Mendez-Gonzalez J, Nadal E, Chen G, Carmona F, Saylos S, et al. A prognostic DNA methylation signature for stage I Non-small-cell lung cancer. J Clin Oncol. 2013;31:4140–47. doi: 10.1200/JCO.2012.48.5516. [DOI] [PubMed] [Google Scholar]
- 15.Pisters KM, Evans WK, Azzoli CG, Kris MG, Smith CA, Desch CE, et al. Cancer Care Ontario and American Society of Clinical Oncology adjuvant chemotherapy and adjuvant radiation therapy for stages I–IIIA resectable non small-cell lung cancer guideline. J Clin Oncol. 2007 Dec 1;25(34):5506–18. doi: 10.1200/JCO.2007.14.1226. Epub 2007 Oct 22. [DOI] [PubMed] [Google Scholar]
- 16.Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw. 2013 Jun 1;11(6):645–53. doi: 10.6004/jnccn.2013.0084. [DOI] [PubMed] [Google Scholar]