Abstract
Motivation: Resource description framework (RDF) is an emerging technology for describing, publishing and linking life science data. As a major provider of bioinformatics data and services, the European Bioinformatics Institute (EBI) is committed to making data readily accessible to the community in ways that meet existing demand. The EBI RDF platform has been developed to meet an increasing demand to coordinate RDF activities across the institute and provides a new entry point to querying and exploring integrated resources available at the EBI.
Availability: http://www.ebi.ac.uk/rdf
Contact: jupp@ebi.ac.uk
1 INTRODUCTION
The European Bioinformatics Institute (EBI) is the largest bioinformatics resource provider in Europe. Our databases are accessible via dedicated interfaces, web services, data download and (in a few cases) direct database access. Modern research in the life sciences necessitates an understanding of data at many different levels: multi-omics, from cells to biological systems, across many different species and studying many different experimental conditions. The biology underpinning these research questions is intrinsically connected, yet data are often collected and stored in technology or domain-specific repositories.
Efforts in the Semantic Web community are already beginning to invest in technology that enables data to be readily integrated (Belleau et al., 2008; Katayama et al., 2010; Marshall et al., 2008). One method used among the Semantic Web community is using the W3C’s resource description framework (RDF) model to represent data. RDF provides a common mechanism for describing data and querying data using SPARQL.
To better serve complex research questions across resources, and to meet an increased demand on the EBI to produce RDF, we have developed an RDF platform. The aim of such a platform is to offer users the ability to ask questions using multiple connected resources that share common identifiers and have a common format (RDF) and query interface (SPARQL). This platform complements other existing data access modes such as our Web site and RESTful web services, but additionally contains explicit links between the different data resources. This enables a single query to be asked across multiple distributed datasets and across a range of biological domains. This approach has been applied for the following EBI resources: Gene Expression Atlas (Kapushesky et al., 2012), ChEMBL (Gaulton et al., 2011), BioModels (Li et al., 2010), Reactome (Matthews et al., 2008), BioSamples (Gostev et al., 2012) and also includes a collaboration with the UniProt Consortium to deliver UniProt RDF (Redaschi and UniProt Consortium, 2009).
2 METHODS
The RDF platform presents a coordinated effort to bring together RDF resources from multiple services and databases at the EBI. The development of the platform began by collecting requirements from both a scientific and a technical perspective. The scientific requirements were gathered as a series of use cases and competency questions collected from research scientists and users of EBI services. In particular, we were looking for questions that required data to be integrated from multiple resources and that are not trivial to answer with our existing infrastructure due to the disparate nature of the data. These questions were used to identify points of integration between resources. The scientific use cases informed the technical requirements on what infrastructure, in terms of both software and hardware, would be needed to deliver a stable and scalable platform. Given RDF technology is still maturing, there are open questions on how to deliver such a platform on this scale; our existing infrastructure is delivered after evaluation of various technologies that will be the subject of another paper.
Data from UniProt, ChEMBL, Reactome and BioModels represents curated knowledge from protein sequence and function, bio-active molecules and their targets, to biochemical pathways and computational models of molecular interactions. The Gene Expression Atlas database provides differential gene expression data from a variety of samples that are highly annotated and curated using the Experimental Factor Ontology (EFO) (Malone et al., 2010). Generating linked RDF for these resources provides a new entry point for exploring the data, such as putting gene expression in the context of protein function, pathways and drug targets. An outline of how resources are connected is shown in Figure 1.
Fig. 1.
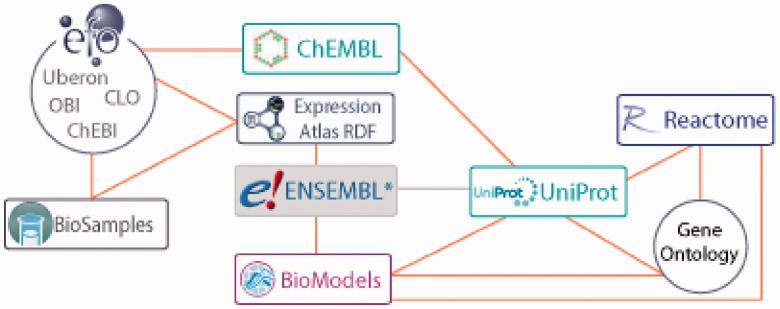
Connections between services (boxes) and ontologies (circles). The graph illustrates how the data are linked within the RDF platform, enabling queries to span all data. Asterisk: ENSEMBL to UniProt (gray line) mappings are included via expression atlas
The graph-based nature of the RDF data model provides a natural fit for explicitly publishing how data are connected. In RDF, resources are identified using uniform resource identifiers (URIs), which provide a web-based global identification system. Guidelines for minting new URIs for EBI resources were established using the new rdf.ebi.ac.uk domain (details can be found at http://www.ebi.ac.uk/rdf/documentation/uris-ebi-data). Canonical URIs are used when existing databases, such as UniProt, already provide stable URIs. In cases where no canonical URIs are provided by external resources, the Identifiers.org registry of scientific identifiers (Juty et al., 2012) was used to provide a referencing URI. As part of the URI strategy, every effort has been made to ensure all EBI RDF datasets only use URIs that can be dereferenced using http, supporting content negotiation for human-orientated HTML views, alongside machine processable versions in various RDF syntaxes.
Using common URI schemes assists data integration with RDF. In addition, ontologies provide a mechanism to semantically describe the data, and the OWL ontology language can be serialized in RDF. The EBI makes extensive use of ontologies to annotate data, however, the richness of these annotations is rarely available in native RDF for exploitation by external applications. The EBI RDF platform adopts a range of common vocabularies and ontologies to annotate data. The ontologies used span common biomedical terminologies such as the Gene Ontology, Chemical Entities of Biological Interest, UBERON, Cell Type Ontology, Biological Pathways Exchange, EFO and more. Additionally, we adopted metadata standards for describing datasets and provenance such as Dublin Core, Data Catalog Vocabulary and Vocabulary of Interlinked Datasets.
3 RESULTS
Complete dumps of the RDF data are available via FTP downloads. These are published in line with existing production and release cycles, ensuring the most up-to-date data are readily available. We are also using triple store technology to index the RDF files and make them available for querying and exploration via SPARQL endpoints and our linked data browser. The underlying infrastructure at the EBI is built on open source triple store technology provided by OpenLink, (http://www.openlinksw.com/), whereas the UniProt data are served by the SIB’s Vital-IT HPC platform using technology from OntoText (http://www.ontotext.com/). We developed LODEStar (http://www.ebi.ac.uk/fgpt/sw/lodestar/) as a generic SPARQL endpoint and linked data browser to provide a consistent interface and some enhanced functionality for querying and browsing EBI-based datasets.
In addition to providing access to the underlying data, an equally important component of the platform is the Web site at http://www.ebi.ac.uk/rdf that provides an entry point to discover all RDF resources being served by the EBI. This site includes documentation on how to find the datasets and provides examples of how to query the data using the SPARQL endpoints (http://www.ebi.ac.uk/rdf/example-sparql-queries). We also provide examples showing developers how they can use the SPARQL API programmatically from common programming environments like Perl, Java and R.
4 CONCLUSION
The EBI RDF platform allows explicit links to be made between datasets using shared semantics from standard ontologies and vocabularies, facilitating a greater degree of data integration. SPARQL provides a standard query language for querying RDF data. Data that have been annotated using ontologies, such as EFO and the Gene Ontology, enable data integration with other community datasets and provides the semantics to perform rich queries. Publishing these datasets as RDF along with their ontologies provides both the syntactic and semantic integration of data long promised by semantic web technologies.
As the trend toward publishing life science data in RDF increases, we anticipate a rise in the number of applications consuming such data. This is evident in efforts such as the Open PHACTS platform (http://www.openphacts.org) and the AtlasRDF-R package (https://github.com/jamesmalone/AtlasRDF-R). Our aim is that the EBI RDF platform enables such applications to be built by releasing production quality services with semantically described RDF to enable pertinent biomedical use cases to be addressed.
ACKNOWLEDGEMENTS
The authors thank the EBI Industry program, Michel Dumontier and staff at OpenLink and OntoText.
Funding: EMBL and European Union grants Diachron (601043) and BioMedBridges (284209). OpenPHACTS (115191). National Institutes of Health grants [1U41HG006104-03] and NCBO [U54-HG004028] and the Swiss Federal Government through the Federal Office of Education and Science.
Conflict of Interest: none declared.
REFERENCES
- Belleau F, et al. Bio2RDF: towards a mashup to build bioinformatics knowledge systems. J. Biomed. Inform. 2008;41:706–716. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Li C, et al. BioModels Database: an enhanced, curated and annotated resource for published quantitative kinetic models. BMC. Syst. Biol. 2010;4:92. doi: 10.1186/1752-0509-4-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulton A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2011;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gostev M, et al. The BioSample Database (BioSD) at the European Bioinformatics Institute. Nucleic Acids Res. 2012;40:D64–D70. doi: 10.1093/nar/gkr937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juty N, et al. Identifiers.org and MIRIAM Registry: community resources to provide persistent identification. Nucleic Acids Res. 2012;40:D580–D586. doi: 10.1093/nar/gkr1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapushesky M, et al. Gene Expression Atlas update–a value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res. 2012;40:D1077–D1081. doi: 10.1093/nar/gkr913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, et al. The 3rd DBCLS BioHackathon: improving life science data integration with Semantic Web technologies. J. Biomed. Semantics. 2010;4:6. doi: 10.1186/2041-1480-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone J, et al. Modeling sample variables with an experimental factor ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall MS, et al. A Prototype Knowledge Base for the Life Sciences. World Wide Web Consortium (W3C) Interest Group Note. 2008. http://www.w3.org/TR/hcls-kb/ (11 January 2014, date last accessed) [Google Scholar]
- Matthews L, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2008;37(Suppl. 1):D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redaschi N Consortium,UniProt. UniProt in RDF: Tackling data integration and distributed annotation with the semantic web. 2009 Available from Nature Precedings http://dx.doi.org/10.1038/npre.2009.3193.1. [Google Scholar]


