Abstract
Summary: Computational diagnosis of amino acid variants in the human exome is the first step in assessing the disruptive impacts of non-synonymous single nucleotide variants (nsSNVs) on human health and disease. The Molecular Evolutionary Genetics Analysis software with mutational diagnosis (MEGA-MD) is a suite of tools developed to forecast the deleteriousness of nsSNVs using multiple methods and to explore nsSNVs in the context of the variability permitted in the long-term evolution of the affected position. In its graphical interface for use on desktops, it enables interactive computational diagnosis and evolutionary exploration of nsSNVs. As a web service, MEGA-MD is suitable for diagnosing variants on an exome scale. The MEGA-MD suite intends to serve the needs for conducting low- and high-throughput analysis of nsSNVs in diverse applications.
Availability: www.megasoftware.net/mega-md and www.mypeg.info
Contact: s.kumar@asu.edu
Scientists routinely use computational methods to evaluate the functional disruptiveness of non-synonymous single nucleotide variants (nsSNVs) because of the lack of high-throughput experimental technology to profile the ever-expanding catalog of variants of unknown effect (Kumar et al., 2011). A large number of computational tools and web resources are available that use a range of methods to diagnose the deleteriousness of nsSNVs (Mah et al., 2011). However, there is a paucity of software tools that facilitate both the functional diagnosis and the exploration of the context of its long-term (inter-specific) evolutionary history of the mutant positions. This is despite the fact that information generated from multispecies alignments form the most powerful measurement of predictive models and is one of the major factors that determine the prediction accuracy (Hicks et al., 2011; Kumar et al., 2012).
We have developed the Molecular Evolutionary Genetics Analysis software with Mutational Diagnosis (MEGA-MD) suite of resources to address this need. MEGA-MD enables researchers to carry out diagnosis of thousands of nsSNVs efficiently and to explore the evolutionary trajectories of mutant positions in a user-friendly interface. In its graphical user interface (GUI), MEGA-MD is a client-server application whose GUI and evolutionary analysis functions are developed by reusing the source code of the MEGA software (Tamura et al., 2013). MEGA-MD accesses a relational database containing mutational diagnoses resident on our servers that contains precomputed diagnoses and associated information for all possible mutations at all amino acid positions in the human exome. In the first version, we have included three primary methods (PolyPhen-2, SIFT and EvoD) (Adzhubei et al., 2010; Kumar et al., 2012; Ng and Henikoff, 2003). The first two are the most popular methods, and the third significantly improves the performance for nsSNVs found at ultra-conserved and at fast-evolving positions (Kumar et al., 2012). The PolyPhen-2 and SIFT diagnoses were obtained from dbNSFP (Liu et al., 2013). We have also included results from a multi-method consensus diagnosis because they have been shown to be more reliable. In this case, we use the evolutionarily balanced versions of PolyPhen-2 and SIFT diagnosis.
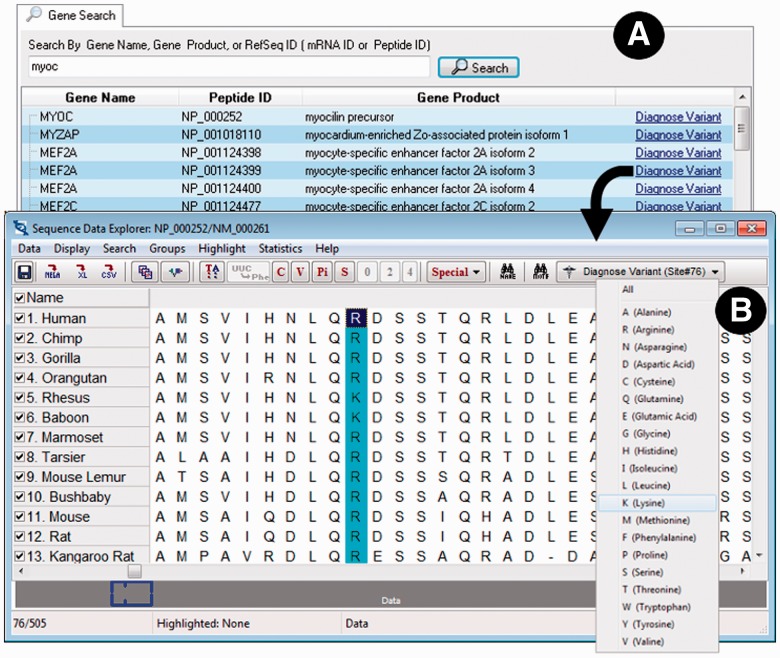
At the start, MEGA-MD asks the user if they would like to interactively specify a protein whose mutations are of interest or load a text file (Fig. 2D) containing a list of variants. If choosing to use the interactive system, the user may enter a gene name, a RefSeq messenger RNA ID, or a protein ID into the search box (Fig. 1A) on the Gene Search tab of the Mutation Explorer window, which results in a table of possibilities to select from. For the selected protein, MEGA-MD automatically retrieves a 46-species protein sequence alignment that comes from the UCSC resource (Fujita et al., 2011), which has been cached in the MD-DB for quick access. This alignment is displayed in a grid (Sequence Data Explorer, Fig. 1B), which also contains the Diagnose Variant command on the top toolbar. For the selected position (e.g. position 76), the user has the option to request diagnosis for a specific variant or all possible variants.
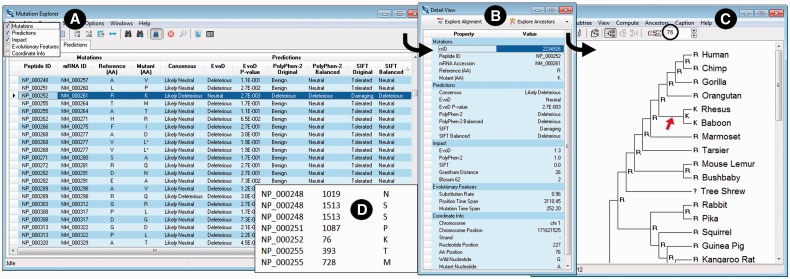
Fig. 2.
Example screenshots displaying results. (A) Mutation Explorer with the Predictions tab selected and diagnoses for many variants displayed; (B) Detail View showing all prediction data for the variant, which is selected on the Prediction Data tab of the Mutation Explorer; (C) Tree Explorer showing the ancestral states in the 46-species reference tree at the amino acid position (position 76 in the example shown) of the variant being investigated; and (D) an example of variants specified in a text file, which can be analyzed en mass in MEGA-MD for high-throughput analysis
Fig. 1.
Elements of the MEGA-MD user interface for interactively specifying variants: (A) the Gene Search tab of the Mutation Explorer displaying a list of genes or proteins to select from. (B) Sequence Data Explorer with an amino acid position selected (highlighted) and drop-down menu for specifying mutant amino acid shown. The Sequence Data Explorer window contains several utilities such as tools for computing compositional characteristics and exporting the alignment to several widely used formats, including FastA, Nexus and MEGA
After the required information is entered into MEGA-MD using one of the two methods described earlier in the text, the system queries the MEGA-MDW (Web version of MEGA-MD) server to diagnose the variants of interest. The results are displayed in a table view (Fig. 2A) on the Predictions tab in the Mutation Explorer’s five column categories (mutations, predictions, impact scores, features and coordinates). This table has capability for searching, sorting, exporting and customizing of columns (e.g. resizing and hiding). For the currently highlighted row in the Mutation Explorer, a Detail View is available, which not only presents an easy-to-read view of all available information for the currently selected variant but also provides buttons to ‘Explore Alignment’ and ‘Explore Ancestors’ (Fig. 2B). Clicking on Explore Alignment produces a display similar to Figure 1B, where the user can view the 46-species alignment associated with the currently selected variant along with the option to explore more predictions (all the predictions accumulate in the Mutation Explorer).
Clicking the Explore Ancestors button provides the user with the option to infer ancestral states for the position where the current amino acid mutation is found. Users can use the maximum likelihood or maximum parsimony approaches and select various analysis options; MEGA-MD automatically uses the 46-species reference phylogeny along with the amino acid alignment. When the ancestral states inference computation is complete, the 46-species tree is displayed in the Tree Explorer window (Fig. 2C). In the example shown, the mutation of interest in humans is an R→K change, which is also found on the ancestral lineage that led to Rhesus and Baboon (marked by red arrow). For this reason, the EvoD diagnosis deems it to be not disruptive (i.e. neutral). However, PolyPhen-2 and SIFT predict it to be deleterious. The three-method consensus result is ‘Likely-deleterious’ as two of three methods produce a deleterious result (Liu and Kumar, 2013).
We have also updated the most recent version of the MEGA software to include the mutational diagnosis functionalities (Tamura et al., 2013), where it is accessed through the ‘Diagnose’ menu. Also, MEGA-MD can be used as a web application through the URL http://www.mypeg.info, which can process tens of thousands of variants quickly and return a comma-delimited result file containing all the information shown in the Mutation Explorer of the GUI version.
In the future, we plan to add results from additional methods of nsSNV diagnosis and more expansive multispecies sequence alignments. In the meantime, we hope that the web and user-interface applications described here will serve the needs of many researchers in further investigating large-scale and individual variants.
ACKNOWLEDGEMENTS
The authors thank Nevin Gerek, Abediyi Banjoko, Ravinder Kanda and Kelly Boccia for valuable advice and help during the development of this software and/or feedback on initial versions of this manuscript.
Funding: US National Institutes of Health (LM010730-03 and HG002096-12 to S.K.).
Conflict of interest: none declared.
REFERENCES
- Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7: 248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39: D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks S, et al. Prediction of missense mutation functionality depends on both the algorithm and sequence alignment employed. Hum. Mutat. 2011;32: 661–668. doi: 10.1002/humu.21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, et al. Phylomedicine: an evolutionary telescope to explore and diagnose the universe of disease mutations. Trends Genet. 2011;27: 377–386. doi: 10.1016/j.tig.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, et al. Evolutionary diagnosis method for variants in personal exomes. Nat. Methods. 2012;9: 855–856. doi: 10.1038/nmeth.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Kumar S. Evolutionary balancing is critical for correctly predicting amino acid variants with functional impact. Mol. Biol. Evol. 2013;30: 1252–1257. doi: 10.1093/molbev/mst037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, et al. dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat. 2013;34: E2393–E2402. doi: 10.1002/humu.22376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah JT, et al. In silico SNP analysis and bioinformatics tools: a review of the state of the art to aid drug discovery. Drug Discov. Today. 2011;16: 800–809. doi: 10.1016/j.drudis.2011.07.005. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31: 3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013;30: 2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]