Abstract
Metastasis is powered by disseminated cancer cells that recreate a full-fledged tumor in unwelcoming tissues, away from the primary site. How cancer cells moving from a tumor into the circulation manage to infiltrate distant organs and initiate metastatic growth is of interest to cancer biologists and clinical oncologists alike. Recent findings have started to define the sources, phenotypic properties, hosting niches, and signaling pathways that support the survival, self-renewal, dormancy and reactivation of cancer cells that initiate metastasis–metastatic stem cells. By dissecting the biology of this process, vulnerabilities are being exposed that could be exploited to prevent metastasis.
Introduction
Metastasis results from disseminated cancer cells that reinitiate a full-fledged tumor. Cancer cells may leave tumors relentlessly, only to perish en masse while attempting to settle in distant tissues. Those that survive the ordeal may remain alive for decades but still fail to form clinically manifest lesions (Braun et al., 2005; Janni et al., 2011). Yet, when metastasis occurs it creates complications that account for the vast majority of deaths from cancer. Cancer cells that succeed at this task possess not only the tumor initiating attributes of tumor-initiating cells, or cancer stem cells (CSCs), but also the ability to exert this capacity under harshly adverse conditions. Metastasis therefore is driven by CSCs at their best, or at their worst, depending on your perspective.
Insights into the identity, behavior and needs of cancer cells that have the capacity to initiate metastasis are coming from three fronts. Firstly, the existence of CSCs originally described in tumors of hematopoietic origin (Bonnet and Dick, 1997; Lapidot et al., 1994) has now been established in many solid tumors including those arising in the brain (Chen et al., 2012; Singh et al., 2004), colon (Dalerba et al., 2007; Merlos-Suarez et al., 2011; O'Brien et al., 2007; Ricci-Vitiani et al., 2007; Schepers et al., 2012), breast (Al-Hajj et al., 2003; Mani et al., 2008; Pece et al., 2010), skin (Driessens et al., 2012; Malanchi et al., 2008), prostate (Wang et al., 2009) and pancreas (Hermann et al., 2007; Li et al., 2007a). These findings have pushed the debate on the nature of cancer stem cells from conjecture to more concretion, though many questions remain. Secondly, the identification of clinically relevant metastasis genes and functions has improved the biological conceptualization of metastasis and its distinct phases (Nguyen et al., 2009a; Valastyan and Weinberg, 2011). Thirdly, high-resolution sequencing of tumor samples and other approaches provide evidence that metastasis relies less on driver mutations than on epigenetic amplification of cell survival and self-renewal mechanisms (Vanharanta and Massagué, 2013).
Building on this progress, here we review the current understanding of sources, lethal challenges, hosting niches, and vital pathways that enable the persistence and progression of metastatic stem cells. At the outset we stress that this is an emerging field and that, because of this, many aspects are derived from models that recapitulate the process imperfectly, or based on inference from clinical data. Nonetheless, the extant evidence provides a useful framework for the analysis of the problem, with the understanding that much work still needs to be done to challenge or support these ideas.
Deadly Seeds Left Behind
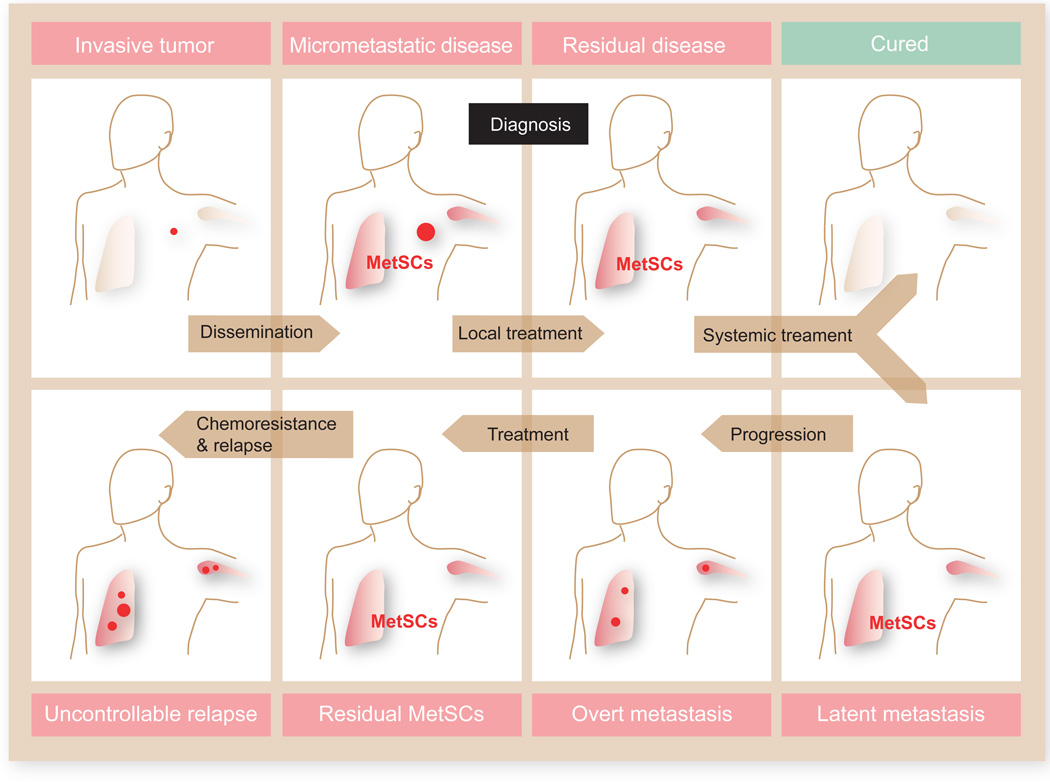
When a surgeon removes a primary tumor mass with perfect marginal clearance the disease does not necessarily go away. At diagnosis a tumor may have already shed thousands of cancer cells, starting when the still incipient lesion broke through basal lamina and malignant cells reached the bloodstream (Figure 1). These disseminated tumor cells (DTCs) are found in bone marrow of breast cancer patients who have no signs of overt metastasis (Braun et al., 2005), have small primary tumors (Klein, 2009; Pantel et al., 2009), or have been treated and are disease-free by every other criteria (Pantel et al., 2009; Pantel and Brakenhoff, 2004).
Figure 1. Deadly seeds left behind in a typical course of metastatic cancer.
At diagnosis a primary tumor (a carcinoma of the lung or breast in this example) may have already seeded distant organs with cancer cells, including cells with tumor-initiating capacity that are defined here as metastatic stem cells (MetSCs). After diagnosis, the primary tumor may be removed by surgery and irradiation, and disseminated cancer cells may be eliminated by systemic chemotherapy, leading to a cure. Alternatively, residual MetSCs may remain in a latent state, eventually giving rise to overt metastasis. New rounds of therapy may then induce regression of the metastatic lesions, but chemoresistant MetSCs selected during each round of treatment may eventually give rise to uncontrollable metastasis. This process is responsible for 90% of deaths from cancer.
Studies on DTCs have been largely based on the analysis of bone marrow samples. However, the presence of bone marrow DTCs is predictive of metastasis not only in bone but also in liver, lung and brain (Braun et al., 2005). This also applies to diseases like colorectal cancer that do not normally metastasize to bone (Pantel and Brakenhoff, 2004). Though little is known about the capacity of other organs to host DTCs, case reports on liver, kidney, or heart transplants that conferred metastatic melanoma to recipients argue that these organs can harbor latent DTCs (Stephens et al., 2000; Strauss and Thomas, 2010).
Systemic anticancer therapy can eliminate DTCs, but often not fully (Becker et al., 2006; Fehm et al., 2006) (Figure 1). The presence of residual DTCs in bone marrow after adjuvant therapy is predictive of subsequent recurrence and poor survival (Janni et al., 2011). Several factors provide DTCs with a chance to persist. In the bone marrow at least, solitary DTCs are largely in a non-proliferative state that is not susceptible to antimitotic therapy (Muller et al., 2005; Naumov et al., 2003). Moreover, DTC populations may differ from the bulk cancer cell population in terms of drug sensitivity (Holohan et al., 2013). DTCs may additionally benefit from stromal signals that protect them from genotoxic stress (Acharyya et al., 2012). Sometimes DTCs thrive while the primary tumor withers, giving rise to metastatic cancers of unknown primary site (Massard et al., 2011).
MetSCs: Definition and Origins
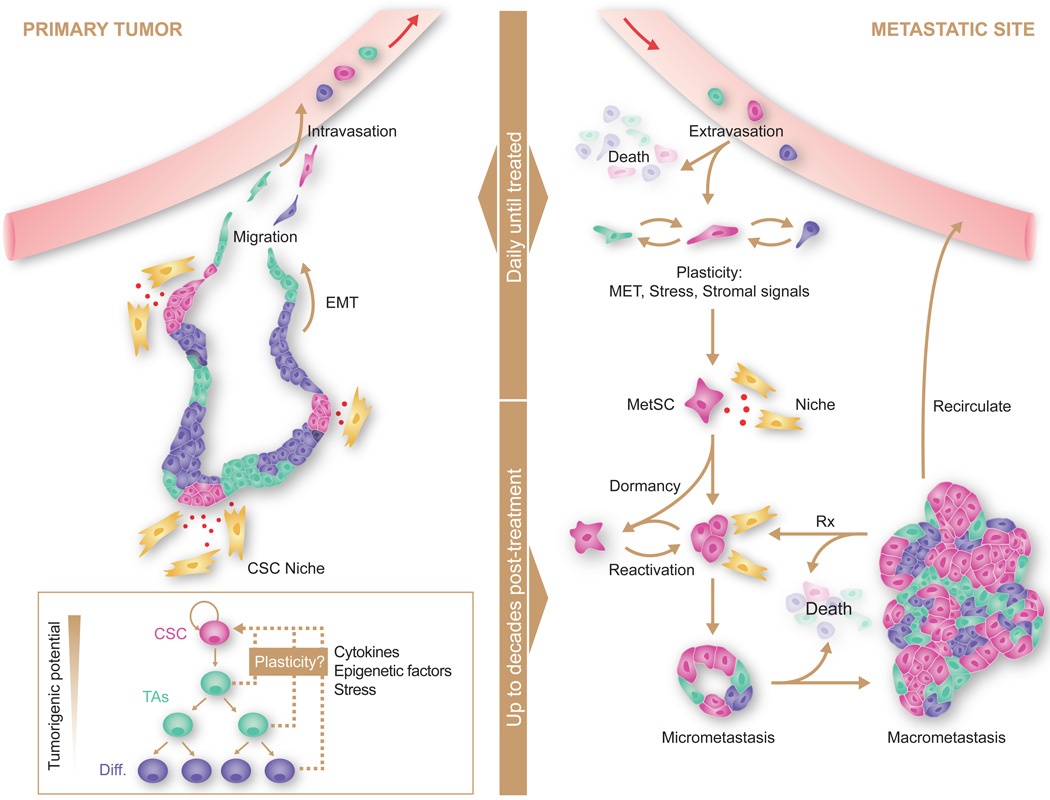
Metastasis involves invasion and intravasation of cancer cells from the primary tumor, dissemination through the circulation, extravasation in different organs, survival on arrival, settlement into latency, reactivation, and overt colonization with generation of a full fledged new tumor (Figure 2). Migrant cancer cells that manage to settle in a distant tissue become DTCs that can give rise to metastasis. This does not make every DTC a potential metastasis-initiating cell. Some DTCs may have progressed too far down a differentiation or senescence path and do not stand a chance of reinitiating tumor growth.
Figure 2. Sources, dissemination, dormancy, and outgrowth of MetSCs.
Several types of cancer display a hierarchical organization with CSCs being the only cell type with long-term self-renewal potential. The progeny of CSCs –the transit amplifying progenitors (TAs) and their differentiated derivatives– are short lived and have a lower tumorigenic potential. Conversion of non-CSCs into CSCs occurs in certain types of cancer and can be triggered by cytokines and stress conditions. CSCs interact with microenvironmental niches that sustain the tumor-perpetuating potential of the cells. Both CSCs and non-CSCs can display migratory behavior at the invasive front of primary tumors, frequently associated with an EMT. Intravasation, circulation, and extravasation of cancer cells in these various states can occur continuously until the primary tumor is removed. Survival of disseminated cancer cells at distant sites is a limiting step, as the vast majority of infiltrating cancer cells die. The survivors that are endowed with tumor-initiating capacity constitute MetSCs. Cells that had previously undergone an EMT must reacquire epithelial traits and co-opt a supportive stromal niche in order to thrive in the new environment. Additionally, disseminated non-CSCs may convert into MetSCs through still poorly understood processes of phenotypic plasticity. MetSCs may generate progeny and give rise to overt metastasis right after infiltrating the host tissue. More frequently, however, disseminated cancer cells enter a dormant state that can last for decades and is largely resistant to current therapies. Upon exit from quiescence, MetSCs may regenerate their lineage in the host organ and release metastatic progeny into the circulation to start secondary lesions in the same or other organs. Treatment of overt metastasis seldom results in eradication of the disease, as residual MetSCs frequently regenerate the tumor after each drug treatment cycle.
With the term Metastatic Stem Cell (MetSC) we refer here to any DTC that is capable of reinitiating macroscopic tumor growth in a distant tissue. This definition is independent of the origin or phenotypic characteristics (e.g. marker genes) of these cells, and does not imply the presence of other functional features (e.g. chemoresistance). MetSCs may already exist in the primary tumor with the necessary traits to overcome the bottlenecks of the metastatic process, or, alternatively, may derive from DTCs that reacquire the competence to initiate tumor growth after a period of indolence (Figure 1). Investigation of these two possible sources by functional analysis of MetSCs in clinical DTC populations remains elusive. So far, studies have been limited to associations between numbers of DTCs and risk of metastasis (Pantel et al., 2009; Pantel et al., 2008). Accurate analysis of the lineage and functional relationships of MetSCs to CSCs in primary tumors and to DTCs in distant tissues awaits the development of better models and technology to track and interrogate these cell populations. However, evidence for the two alternative sources of MetSCs can be found at least in model systems.
MetSCs by Birthright
It is increasingly clear that the cell heterogeneity characteristic of many cancer types results from a hierarchical organization that resembles that of the tissue of origin (Figure 2). CSCs comprise the apex of this hierarchy and appear to be the phenotypic and functional equivalents of normal stem cells harboring oncogenic mutations. CSCs in primary tumors self-renew their own population and can generate short-lived progeny (i.e. transit amplifying cells and more differentiated descendants). Recent lineage tracing experiments in mouse models provide genetic evidence that primary tumors of the brain, colon, and skin comply with this organization (Chen et al., 2012; Driessens et al., 2012; Nakanishi et al., 2013; Schepers et al., 2012). It remains to be determined whether metastases arising from these tumors retain a similar hierarchy. Nevertheless, because the long-term growth capabilities of these and other cancers relies on CSCs, the idea that MetSCs in these cases may be primary CSCs that resume their regenerative potential at metastatic sites is of interest (Figure 2).
Circumstantial evidence in favor of this idea comes from the clinic. High expression of adult stem cell markers in primary tumors is associated with poor prognosis and metastatic relapse (Dalerba et al., 2011; Merlos-Suarez et al., 2011; Pece et al., 2010). Cell populations capable of generating metastasis when transplanted into mice can be isolated from primary tumor samples using stem cell marker genes (Hermann et al., 2007; Malanchi et al., 2012; Pang et al., 2010). Cancer cells expressing stem cell markers have been detected in the blood of breast cancer patients, and when inoculated into immunodeficient mice these cells generated bone, liver and lung metastases (Baccelli et al., 2013).
Perhaps the most compelling evidence for a lineage relationship between MetSCs, primary tumor CSCs and normal stem cells comes from colorectal cancer. Studies with genetic mouse models indicate that upon acquiring activating mutations in the WNT pathway intestinal stem cells generate adenomas (Barker et al., 2009). Lineage-tracing analysis shows that a population of stem cells that resembles those present in normal intestinal mucosa sustains the long-term growth of these benign lesions while generating short-lived progeny that undergo differentiation (Kozar et al., 2013; Nakanishi et al., 2013; Schepers et al., 2012). This hierarchical organization appears to be retained in late stage colorectal cancers (Dalerba et al., 2011; Merlos-Suarez et al., 2011; Vermeulen et al., 2008) and even liver metastases contain stem- and differentiated-like tumor cells (Merlos-Suarez et al., 2011). Clonal analysis of human colorectal cancer samples upon lentiviral marking of tumor cell populations demonstrates that metastases arise from primary tumor cells that display long-term self-renewal capacity and are quiescent and resistant to chemotherapy (Dieter et al., 2011; Kreso et al., 2013).
Certain cancers such as melanoma do not appear to rely on a hierarchical organization (Meacham and Morrison, 2013; Quintana et al., 2010). However, these tumors still contain MetSCs as defined here, as cells that hijack stem cell traits that facilitate regeneration of the disease at a distant site.
MetSCs by Reeducation
An alternative route to MetSC status is based on regaining tumor-initiating capacity through phenotypic plasticity (Figure 2). This possibility is suggested by several observations. In cell line xenograft models, breast cancer stem-like cells can arise from non-stem populations as a result of stochastic transitions between both states (Gupta et al., 2011). Certain cytokines can stimulate the expression of CSC features. For example, hepatocyte growth factor from stromal fibroblasts potentiates WNT/β-catenin signaling to enhance the stem cell potential of colorectal cancer cells (Vermeulen et al., 2010). Transforming growth factor β (TGF-β) enhances the CSC potential in glioblastoma (Anido et al., 2010) and collaborates with WNT to promote CSC potential in breast cancer cells (Scheel et al., 2011). Given the harsh conditions that metastatic cells face, the recent finding that environmental stresses such as acidic pH can reprogram pluripotency (Obokata et al., 2014a; Obokata et al., 2014b) is potentially relevant to the generation of MetSCs from non-stem cells.
Gains in stem-like features are sometimes accompanied by an epithelial to mesenchymal transition (EMT) (Meacham and Morrison, 2013; Nieto, 2013; Thiery et al., 2009; Valastyan and Weinberg, 2011) (Figure 2). EMT is a vital process during embryogenesis, particularly in steps that involve tissue invasion such as gastrulation or neural crest migrations (Nieto, 2013). At its core, the EMT is driven by a network of transcription factors including Snail1~3, ZEB1 and 2 (zinc-finger E-box binding factor), Twist and others (Nieto, 2013). Epigenetic changes underlie the EMT (Tam and Weinberg, 2013). In cancers arising from epithelial tissues, the carcinoma cells are embedded within glandular structures that restrict their migration. In skin and breast carcinoma models, tumor-derived TGF-β stimulates EMT (Oft et al., 1996; Xu et al., 2009). By undergoing an EMT carcinoma cells at the tumor invasive front lose cell-to-cell adhesion and apical-basal polarity and gain migratory behavior to surpass these barriers. Remarkably, the enforced expression of EMT transcription factors in breast and pancreatic cancer cells additionally confers stem-like features upon them (Mani et al., 2008; Scheel et al., 2011; Wellner et al., 2009). In addition, EMT markers and stem cell markers are co-expressed in circulating tumor cells from patients with metastasis (Aktas et al., 2009; Baccelli et al., 2013; Yu et al., 2013; Yu et al., 2012).
To be clear though, CSCs exist both in epithelial and mesenchymal states (Liu et al., 2014). Although EMT favors the emigration of carcinoma cells from the primary tumor, it inhibits cell proliferation and interferes with the initiation of metastatic outgrowth (Ocana et al., 2012; Stankic et al., 2013; Tsai et al., 2012) (Celia-Terrassa et al., 2012). As we discuss later, MetSCs that have undergone an EMT must reacquire an epithelial phenotype in order to resume growth at the metastatic site.
Sources of Metastatic Traits
Regardless of their origin, cancer stem cells require additional traits in order to successfully express metastasis-initiating potential. For example, xenotrasplantation studies of stage IIIB/C human melanomas showed that most samples contained cells that could initiate subcutaneous lesions in mice. However, distant metastasis occurred only with transplanted tumors from patients with metastatic melanoma. These differences correlated with the presence of circulating melanoma cells in the blood of the recipient mice, consistent with the idea that CSCs require specific traits, such as an ability to enter into and survive in the circulation, in order to metastasize (Quintana et al., 2012).
The question of whether metastatic traits are directly conferred by mutations in specific “metastasis genes” has long been debated. Mutations affecting key cell proliferation, survival, and self-renewal pathways clearly act as “drivers” of tumor initiation (Vogelstein et al., 2013). Further tumor progression is accompanied with widespread genetic heterogeneity in the cancer cell population (Burrell et al., 2013). A reasonable expectation would be that metastasis-driving mutations are contained within this heterogeneity (Fidler, 2003). However, large-scale genome sequencing of human tumors found little evidence for recurrent metastasis-restricted mutations (Bozic et al., 2010; Yachida et al., 2010), other than mutations in classic initiator oncogenes that are further enriched in metastatic lesions (Campbell et al., 2010; Ding et al., 2010). Metastasis therefore cannot easily be ascribed to recurrent mutations in “metastasis-driver” genes.
Mutations in epigenetic control pathways are another matter. Mutations in epigenetic regulators or in metabolic pathways that support their function (such as that controlled by isocitrate dehydrogenases IDH1 and 2) can lock transcriptomic outputs in altered states that favor the emergence and selection of tumor progression traits. As this literature is not the primary focus of this review, we will refer reader to recent reviews for further reading on these topics (Shen and Laird, 2013; Ward and Thompson, 2012). In the context of metastatic progression the pleiotropic impact of these mutations on transcriptional output provides cancer cell populations with opportunities to evolve under the selective pressure of invaded microenvironments. Such mutations likely play a prominent role in favoring the emergence of metastatic traits (Vanharanta and Massagué, 2013). The mutant genes are not direct oncogenic drivers in the classical sense but they rather channel the effect through imbalances in cell growth, invasiveness and/or self-renewal pathways. For instance, in renal cell carcinomas driven by the VHL-HIF2α pathway, alterations in methylation of histone H3K27 and DNA expand the transcriptional output of this pathway to facilitate the expression of CXCR4 and CYTIP. These two HIF2α target genes are not required for primary tumor formation but are powerful enhancers of metastasis (Vanharanta et al., 2013). Suppressed expression of the differentiation factors Nkx2-1, GATA6 and HOPX, probably as a result of epigenetic silencing, enhances metastasis in non-small cell lung carcinoma (Cheung et al., 2013; Winslow et al., 2011). Alterations in mRNA processing, noncoding RNAs and the translational machinery can also alter the output of cancer cell pathways to favor the emergence of metastatic traits (Di Leva et al., 2013; Guo et al., 2012; Moore and Proudfoot, 2009; Pencheva and Tavazoie, 2013; Valastyan et al., 2011). Future genome-wide sequencing studies may yet reveal mutations that directly and specifically drive metastasis and not primary tumor formation. However, the evidence to date favors the conclusion that mutations giving rise to pleiotropic epigenetic alterations are a major source of selectable metastatic traits.
Metastatic Selection in Primary tumors
Metastasis-seeding traits become relevant as soon as cancer cells reach distant organs. The traits that give cancer cells an edge at surviving on arrival in distant organs must therefore be acquired before dissemination. That such traits are present in primary tumors is suggested by the repeated identification of mediators of dissemination and metastatic seeding among gene expression signatures that predict relapse in primary tumors (Bos et al., 2009; Calón et al., 2012; Cheung et al., 2013; Guo et al., 2012; Hu et al., 2009; Minn et al., 2005; Nguyen et al., 2009b; Tavazoie et al., 2008; Vanharanta et al., 2013; Zhang et al., 2013a). These traits are distinct from those that mediate overt metastatic colonization months or years later (Eisinger-Mathason et al., 2013; Minn et al., 2005; Nguyen et al., 2009a; Valastyan and Weinberg, 2011) Colonization traits, such as the ability of DTCs in the bone marrow to coopt osteoclasts for osteolytic metastasis (Guise, 2010; Kang et al., 2003; Waning et al., 2013) are not required for the initial survival of DTCs. Their acquisition can probably wait.
A likely site for selection of metastatic traits in primary tumors is at the invasive front, the intersection of an advancing tumor mass with the surrounding stroma. Cancer cells that have overcome their own cell-autonomous oncogenic stresses engage in invasive behavior that exposes them to a reactive stroma, hypoxia, and immune surveillance (Figure 3A). The invasive front is rich in tumor-associated macrophages (TAMs), myeloid progenitor cells, cancer-associated fibroblasts (CAFs), and newly generated blood vessels (Joyce and Pollard, 2009). Wnt, Notch, TNF-α, TGF-β and hedgehog cytokines derived from the stroma are present in invasive fronts. This milieu is thought to support the survival and fitness of CSCs (Takebe et al., 2011) and may select for traits that are advantageous to cancer cells not only locally but also in distant tissues.
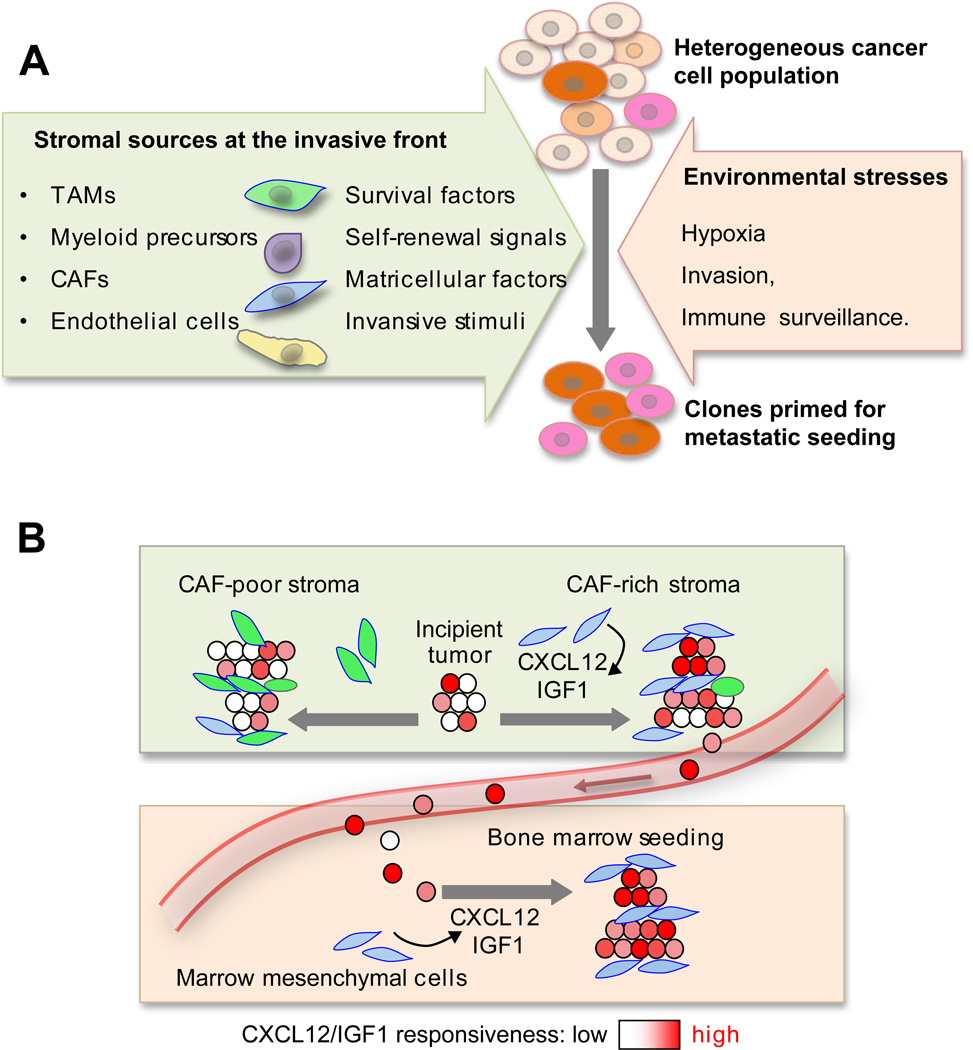
Figure 3. Selection of metastatic traits in primary tumors.
A. Cancer cells at the invasive front of primary tumors are exposed to the stresses of invading surrounding tissue, hypoxia, and immune surveillance. Various stromal cell types produce signals that cancer cells can use for survival, self-renewal, invasiveness and migration. Under selective pressure, these signals skew the heterogeneous cancer cell population towards a preponderance of clones that are primed for survival also under stress of infiltrating distant tissues. B. When the stroma of a primary tumor is rich in cell and signals that resemble those of a particular distant tissue, cancer cell clones selected in the primary tumor may be primed to thrive in that particular tissue. An example is provided by the ability of a CAF-rich stroma in breast tumors to select for cancer cell clones that are fittest to respond to the CAF-derived factors CXCL12 and IGF1, and thereby primed for survival in the CXCL12- and IGF1-rich environment of the bone marrow. [Image adapted from (Zhang et al., 2013b)].
In addition to exerting selection for general metastasis supporting traits, the primary tumor stroma can also select for organ-specific seeding traits. This specificity was recently shown in the case of bone metastatic breast cancer (Zhang et al., 2013b). A CAF-rich stroma in breast tumors produces CXCL12/SDF1 and IGF1, which select for Src hyperactive cancer clones that are superior at responding to these signals with activation of the PI3K-AKT survival pathway. Src-high clones are thereby primed for seeding the bone marrow where local sources of CXCL12 and IGF1 provide them with a higher chance of survival (Figure 3B). As a corollary to these findings, CAF content, CXCL12/IGF1 signaling, and high Src activity in breast tumors all predict an increased likelihood of bone relapse in breast cancer patients (Zhang et al., 2013b; Zhang et al., 2009).
Cancer cells may leave a primary tumor early and evolve separately from the tumor. It has been proposed that the parallel evolution of early disseminated cancer cells over a period of indolence affords these cells a superior adaptation to their metastatic microenvironment and a leading role in metastatic relapse (Klein, 2009). Cancer cell entry into the circulation and lodging in distant organs can certainly occur after minimal genetic changes (Podsypanina et al., 2008; Schardt et al., 2005). However, large-scale genome sequencing studies have shown more similarities than differences between primary tumors and their metastases, suggesting that most of the genetic changes required for metastasis accumulate in primary tumors (Yachida et al., 2010). Actively growing cancer cells in primary tumors may be more likely to undergo variation for the selection of metastatic traits than their precociously dispersed, indolent comrades.
Metastasis Patterns and Probabilities
The spectrum of organs affected by metastasis and the latency period between diagnosis and relapse depend on the type of cancer (Hess et al., 2006; Jones et al., 2008). For example, breast cancer, lung cancer, and skin melanoma typically relapse in multiple organs (bones, lungs, liver, brain), whereas prostate carcinoma relapses most aggressively in bone, ocular melanoma in liver, and sarcomas in lung. Metastasis typically has a long latency period in melanoma, sarcoma, prostate cancer, and luminal breast cancer, but not in lung cancer or basal breast cancer.
Metastasis is clearly a function of many variables, and in the process many mediators intervene that increase an always very small probability of a cancer cell to complete the process. The dissemination of cancer cells to distant tissues is influenced by circulation patterns, as is particularly manifest in intestinal cancers. Colorectal cancer cells enter the mesenteric circulation that feeds the liver capillary network and provides an ample opportunity to infiltrate the hepatic parenchyma. Colorectal cancer metastasis occurs mostly in the liver, with lung relapse a distant second. Beyond the passive role of circulation patterns, cancer cell dissemination is actively influenced by cancer cell-autonomous functions (e.g., invadopodia formation), paracrine factors (e.g. VEGF and EGF family members), proteases (e.g., metalloproteinases, cathepsins), recruitment of stromal components (e.g. tumor-associated macrophages), and interactions with blood platelets (Bos et al., 2009; Chabottaux et al., 2009; Chen et al., 2011a; Gay and Felding-Habermann, 2011; Gocheva et al., 2010; Greenberg et al., 2008; Gupta et al., 2007; Kim et al., 2009; Labelle et al., 2011; Li et al., 2010; Padua et al., 2008; Reymond et al., 2013; Rolny et al., 2011; Sonoshita et al., 2011; Stockmann et al., 2008; Wyckoff et al., 2004; Wyckoff et al., 2007; Zijlstra et al., 2008).
Though most of these mediators of cancer cell dissemination have yet to be specifically investigated in CSCs, their expression in primary tumors is associated with metastatic relapse in the clinic. The association suggests that these mediators are also relevant to the dissemination of MetSCs or to their establishment as viable seeds in distant host tissues.
The Metastasis Seeding Bottleneck
The organ distribution pattern of metastatic disease depends not only on the likelihood that MetSCs will reach in distant organs but also on the probability that these cells will survive there and eventually initiate metastatic outgrowth. On a per cell basis, this probability is invariably low. The cellular composition, vascularity, immune surveillance and inflammatory status of the infiltrated tissues differ sharply from those of the primary tumor where cancer clonal selection originally took place. Both experimental evidence and clinical observation suggest that circulating cancer cells suffer massive attrition upon infiltrating distant tissues (Chambers et al., 2002; Fidler, 2003; Nguyen et al., 2009a). Most cancer cells injected intravenously to flood the lungs of mice undergo apoptosis within two days (Wong et al., 2001). Most melanoma cells injected intra-portally to flood the liver fail to form micrometastases, and only 0.02% formed macrometastases (Cameron et al., 2000; Luzzi et al., 1998). In experimental systems, circulating cancer cells do worse in metastatic sites than on re-infiltrating the tumor of origin, a phenomenon called “tumor self-seeding” (Kim et al., 2009). Thus, no tissue ‘soil’ may be welcoming to metastatic seeds, though certain soils may be less hostile than others.
That only a tiny fraction of disseminated cancer cells initiate metastatic outgrowth cannot be explained by a scarcity of CSCs in the population. The frequency of tumor initiating cells in different types of cancer may have been underestimated due to biases imposed by the experimental models (Quintana et al., 2008). Quantitative lineage tracing clonal analysis is needed to assess the abundance of CSCs in different tumor types (Clevers, 2011; Meacham and Morrison, 2013) (Beck and Blanpain, 2013). However, in experimental models at least, infiltrating distant organs is clearly a traumatic experience for CSCs (Figure 2). Colorectal CSCs can readily reach the liver only to suffer massive apoptosis (Calón et al., 2012), and breast CSCs that infiltrate the lung parenchyma are eliminated within days (Malanchi et al., 2012).
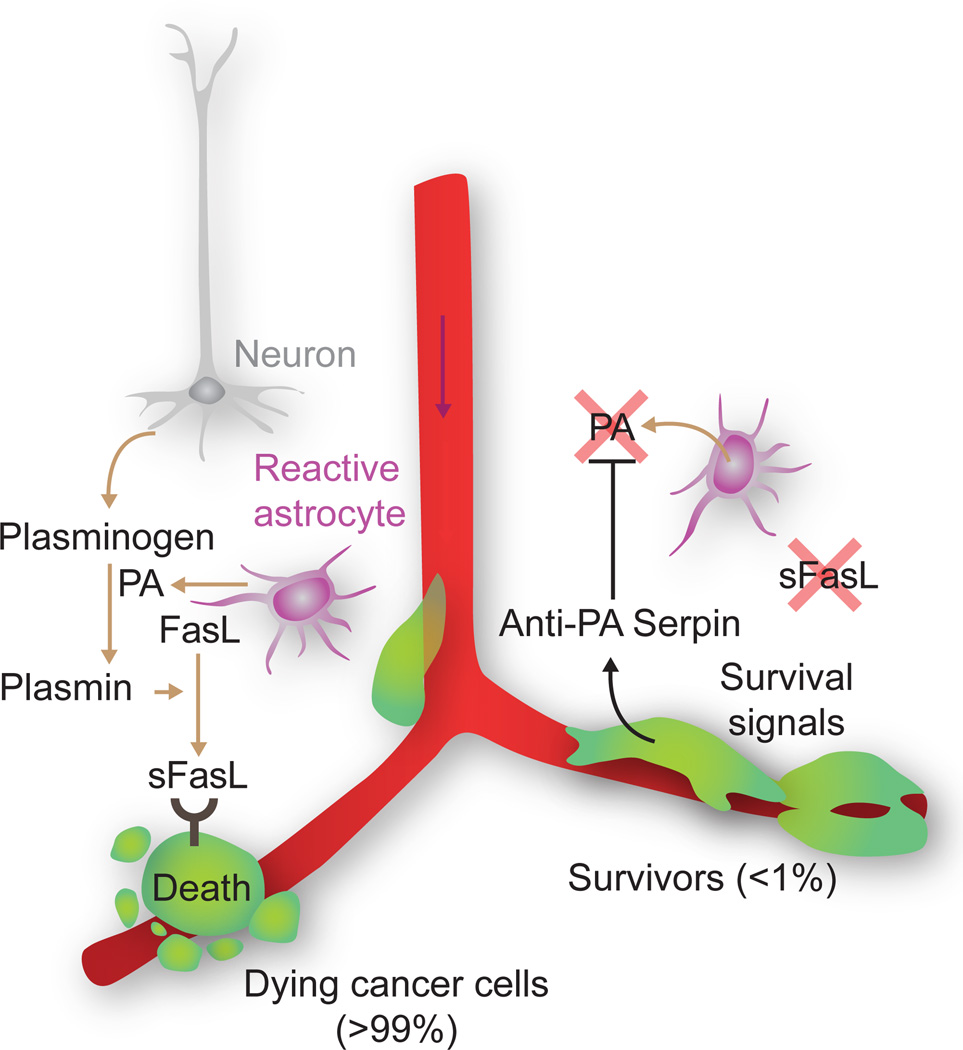
Remarkably little is known about what kills cancer cells upon infiltration of distant organs. A scarcity of survival signals in the host parenchyma, lack of a supportive stroma, and overexposure to innate immunity are potential causes (Chambers et al., 2002; Fidler, 2003; Nguyen et al., 2009a; Schreiber et al., 2011). Human bone metastasis lesions express the cytokine TRAIL, which is lethal to breast cancer cells (Zhang et al., 2009). Cancer cell death on arrival is a general phenomenon in all organs and is particularly acute in the brain (Heyn et al., 2006; Kienast et al., 2010; Perera et al., 2012; Steeg et al., 2011). Recent work in models of brain metastasis from breast and lung cancers has shown that the brain stroma takes a very active role in killing the infiltrating cancer cells (Valiente et al., 2014). In this context, cancer cells exiting from brain capillaries are confronted by reactive astrocytes that generate plasmin to mobilize the killer cytokine FasL against the infiltrators. The metastatic cells can fend off this attack by producing serpin inhibitors of plasmin generation (Figure 4).
Figure 4. A reactive stroma kills infiltrating cancer cells.
Astrocytes in the brain stroma react to infiltrating breast or lung cancer cells by expressing plasminogen activator (PA). PA generates plasmin that cleaves and mobilizes membrane-bound Fas ligand (FasL) from astrocytes. The cancer cells succumb to Fas-mediated apoptosis, but can avert this fate by expressing PA inhibitory serpins. Neuroserpin is normally expressed only in neurons, which use it for protection against astrocytes reacting to brain injury. [Image adapted from (Valiente et al., 2014)].
Metastatic Niches
The need to find supportive sites is thought to be important for disseminated cancer cells. Stem cells in adult tissues reside in specific sites or “niches”, the cellular and molecular components of which regulate the self-renewal potential of stem cells and their access to differentiation cues. The location and constitution of stem cell niches have been defined in various tissues, including the intestinal epithelium, hematopoietic bone marrow, epidermis, and brain (Clevers, 2013; Hsu and Fuchs, 2012; Moore and Lemischka, 2006; Morrison and Spradling, 2008). In primary tumors cancer cells may interact with these native stem cell niches, but such interactions will cease as cancer cells leave the tumor (Figure 2). Growing evidence indicates that the survival and fitness of metastasis-initiating DTCs depend on specific components of the host environment that play the part of a niche for these cells.
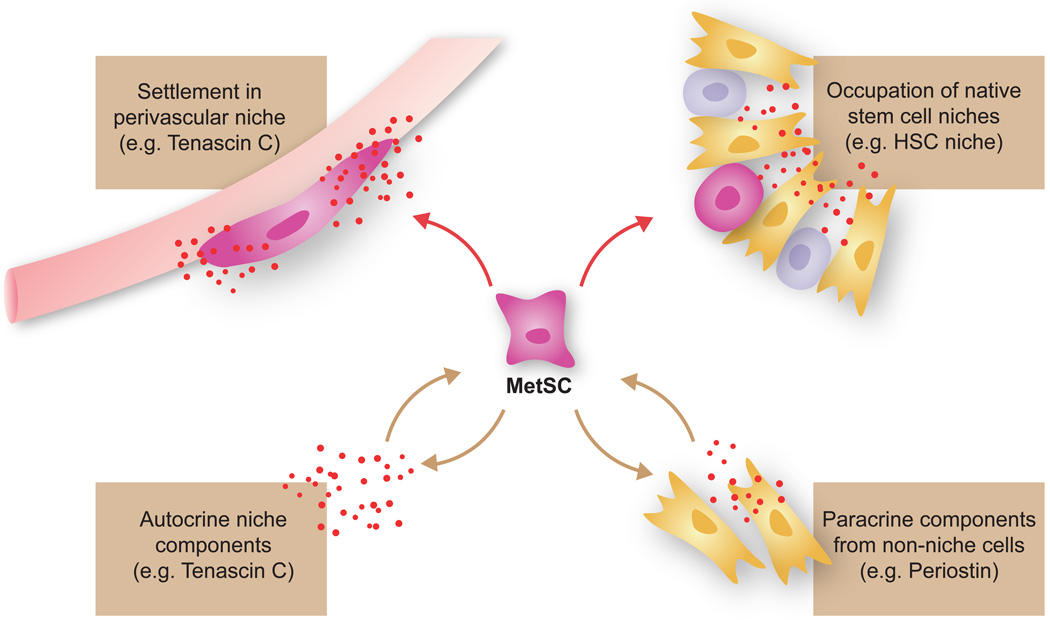
We use the term “metastatic niche” broadly here to designate the specific locations, stromal cell types, diffusible signals, and extracellular matrix proteins that support the survival and self-renewal of disseminated MetSCs. Three distinct sources of metastatic niche functionality can be envisioned (Figure 5): (i) native stem cell niches that metastatic cells may occupy in the host tissues; (ii) niche functions provided by stromal cells not belonging to stem cell niches; and, (iii) stem cell niche components that the cancer cells themselves may produce. Model systems have provided experimental evidence for these sources and a basis to consider their contributions in human disease.
Figure 5. Three possible sources of metastatic niche support.
Disseminated cancer cells can obtain stem cell niche support by occupying native stem cell niches including perivascular sites, by recruiting stromal cells that produce stem cell niche-like components, or by producing niche components themselves.
Niche locations
Cancer cells that infiltrate distant organs may lodge in random locations of the invaded parenchyma. However, recent work provides evidence that DTCs can occupy native stem cell niches of the host tissue. Prostate cancer cells showed affinity for the hematopoietic stem cell niche within the bone marrow, where they may benefit from cues that enhance stem cell properties and deter differentiation (Shiozawa et al., 2011).
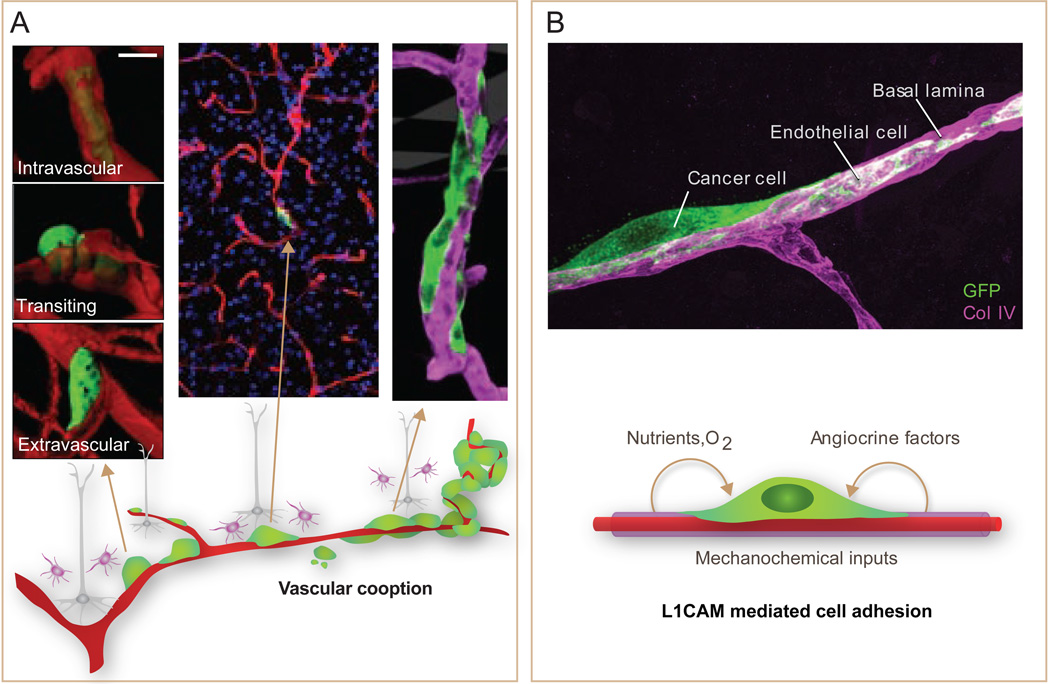
Another location where cancer cells initiate metastatic outgrowth is around blood capillaries –the perivascular niche. This niche has been studied as a preferred residence for glioma stem cells that supplies these cells with hedgehog, Notch and PI3K activating signals (Charles and Holland, 2010; Hambardzumyan et al., 2008). Breast cancer, lung cancer, and melanoma cells that infiltrate the brain conspicuously place themselves around capillaries (Carbonell et al., 2009; Kienast et al., 2010). The cells proliferate over the coopted vessels forming a furrow that eventually becomes multilayered and remodels the capillary network at the core of the expanding lesion (Figure 6). Recent work showed that brain metastasis-initiating cells express the Ig family cell adhesion molecule L1CAM and use it to stretch over the perivascular basal lamina (Valiente et al., 2014). L1CAM is normally expressed only in neurons for axon guidance (Maness and Schachner, 2007). Its expression in many types of cancer is associated with poor prognosis (Doberstein et al., 2011; Schroder et al., 2009), raising the possibility of a role for L1CAM in metastasis to other organs besides the brain.
Figure 6. The perivascular niche for metastasis initiation.
A. Metastasis-initiating cells (green) exiting from brain capillaries (red) remain tightly associated with the vessels, adhering to and stretching over their abluminal surface. This interaction is required for metastatic outgrowth. Outgrowth occurs forming a furrow over the capillary and later a multilayered cell colony. B. The axonal guidance cell adhesion receptor L1CAM is aberrantly expressed in tumors, and its expression is associated with relapse. L1CAM in metastasis-initiating cells (green) that infiltrate the brain mediates their adhesion to capillary basal lamina (magenta). Besides providing MetSCs with mechanochemical cues, vascular cooption facilitates their access to oxygen and nutrients from the blood supply and to factors from the endothelium and surrounding stroma. [Adapted from (Valiente et al., 2014)].
Niche cells and cytokines
Stem cell niches are sources of developmental and self-renewal signals including Wnt, Notch, TGF-β family, CXCL12/SDF1, hedgehog signals (Clevers, 2013; Hsu and Fuchs, 2012; Moore and Lemischka, 2006; Morrison and Spradling, 2008). Perhaps not surprisingly, gene expression profiles of metastatic samples overlap in the repertoire of activated pathways with those of adult stem cell niches (Takebe et al., 2011). A source of these signals in the bone marrow are mesenchymal cells that produce CXCL12/SDF1 for hematopoietic stem cell maintenance. The cognate chemokine receptor CXCR4 is frequently overexpressed in bone metastatic cells and provides these cells with chemotaxis and PI3K-mediated survival signals (Muller et al., 2001; Zlotnik et al., 2011) (Figure 7).
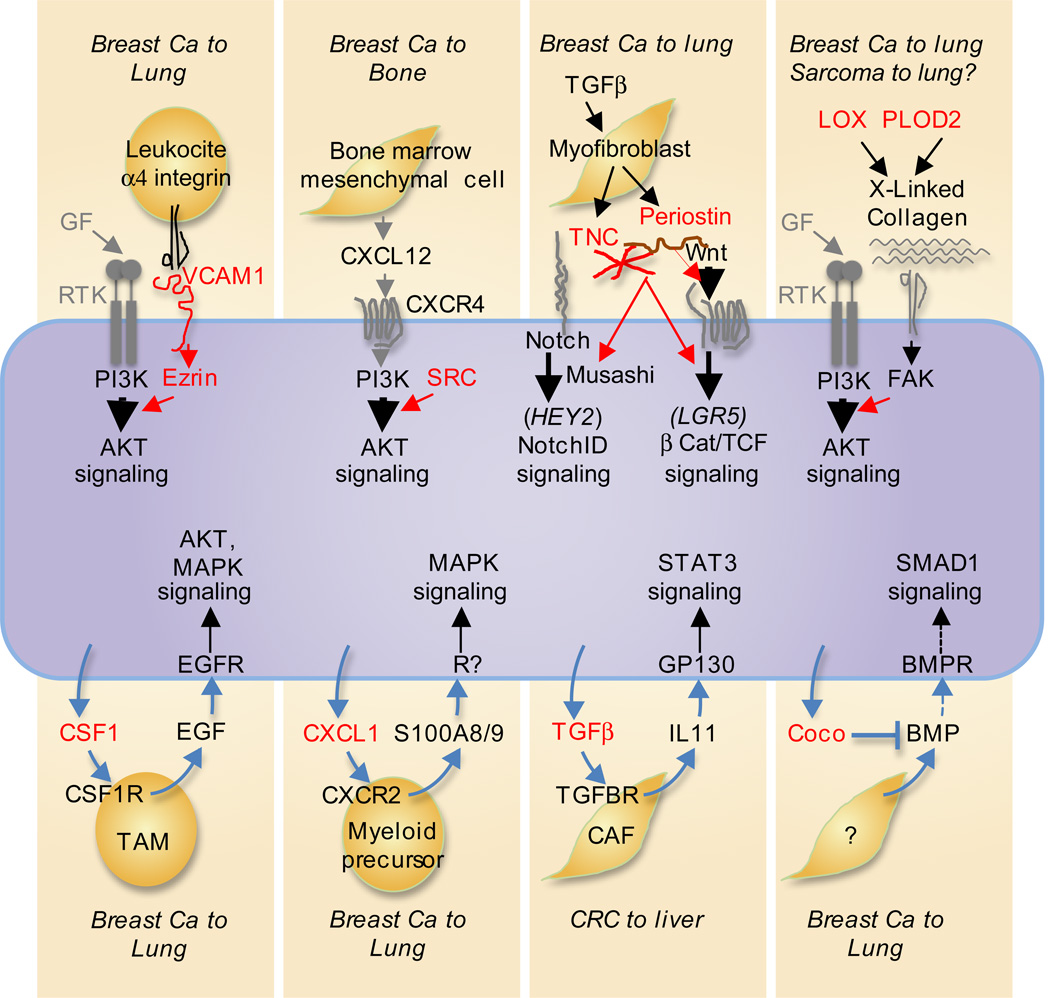
Figure 7. Pathway amplifiers and paracrine loops for MetSC support.
Many of the known traits that promote metastatic seeding involve gene products (red) that the cancer cells express to amplify their own responsiveness to vital stromal cues. The cues that activate these pathways include receptor tyrosine kinase (RTK) growth factor ligands (GF), chemokines like CXCL12, and Wnt, and Notch ligands. Examples amplifiers include cell adhesion receptors like the VCAM1-Ezrin complex that is engaged by tumor leukocyte integrins; the tyrosine kinase Src that amplifies PI3K-AKT activation by CXCR4 and IGF1R (not shown); the cancer cell- and myofibroblast-derived ECM components tenascin C (TNC) and periostin that enhance Wnt access to their receptors (periostin) or amplify signaling by the Wnt and Notch pathways (TNC); and, the collagen crosslinking enzymes LOX and PLOD2 that stiffen the ECM for integrin/focal adhesion kinase (FAK) mediated amplification of RTK signaling. Various cancer cell-derived cytokines (bottom, red) provide additional support by recruiting stromal cells that secrete activators of AKT, MAPK and STAT3 in incipient metastatic lesions. MetSC-derived BMP blockers like Coco protect self-renewal by inhibiting Smad1 signaling.
Perivascular niches may support MetSCs not only by supplying attachment, oxygen and nutrients but also paracrine factors from the activated endothelium, in what is called “angiocrine” stimulation (Butler et al., 2010). Interestingly, Jagged-1 expression in endothelial cells promotes the CSC phenotype in colorectal cancer cells (Lu et al., 2013). miR-126 initially identified as a metastasis-suppressor microRNA (Tavazoie et al., 2008) was shown to inhibit endothelial cell recruitment and thereby suppress lung metastasis (Png et al., 2012). Endothelial cells also express various extracellular matrix (ECM) components that promote metastatic functions in tissue culture (Ghajar et al., 2013). As metastatic lesions grow, the cancer cells recruit TAMs, myeloid precursors, and mesenchymal cells that establish paracrine loops feeding back to the cancer cells with various survival and self-renewal factors (Acharyya et al., 2012; Calón et al., 2012; Joyce and Pollard, 2009; Vermeulen et al., 2010) (Figure 7).
Primary tumors can systemically influence the microenvironment of distant organs, specially the lungs, to establish a “pre-metastatic niche” (Kaplan et al., 2005). In mouse models, breast, lung, and gastrointestinal tumors secrete inflammatory cytokines and ECM remodeling enzymes into the circulation. These factors can induce changes in the lung parenchyma microenvironment that enhance metastasis initiation as circulating cancer cells arrive in these locations (Kaplan et al., 2005; Yamamoto et al., 2008).
Niche extracellular matrix
The ECM components tenascin C (TNC) and periostin have been shown to play important roles in the metastatic niche in mouse models of cancer. TNC is a hexameric glycoprotein that is found in stem cell niches and it supports stem cell functions (von Holst, 2008). TNC expression in breast tumors is associated with increased risk of lung metastasis (Minn et al 2005). In xenotransplantation models, breast cancer cells that express high levels of TNC have a distinct advantage at initiating metastases after extravasating in the lungs (Oskarsson et al., 2011). TNC regulates Musashi and other as yet unknown factors to enhance Notch and Wnt signaling in the cancer cells (Figure 7). By expressing their own TNC, breast cancer cells have a higher probability of surviving during micrometastatic outgrowth. Myofibroblasts and S100A4+ fibroblasts eventually migrate into the growing lesion to provide additional sources of TNC (O'Connell et al., 2011; Oskarsson et al., 2011).
Like TNC, periostin is present in stem cell niches and is required for the initiation of lung metastasis by breast cancer cells in mice (Malanchi et al., 2012). Recruited myofibroblasts are a source of periostin in response to tumor-derived TGF-β Periostin binds and presents stromal Wnt ligands to the cancer cells (Figure 7). TNC and periostin thus enhance WNT and Notch signaling to promote the fitness of MetSCs during the initiation of metastatic colonization. TNC and periostin bind to cell surface integrins and bind tightly to each other (Kii et al JBC 2010). The physical interaction of TNC and periostin in the ECM may underlie a functional cooperation of these two proteins in stem cell niches (Oskarsson and Massagué, 2012).
In the course of cancer progression, tumors exhibit increased stiffness and the ECM is a key mediator of this tissue tension. Cancer cells respond to tensile forces imposed by a stiff ECM with activation of focal adhesion kinase (FAK) and PI3K-AKT, and with invasive behavior (Levental et al., 2009) (Figure 7). One enhancer of ECM stiffness is lysyl oxidase (LOX), a collagen crosslinking enzyme that is induced by hypoxia. LOX expression contributes to tissue stiffness, myeloid cell attraction for tumor growth. By acting systemically, LOX can additionally contribute to the formation of pre-metastatic niche (Erler et al., 2009; Erler et al., 2006). In a mouse model of undifferentiated pleomorphic sarcoma, a particularly aggressive subtype of sarcomas, hypoxia-activated HIF-1α induced the expression of procollagen lysyl oxidase PLOD2, yet another enzyme that stabilizes collagen cross-links (Eisinger-Mathason et al., 2013). PLOD2 promoted metastatic dissemination and is clinically associated with poor prognosis. PLOD2 and the prolyl hydroxylases P4HA1 and P4HA2 have been also implicated in breast cancer metastasis (Gilkes et al., 2013a; Gilkes et al., 2013b).
Other ECM components are also noteworthy. In mouse models, binding of the ECM glycosaminoglycan hyaluronan to its cell surface receptor CD44 inhibits apoptosis of breast cancer cells during lung colonization (Yu et al., 1997). CD44 also functions as a periostin receptor that enhances the aggressiveness of glioma stem cells in the perivascular niche (Pietras et al., 2014). CD44 activity promotes the migration and metastasis of colorectal cancer stem cells (Todaro et al., 2014). Moreover, hyaluronan synthase-2 (HAS2), the key enzyme in hyaluronan biosynthesis, is a significant inducer of metastasis in mouse models for breast cancer (Li et al., 2007b; Okuda et al., 2012). High levels of hyaluronan predict poor clinical outcome in breast cancer (Auvinen et al., 2013). The role of the ECM in metastasis is currently under systematic analysis and likely to expand in the future (Reticker-Flynn et al., 2012).
Life in The Niche
The current knowledge about the locations and composition of the niches where disseminated MetSCs reside is still rudimentary. However, there is a general understanding of the functions that these locations need to support, and some insights into the underlying molecular mechanisms.
Survival on arrival
Most circulating cancer cells that infiltrate a distant tissue die, and this applies to CSCs as well (Calón et al., 2012). To survive, disseminated cancer cells must avoid exposure to lethal signals from the reactive stroma, up-regulate cell survival and anti-apoptotic pathways to counter the effect of such signals, or both. One example of the former is the cited case of brain metastatic cells that express serpins to prevent the mobilization of FasL by plasmin from reactive astrocytes (Valiente et al., 2014).
The PI3K-AKT pathway is a critical survival input for disseminated cancer cells. Src activity amplifies the PI3K-AKT response of breast cancer cells to stromal CXCL12/SDF-1 and IGF1 in the bone marrow (Zhang et al., 2009) (Figure 7). Src activation is achieved by interaction with the estrogen receptor in luminal breast cancer cells, and is selected for by a CAF-rich stroma in basal subtype tumors (Zhang et al., 2013b). The endothelial cell adhesion molecule VCAM1 amplifies the PI3K-AKT response of breast cancer cells in the lungs (Figure 7). Alpha-4 integrins on pulmonary macrophages engage VCAM1 on the cancer cells to trigger Ezrindependent amplification of PI3K activation (Chen et al., 2011b). In these examples, Src and VCAM1 amplify the ability of metastatic cells to make the most of limited PI3K-AKT activating signals in the host stroma. Src activity in breast tumor clinical samples is associated with bone relapse (Zhang et al., 2009) and VCAM1 expression with lung relapse (Minn et al 2005; Chen et al 2011).
Interactions between the receptor tyrosine kinases EGFR and Met with ECM-binding integrins enhance metastatic colonization in model systems (Barkan et al., 2010; Desgrosellier and Cheresh, 2010). Results from animal models argue for significant roles of NF-kB signaling in metastasis of colon, lung and breast cancers (Luo et al., 2004; Maeda et al., 2009; Park et al., 2007). JAK-STAT3 signaling promotes metastasis in melanoma, and in breast, prostate, and pancreatic carcinomas (Abdulghani et al., 2008; Barbieri et al., 2010; Wei et al., 2003; Xie et al., 2006). An inherent limitation is that many of these studies were based on the analysis of general metastatic cell populations, not MetSCs specifically. However, an example of STAT-driven survival of MetSCs was described in liver metastasis of colorectal cancer. MetSCs infiltrating the liver or lungs produced TGF-β that stimulated stromal fibroblasts to produce interleukin-11. In turn, interleukin-11 activated pro-survival signaling through GP130/Stat3 in the cancer cells and facilitated the initiation of liver and lung metastasis (Calón et al., 2012) (Figure 7).
Preserving stemness
Ensuring the self-renewal capacity (‘stemness’) of disseminated cancer cells, and not just their survival, is a key role of metastasis niches. Notch and Wnt promote self-renewal within stem cell niches in the bone marrow (Moore and Lemischka, 2006), the intestinal mucosa (Clevers, 2013), the skin (Hsu and Fuchs, 2012) and the brain (Hambardzumyan et al., 2008), roles that are mirrored in the regulation of MetSCs in metastatic niches. The involvement of the Wnt and Notch pathways in supporting the metastasis-initiating capacity of disseminated breast cancer cells, and the role of ECM molecules like periostin and TNC in this process, have interesting parallels with the involvement of the PI3K-AKT pathway in preserving the survival of MetSCs (Zhang et al., 2009) (Chen et al., 2011b). Periostin and TNC in the case on Wnt and Notch signaling, like Src and VCAM1 in the case of PI3K-AKT signaling, act as amplifiers of the ability of MetSCs to respond to limiting levels of stromal Wnt and Notch ligands for activation of vital self-renewal pathways.
Reversing EMT
The motility and invasiveness of cancer cells that undergo an EMT may favor their dispersion to distant organs. Paradoxically, carcinoma metastases in patients typically present an epithelial cytology. Recent evidence from experimental systems shows that cancer cells that underwent an EMT for metastatic dissemination must undergo the reverse process, a mesenchymal-epithelial transition (MET), in order to start metastatic colonization (refer to Figure 2). In a model of squamous cell carcinoma, expression of the EMT master regulator Twist enhanced cancer cell dissemination, but the formation of overt metastases required a loss of Twist expression and reacquisition of epithelial traits (Tsai et al., 2012). Similarly, expression of the EMT transcription factor Prrx-1 promoted tumor dissemination of breast cancer cells whereas the subsequent down-regulation of Prrx-1 induced an MET that facilitated metastatic colonization without suppressing stem cell traits (Ocana et al., 2012). Id1promotes metastatic colonization in breast cancer cells (Gupta et al 2007b) and functions downstream of TGF-β to suppress Twist expression in basal breast cancer cells that infiltrate the lung parenchyma (Stankic et al., 2013). What prompts these cells to respond to TGF-β with an EMT right before extravasation (Labelle et al., 2011) and with MET right after, is unknown.
Entering and exiting dormancy
Metastasis can remain latent for years after the removal of a primary tumor. Despite the clinical importance of this phenomenon, little is known about how and why DTCs enter a dormant state, the different forms that this state may take, the cell-autonomous and stromal signals that induce and sustain it, and what leads to the eventual exit from latency for metastatic outgrowth. Most of the mouse models used for the study of metastasis lack a prolonged latency period. This is in contrast to metastasis in the clinic, where latency is measured in months to years. For these reasons, it remains unclear whether the metastatic niche components and support pathways discussed above are required before, during, and/or after MetSCs pass through a period of quiescence. Not much can be said about these questions that is based on hard evidence.
Most DTCs detected in bone marrow are proliferatively quiescent, or ‘dormant’ (Muller et al., 2005; Pantel et al., 1993). Although entry into G0 has been regarded as a failure of cancer cells to proceed with their tumor propagating potential, in fact it may represent a defense under adverse conditions (Barkan et al., 2010; Goss and Chambers, 2010; Klein, 2011). Various signals and pathways have been implicated in the balance between dormancy and proliferation in experimental systems in the few available models. Two mitogen activated kinases (MAPKs), namely of p38 and ERK, respectively control the switch of DTCs between dormancy and active growth (Aguirre-Ghiso et al., 2004). Urokinase receptor (uPAR) induces ERK via alpha5-beta1 integrins and inhibits p38 (Sosa et al., 2011). BMP signals in the lung parenchyma have been proposed to enforce latency in breast cancer cells by suppressing self-renewal and promoting differentiation, with metastatic progression being triggered by Coco or related BMP sequestering antagonists (Gao et al., 2012) (Figure 7).
During metastatic latency, cancer cells somehow are able to evolve and acquire a full complement of metastasis colonization functions that they did not express before. It is difficult to envision how this progression could occur in DTCs that remain in a state of replicative quiescence. Although DTCs in bone marrow look quiescent, the overall DTC population is not static. Circulating cancer cells can be detected in blood in the apparent absence of active metastatic disease. If not in the bone marrow at least in other tissues DTCs may be constantly exiting and re-entering a dormant state, undergoing further selection for colonization traits during the active interludes (refer to Figure 2). Transition between quiescent and proliferative states is a property of adult stem cells that may be hijacked by MetSCs.
Towards overt colonization
Metastatic cells can aggressively overtake a tissue after they acquire specialized functions for cooption of local stromal components. The clearest example is provided by osteolytic bone metastasis of breast cancer, by far the most extensively characterized overt metastatic process (Figure 8). Numerous molecular mediators have been identified that support the ability of bone metastatic cells to mobilize and activate osteoclasts. The subjugated osteoclasts aggressively resorb bone matrix, making room for the growing tumor. Furthermore, osteolysis causes the release of matrix-stored TGF-β and other growth factors that further stimulate cancer cells in a feed-forward cycle of tissue destruction and tumor expansion (Ell and Kang, 2012; Weilbaecher et al., 2011). Cancer cell derived factors including parathyroid hormone-related protein, interleukins 1, 6, 8 and 11, and TNF-α, proteases including MMP1 and ADAMTS1, the Notch ligand Jagged1, and cell adhesion molecules VCAM1 and ICAM1, many of which induced by bone-derived TGF-β, all converge on osteoclast activation (Ell et al., 2013; Lu et al., 2011; Sethi et al., 2011; Weilbaecher et al., 2011).
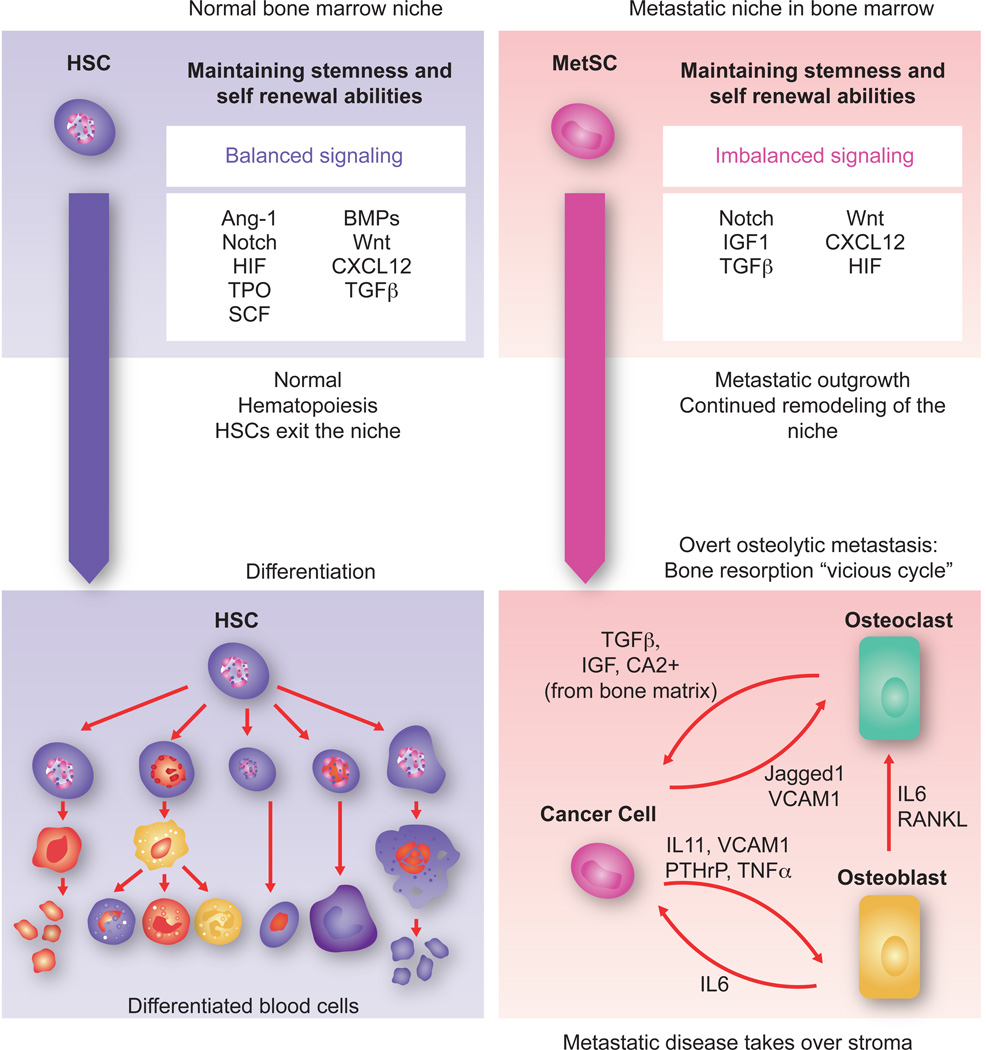
Figure 8. Towards overt colonization.
The hematopoietic stem cell (HSC) niche in the bone marrow is a highly regulated microenvironment that balances stem cell self-renewal and differentiation signals. Within the niche, factors like Ang-1, SCF, TPO, HIF1α and TGF-β maintain stem cell quiescence while Notch and Wnt promote self-renewal and proliferation. BMPs induce expansion of osteoblasts thereby indirectly supporting HSCs. The CXCL12- CXCR4 signaling axis is important for HSC retention in the niche. In the context of bone metastasis, breast cancer MetSCs coopt these cues for survival and to overtakes the host stroma. In the osteolytic process that follows, the cancer cells stimulate osteoclast differentiation and activation through cancer cell-derived cytokines such as IL11 and TNF-α, parathyroid hormone-related protein (PTHrP), MMP1, VCAM1, and the Notch ligand Jagged1. Some of these mediators act directly on osteoclast precursors to stimulate their maturation. Others act by stimulating osteoblasts to produce IL6 and RANKL, which in turn stimulate osteoclast differentiation and activity. Osteoclast mediated bone matrix resorption releases TGF-β and IGF1, which in turn up-regulate the expression IL11, PTHrP, Jagged1 and other metastasis mediators in the cancer cells to create a vicious cycle of relentless bone destruction.
Osteoclast mobilization traits provide a selective advantage in the context of bone metastasis but not in primary tumors or other tissues where there are no osteoclasts to activate or bone to resorb. Indeed, in experimental models, the acquisition of some of these traits has been shown to occur after metastatic cells remained latent for many months, became reactivated, formed micrometastases in the bone marrow, and were faced with the need to activate osteoclasts in order to resorb bone and make room (Lu et al., 2011). This admittedly limited set of available examples suggest that progression towards overt colonization involves the acquisition of a final set of metastatic traits by the progeny of MetSCs that survived arrival and latency in the host metastatic tissue.
Perspectives.
Recent work in this field has aimed at better conceptualizing and mechanistically explaining the biology of MetSCs. Residual disease remains challenging to properly model for study, but it offers a singular opportunity to develop therapeutic interventions for the suppression of metastasis. Some perspectives on the origins of metastatic traits have been summarized elsewhere (Vanharanta and Massagué, 2013). Here we highlight several salient questions about the biology of metastasis-initiating cells that seem worthy of attention. They are,
The phenotypic properties and signaling pathways required for metastatic outgrowth of disseminated cancer cells are increasingly being elucidated and overlap with those of normal stem cells. While the relationship between normal stem cells and CSCs is gaining clarity in many tumor types, the relationship between CSCs and MetSCs is still enigmatic at present. However, the problem may be poised for rapid progress.
A basic conceptual framework for the traits that allow CSC infiltration and seeding of distant organs is in place. However, many questions remain about how (genetically or epigenetically?), when (in the incipient tumor or in the advanced?), and where (at the invasive front or in a distant organ?) these traits are selected to generate MetSCs.
The source of MetSCs may depend on the type of cancer. For each type of cancer, a combination of lineage tracing techniques and single-cell analysis in animal models is needed in order to reveal the relative contribution of mechanisms that preserve a CSC phenotype of origin versus mechanisms that reestablish a stem cell phenotype to generate MetSCs in disseminated cancer cell populations.
Metastasis is an intrinsically inefficient process. The evidence suggests that most CSCs that try will fail. While our knowledge of how the survivors are saved is increasing, very little is known about what kills the majority of disseminated cancer cells. More information on how the reactive stroma repels infiltrating cancer cells could yield clues for how to leverage these mechanisms for therapeutic benefit.
Despite the clinical importance of metastatic latency, little is known about what induces MetSCs to enter a dormant state, and what allows them to remain viable for years in this state. Better models are needed in order to determine what MetSC supporting pathways are important before, during, or after the latency phase of metastasis. Killing latent MetSCs by depriving them of this support seems more attractive therapeutically than attempting to perpetually prevent their exit from dormancy.
Acknowledgements
We thank E. Sancho, S. Malladi and X. Jin for useful input. Work on metastasis by TO was supported by the BioRN Spitzencluster “Molecular and Cell Based Medicine” from the German Bundesministerium für Bildung und Forschung, and the Dietmar Hopp Foundation. EB was supported by the European Research Council, Ministerio de Economia y Competitividad, The Marcelino Botin Foundation, and The Josef Steiner Foundation. JM was supported by HHMI, the National Cancer Institute, the Department of Defense, the BBVA Foundation, and the MSKCC Metastasis Research Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdulghani J, Gu L, Dagvadorj A, Lutz J, Leiby B, Bonuccelli G, Lisanti MP, Zellweger T, Alanen K, Mirtti T, et al. Stat3 promotes metastatic progression of prostate cancer. Am J Pathol. 2008;172:1717–1728. doi: 10.2353/ajpath.2008.071054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N, Seshan VE, et al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Ghiso JA, Ossowski L, Rosenbaum SK. Green fluorescent protein tagging of extracellular signal-regulated kinase and p38 pathways reveals novel dynamics of pathway activation during primary and metastatic growth. Cancer Res. 2004;64:7336–7345. doi: 10.1158/0008-5472.CAN-04-0113. [DOI] [PubMed] [Google Scholar]
- Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11:R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, Prieto-Sanchez RM, Barba I, Martinez-Saez E, Prudkin L, et al. TGF-beta Receptor Inhibitors Target the CD44(high)/Id1(high) Glioma-Initiating Cell Population in Human Glioblastoma. Cancer Cell. 2010;18:655–668. doi: 10.1016/j.ccr.2010.10.023. [DOI] [PubMed] [Google Scholar]
- Auvinen P, Tammi R, Kosma VM, Sironen R, Soini Y, Mannermaa A, Tumelius R, Uljas E, Tammi M. Increased hyaluronan content and stromal cell CD44 associate with HER2 positivity and poor prognosis in human breast cancer. Int J Cancer. 2013;132:531–539. doi: 10.1002/ijc.27707. [DOI] [PubMed] [Google Scholar]
- Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bauerle T, Wallwiener M, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- Barbieri I, Quaglino E, Maritano D, Pannellini T, Riera L, Cavallo F, Forni G, Musiani P, Chiarle R, Poli V. Stat3 is required for anchorage-independent growth and metastasis but not for mammary tumor development downstream of the ErbB-2 oncogene. Mol Carcinog. 2010;49:114–120. doi: 10.1002/mc.20605. [DOI] [PubMed] [Google Scholar]
- Barkan D, Green JE, Chambers AF. Extracellular matrix: a gatekeeper in the transition from dormancy to metastatic growth. Eur J Cancer. 2010;46:1181–1188. doi: 10.1016/j.ejca.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- Beck B, Blanpain C. Unravelling cancer stem cell potential. Nat Rev Cancer. 2013;13:727–738. doi: 10.1038/nrc3597. [DOI] [PubMed] [Google Scholar]
- Becker S, Becker-Pergola G, Wallwiener D, Solomayer EF, Fehm T. Detection of cytokeratin-positive cells in the bone marrow of breast cancer patients undergoing adjuvant therapy. Breast Cancer Res Treat. 2006;97:91–96. doi: 10.1007/s10549-005-9095-6. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massagué J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Vogl FD, Naume B, Janni W, Osborne MP, Coombes RC, Schlimok G, Diel IJ, Gerber B, Gebauer G, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353:793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- Butler JM, Kobayashi H, Rafii S. Instructive role of the vascular niche in promoting tumour growth and tissue repair by angiocrine factors. Nat Rev Cancer. 2010;10:138–146. doi: 10.1038/nrc2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calón A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Cespedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, et al. Dependency of colorectal cancer on a TGF-beta-driven program in stromal cells for metastasis initiation. Cancer Cell. 2012;22:571–584. doi: 10.1016/j.ccr.2012.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron MD, Schmidt EE, Kerkvliet N, Nadkarni KV, Morris VL, Groom AC, Chambers AF, MacDonald IC. Temporal progression of metastasis in lung: cell survival, dormancy, and location dependence of metastatic inefficiency. Cancer Res. 2000;60:2541–2546. [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature. 2010;467:1109–1113. doi: 10.1038/nature09460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell WS, Ansorge O, Sibson N, Muschel R. The vascular basement membrane as "soil" in brain metastasis. PLoS One. 2009;4:e5857. doi: 10.1371/journal.pone.0005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celia-Terrassa T, Meca-Cortes O, Mateo F, de Paz AM, Rubio N, Arnal-Estape A, Ell BJ, Bermudo R, Diaz A, Guerra-Rebollo M, et al. Epithelial-mesenchymal transition can suppress major attributes of human epithelial tumor-initiating cells. J Clin Invest. 2012;122:1849–1868. doi: 10.1172/JCI59218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabottaux V, Ricaud S, Host L, Blacher S, Paye A, Thiry M, Garofalakis A, Pestourie C, Gombert K, Bruyere F, et al. Membrane-type 4 matrix metalloproteinase (MT4-MMP) induces lung metastasis by alteration of primary breast tumour vascular architecture. J Cell Mol Med. 2009;13:4002–4013. doi: 10.1111/j.1582-4934.2009.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- Charles N, Holland EC. The perivascular niche microenvironment in brain tumor progression. Cell Cycle. 2010;9:3012–3021. doi: 10.4161/cc.9.15.12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yao Y, Gong C, Yu F, Su S, Liu B, Deng H, Wang F, Lin L, Yao H, et al. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell. 2011a;19:541–555. doi: 10.1016/j.ccr.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang XH, Massagué J. Macrophage binding to receptor VCAM-1 transmits survival signals in breast cancer cells that invade the lungs. Cancer Cell. 2011b;20:538–549. doi: 10.1016/j.ccr.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WK, Zhao M, Liu Z, Stevens LE, Cao PD, Fang JE, Westbrook TF, Nguyen DX. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23:725–738. doi: 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. ProcNatlAcadSciUSA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in Cancer. Annu Rev Pathol. 2013 doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieter SM, Ball CR, Hoffmann CM, Nowrouzi A, Herbst F, Zavidij O, Abel U, Arens A, Weichert W, Brand K, et al. Distinct types of tumor-initiating cells form human colon cancer tumors and metastases. Cell Stem Cell. 2011;9:357–365. doi: 10.1016/j.stem.2011.08.010. [DOI] [PubMed] [Google Scholar]
- Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein K, Wieland A, Lee SB, Blaheta RA, Wedel S, Moch H, Schraml P, Pfeilschifter J, Kristiansen G, Gutwein P. L1-CAM expression in ccRCC correlates with shorter patients survival times and confers chemoresistance in renal cell carcinoma cells. Carcinogenesis. 2011;32:262–270. doi: 10.1093/carcin/bgq249. [DOI] [PubMed] [Google Scholar]
- Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger-Mathason TS, Zhang M, Qiu Q, Skuli N, Nakazawa MS, Karakasheva T, Mucaj V, Shay JE, Stangenberg L, Sadri N, et al. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 2013;3:1190–1205. doi: 10.1158/2159-8290.CD-13-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ell B, Kang Y. SnapShot: Bone Metastasis. Cell. 2012;151:690–690. e691. doi: 10.1016/j.cell.2012.10.005. [DOI] [PubMed] [Google Scholar]
- Ell B, Mercatali L, Ibrahim T, Campbell N, Schwarzenbach H, Pantel K, Amadori D, Kang Y. Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell. 2013;24:542–556. doi: 10.1016/j.ccr.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A, Le QT, Giaccia AJ. Hypoxia-induced lysyl oxidase is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15:35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidase is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- Fehm T, Becker S, Becker-Pergola G, Sotlar K, Gebauer G, Durr-Storzer S, Neubauer H, Wallwiener D, Solomayer EF. Presence of apoptotic and nonapoptotic disseminated tumor cells reflects the response to neoadjuvant systemic therapy in breast cancer. Breast Cancer Res. 2006;8:R60. doi: 10.1186/bcr1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: the 'seed and soil' hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Gao H, Chakraborty G, Lee-Lim AP, Mo Q, Decker M, Vonica A, Shen R, Brogi E, Brivanlou AH, Giancotti FG. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell. 2012;150:764–779. doi: 10.1016/j.cell.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay LJ, Felding-Habermann B. Contribution of platelets to tumour metastasis. Nat Rev Cancer. 2011;11:123–134. doi: 10.1038/nrc3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghajar CM, Peinado H, Mori H, Matei IR, Evason KJ, Brazier H, Almeida D, Koller A, Hajjar KA, Stainier DY, et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15:807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL. Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. J Biol Chem. 2013a;288:10819–10829. doi: 10.1074/jbc.M112.442939. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gilkes DM, Bajpai S, Wong CC, Chaturvedi P, Hubbi ME, Wirtz D, Semenza GL. Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol Cancer Res. 2013b;11:456–466. doi: 10.1158/1541-7786.MCR-12-0629. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gocheva V, Wang HW, Gadea BB, Shree T, Hunter KE, Garfall AL, Berman T, Joyce JA. IL-4 induces cathepsin protease activity in tumor-associated macrophages to promote cancer growth and invasion. Genes Dev. 2010;24:241–255. doi: 10.1101/gad.1874010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss PE, Chambers AF. Does tumour dormancy offer a therapeutic target? Nat Rev Cancer. 2010;10:871–877. doi: 10.1038/nrc2933. [DOI] [PubMed] [Google Scholar]
- Greenberg JI, Shields DJ, Barillas SG, Acevedo LM, Murphy E, Huang J, Scheppke L, Stockmann C, Johnson RS, Angle N, Cheresh DA. A role for VEGF as a negative regulator of pericyte function and vessel maturation. Nature. 2008;456:809–813. doi: 10.1038/nature07424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guise T. Examining the metastatic niche: targeting the microenvironment. Semin Oncol. 2010;37(Suppl 2):S2–S14. doi: 10.1053/j.seminoncol.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Guo W, Keckesova Z, Donaher JL, Shibue T, Tischler V, Reinhardt F, Itzkovitz S, Noske A, Zurrer-Hardi U, Bell G, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta GP, Nguyen DX, Chiang AC, Bos PD, Kim JY, Nadal C, Gomis RR, Manova-Todorova K, Massagué J. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature. 2007;446:765–770. doi: 10.1038/nature05760. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–644. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- Hambardzumyan D, Becher OJ, Holland EC. Cancer stem cells and survival pathways. Cell Cycle. 2008;7:1371–1378. doi: 10.4161/cc.7.10.5954. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Hess KR, Varadhachary GR, Taylor SH, Wei W, Raber MN, Lenzi R, Abbruzzese JL. Metastatic patterns in adenocarcinoma. Cancer. 2006;106:1624–1633. doi: 10.1002/cncr.21778. [DOI] [PubMed] [Google Scholar]
- Heyn C, Ronald JA, Ramadan SS, Snir JA, Barry AM, MacKenzie LT, Mikulis DJ, Palmieri D, Bronder JL, Steeg PS, et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn Reson Med. 2006;56:1001–1010. doi: 10.1002/mrm.21029. [DOI] [PubMed] [Google Scholar]
- Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13:714–726. doi: 10.1038/nrc3599. [DOI] [PubMed] [Google Scholar]
- Hsu YC, Fuchs E. A family business: stem cell progeny join the niche to regulate homeostasis. Nat Rev Mol Cell Biol. 2012;13:103–114. doi: 10.1038/nrm3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, Reiss M, Au JL, Haffty BG, Kang Y. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janni W, Vogl FD, Wiedswang G, Synnestvedt M, Fehm T, Juckstock J, Borgen E, Rack B, Braun S, Sommer H, et al. Persistence of disseminated tumor cells in the bone marrow of breast cancer patients predicts increased risk for relapse--a European pooled analysis. Clin Cancer Res. 2011;17:2967–2976. doi: 10.1158/1078-0432.CCR-10-2515. [DOI] [PubMed] [Google Scholar]
- Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc Natl Acad Sci U S A. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordon-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, Winkler F. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med. 2010;16:116–122. doi: 10.1038/nm.2072. [DOI] [PubMed] [Google Scholar]
- Kim MY, Oskarsson T, Acharyya S, Nguyen DX, Zhang XH, Norton L, Massagué J. Tumor self-seeding by circulating cancer cells. Cell. 2009;139:1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CA. Parallel progression of primary tumours and metastases. Nat Rev Cancer. 2009;9:302–312. doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- Klein CA. Framework models of tumor dormancy from patient-derived observations. Curr Opin Genet Dev. 2011;21:42–49. doi: 10.1016/j.gde.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Kozar S, Morrissey E, Nicholson AM, van der Heijden M, Zecchini HI, Kemp R, Tavare S, Vermeulen L, Winton DJ. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell. 2013;13:626–633. doi: 10.1016/j.stem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Kreso A, O'Brien CA, van Galen P, Gan OI, Notta F, Brown AM, Ng K, Ma J, Wienholds E, Dunant C, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science. 2013;339:543–548. doi: 10.1126/science.1227670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labelle M, Begum S, Hynes RO. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell. 2011;20:576–590. doi: 10.1016/j.ccr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, Fong SF, Csiszar K, Giaccia A, Weninger W, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Dawson JC, Forero-Vargas M, Spence HJ, Yu X, Konig I, Anderson K, Machesky LM. The actin-bundling protein fascin stabilizes actin in invadopodia and potentiates protrusive invasion. Curr Biol. 2010;20:339–345. doi: 10.1016/j.cub.2009.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007a;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Li Y, Li L, Brown TJ, Heldin P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. Int J Cancer. 2007b;120:2557–2567. doi: 10.1002/ijc.22550. [DOI] [PubMed] [Google Scholar]
- Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, Martin-Trevino R, Shang L, McDermott SP, Landis MD, et al. Breast Cancer Stem Cells Transition between Epithelial and Mesenchymal States Reflective of their Normal Counterparts Stem Cell Reports. 2014;2:1–14. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Ye X, Fan F, Xia L, Bhattacharya R, Bellister S, Tozzi F, Sceusi E, Zhou Y, Tachibana I, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23:171–185. doi: 10.1016/j.ccr.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Mu E, Wei Y, Riethdorf S, Yang Q, Yuan M, Yan J, Hua Y, Tiede BJ, Haffty BG, et al. VCAM-1 promotes osteolytic expansion of indolent bone micrometastasis of breast cancer by engaging alpha4beta1-positive osteoclast progenitors. Cancer Cell. 2011;20:701–714. doi: 10.1016/j.ccr.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JL, Maeda S, Hsu LC, Yagita H, Karin M. Inhibition of NF-kappaB in cancer cells converts inflammation-induced tumor growth mediated by TNFalpha to TRAIL-mediated tumor regression. Cancer Cell. 2004;6:297–305. doi: 10.1016/j.ccr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- Luzzi KJ, MacDonald IC, Schmidt EE, Kerkvliet N, Morris VL, Chambers AF, Groom AC. Multistep nature of metastatic inefficiency: dormancy of solitary cells after successful extravasation and limited survival of early micrometastases. Am J Pathol. 1998;153:865–873. doi: 10.1016/S0002-9440(10)65628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Hikiba Y, Sakamoto K, Nakagawa H, Hirata Y, Hayakawa Y, Yanai A, Ogura K, Karin M, Omata M. Ikappa B kinasebeta/nuclear factor-kappaB activation controls the development of liver metastasis by way of interleukin-6 expression. Hepatology. 2009;50:1851–1860. doi: 10.1002/hep.23199. [DOI] [PubMed] [Google Scholar]
- Malanchi I, Peinado H, Kassen D, Hussenet T, Metzger D, Chambon P, Huber M, Hohl D, Cano A, Birchmeier W, Huelsken J. Cutaneous cancer stem cell maintenance is dependent on beta-catenin signalling. Nature. 2008;452:650–653. doi: 10.1038/nature06835. [DOI] [PubMed] [Google Scholar]
- Malanchi I, Santamaria-Martinez A, Susanto E, Peng H, Lehr HA, Delaloye JF, Huelsken J. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2012;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- Maness PF, Schachner M. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat Neurosci. 2007;10:19–26. doi: 10.1038/nn1827. [DOI] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massard C, Loriot Y, Fizazi K. Carcinomas of an unknown primary origin--diagnosis and treatment. Nat Rev Clin Oncol. 2011;8:701–710. doi: 10.1038/nrclinonc.2011.158. [DOI] [PubMed] [Google Scholar]
- Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501:328–337. doi: 10.1038/nature12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, Sevillano M, Hernando-Momblona X, da Silva-Diz V, Munoz P, et al. The Intestinal Stem Cell Signature Identifies Colorectal Cancer Stem Cells and Predicts Disease Relapse. Cell Stem Cell. 2011 doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Muller V, Stahmann N, Riethdorf S, Rau T, Zabel T, Goetz A, Janicke F, Pantel K. Circulating tumor cells in breast cancer: correlation to bone marrow micrometastases, heterogeneous response to systemic therapy and low proliferative activity. Clin Cancer Res. 2005;11:3678–3685. doi: 10.1158/1078-0432.CCR-04-2469. [DOI] [PubMed] [Google Scholar]
- Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45:98–103. doi: 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- Naumov GN, Townson JL, MacDonald IC, Wilson SM, Bramwell VH, Groom AC, Chambers AF. Ineffectiveness of doxorubicin treatment on solitary dormant mammary carcinoma cells or late-developing metastases. Breast Cancer Res Treat. 2003;82:199–206. doi: 10.1023/B:BREA.0000004377.12288.3c. [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009a;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massagué J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009b;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342 doi: 10.1126/science.1234850. 1234850. [DOI] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- O'Connell JT, Sugimoto H, Cooke VG, MacDonald BA, Mehta AI, LeBleu VS, Dewar R, Rocha RM, Brentani RR, Resnick MB, et al. VEGF-A and Tenascin-C produced by S100A4+ stromal cells are important for metastatic colonization. Proc Natl Acad Sci U S A. 2011;108:16002–16007. doi: 10.1073/pnas.1109493108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obokata H, Sasai Y, Niwa H, Kadota M, Andrabi M, Takata N, Tokoro M, Terashita Y, Yonemura S, Vacanti CA, Wakayama T. Bidirectional developmental potential in reprogrammed cells with acquired pluripotency. Nature. 2014a;505:676–680. doi: 10.1038/nature12969. [DOI] [PubMed] [Google Scholar]
- Obokata H, Wakayama T, Sasai Y, Kojima K, Vacanti MP, Niwa H, Yamato M, Vacanti CA. Stimulus-triggered fate conversion of somatic cells into pluripotency. Nature. 2014b;505:641–647. doi: 10.1038/nature12968. [DOI] [PubMed] [Google Scholar]
- Ocana OH, Corcoles R, Fabra A, Moreno-Bueno G, Acloque H, Vega S, Barrallo-Gimeno A, Cano A, Nieto MA. Metastatic colonization requires the repression of the epithelial-mesenchymal transition inducer Prrx1. Cancer Cell. 2012;22:709–724. doi: 10.1016/j.ccr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Oft M, Peli J, Rudaz C, Schwarz H, Beug H, Reichmann E. TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 1996;10:2462–2477. doi: 10.1101/gad.10.19.2462. [DOI] [PubMed] [Google Scholar]
- Okuda H, Kobayashi A, Xia B, Watabe M, Pai SK, Hirota S, Xing F, Liu W, Pandey PR, Fukuda K, et al. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem-like cells with macrophages and stromal cells. Cancer Res. 2012;72:537–547. doi: 10.1158/0008-5472.CAN-11-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T, Acharyya S, Zhang XH, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massagué J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T, Massagué J. Extracellular matrix players in metastatic niches. Embo J. 2012;31:254–256. doi: 10.1038/emboj.2011.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padua D, Zhang XH, Wang Q, Nadal C, Gerald WL, Gomis RR, Massagué J. TGFbeta primes breast tumors for lung metastasis seeding through angiopoietin-like 4. Cell. 2008;133:66–77. doi: 10.1016/j.cell.2008.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang R, Law WL, Chu AC, Poon JT, Lam CS, Chow AK, Ng L, Cheung LW, Lan XR, Lan HY, et al. A subpopulation of CD26+ cancer stem cells with metastatic capacity in human colorectal cancer. Cell Stem Cell. 2010;6:603–615. doi: 10.1016/j.stem.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Pantel K, Alix-Panabieres C, Riethdorf S. Cancer micrometastases. Nat Rev Clin Oncol. 2009;6:339–351. doi: 10.1038/nrclinonc.2009.44. [DOI] [PubMed] [Google Scholar]
- Pantel K, Brakenhoff RH. Dissecting the metastatic cascade. Nat Rev Cancer. 2004;4:448–456. doi: 10.1038/nrc1370. [DOI] [PubMed] [Google Scholar]
- Pantel K, Brakenhoff RH, Brandt B. Detection, clinical relevance and specific biological properties of disseminating tumour cells. Nature Reviews Cancer. 2008;8:329–340. doi: 10.1038/nrc2375. [DOI] [PubMed] [Google Scholar]
- Pantel K, Schlimok G, Braun S, Kutter D, Lindemann F, Schaller G, Funke I, Izbicki JR, Riethmuller G. Differential expression of proliferation-associated molecules in individual micrometastatic carcinoma cells. J Natl Cancer Inst. 1993;85:1419–1424. doi: 10.1093/jnci/85.17.1419. [DOI] [PubMed] [Google Scholar]
- Park BK, Zhang H, Zeng Q, Dai J, Keller ET, Giordano T, Gu K, Shah V, Pei L, Zarbo RJ, et al. NF-kappaB in breast cancer cells promotes osteolytic bone metastasis by inducing osteoclastogenesis via GM-CSF. Nat Med. 2007;13:62–69. doi: 10.1038/nm1519. [DOI] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera M, Ribot EJ, Percy DB, McFadden C, Simedrea C, Palmieri D, Chambers AF, Foster PJ. In Vivo Magnetic Resonance Imaging for Investigating the Development and Distribution of Experimental Brain Metastases due to Breast Cancer. Transl Oncol. 2012;5:217–225. doi: 10.1593/tlo.12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras A, Katz AM, Ekström EJ, Wee B, Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT, Holland EJ. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.01.005. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png KJ, Halberg N, Yoshida M, Tavazoie SF. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- Podsypanina K, Du YC, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Piskounova E, Shackleton M, Weinberg D, Eskiocak U, Fullen DR, Johnson TM, Morrison SJ. Human melanoma metastasis in NSG mice correlates with clinical outcome in patients. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3004599. 159ra149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Foster HR, Fullen DR, Sabel MS, Johnson TM, Morrison SJ. Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell. 2010;18:510–523. doi: 10.1016/j.ccr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reticker-Flynn NE, Malta DF, Winslow MM, Lamar JM, Xu MJ, Underhill GH, Hynes RO, Jacks TE, Bhatia SN. A combinatorial extracellular matrix platform identifies cell-extracellular matrix interactions that correlate with metastasis. Nat Commun. 2012;3:1122. doi: 10.1038/ncomms2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond N, d'Agua BB, Ridley AJ. Crossing the endothelial barrier during metastasis. Nat Rev Cancer. 2013;13:858–870. doi: 10.1038/nrc3628. [DOI] [PubMed] [Google Scholar]
- Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De MR. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- Rolny C, Mazzone M, Tugues S, Laoui D, Johansson I, Coulon C, Squadrito ML, Segura I, Li X, Knevels E, et al. HRG inhibits tumor growth and metastasis by inducing macrophage polarization and vessel normalization through downregulation of PlGF. Cancer Cell. 2011;19:31–44. doi: 10.1016/j.ccr.2010.11.009. [DOI] [PubMed] [Google Scholar]
- Schardt JA, Meyer M, Hartmann CH, Schubert F, Schmidt-Kittler O, Fuhrmann C, Polzer B, Petronio M, Eils R, Klein CA. Genomic analysis of single cytokeratin-positive cells from bone marrow reveals early mutational events in breast cancer. Cancer Cell. 2005;8:227–239. doi: 10.1016/j.ccr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Scheel C, Eaton EN, Li SH, Chaffer CL, Reinhardt F, Kah KJ, Bell G, Guo W, Rubin J, Richardson AL, Weinberg RA. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145:926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- Schroder C, Schumacher U, Fogel M, Feuerhake F, Muller V, Wirtz RM, Altevogt P, Krenkel S, Janicke F, Milde-Langosch K. Expression and prognostic value of L1-CAM in breast cancer. Oncol Rep. 2009;22:1109–1117. doi: 10.3892/or_00000543. [DOI] [PubMed] [Google Scholar]
- Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Laird PW. Interplay between the cancer genome and epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozawa Y, Pedersen EA, Havens AM, Jung Y, Mishra A, Joseph J, Kim JK, Patel LR, Ying C, Ziegler AM, et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J Clin Invest. 2011;121:1298–1312. doi: 10.1172/JCI43414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Sonoshita M, Aoki M, Fuwa H, Aoki K, Hosogi H, Sakai Y, Hashida H, Takabayashi A, Sasaki M, Robine S, et al. Suppression of colon cancer metastasis by Aes through inhibition of Notch signaling. Cancer Cell. 2011;19:125–137. doi: 10.1016/j.ccr.2010.11.008. [DOI] [PubMed] [Google Scholar]
- Sosa MS, Avivar-Valderas A, Bragado P, Wen HC, Aguirre-Ghiso JA. ERK1/2 and p38alpha/beta signaling in tumor cell quiescence: opportunities to control dormant residual disease. Clin Cancer Res. 2011;17:5850–5857. doi: 10.1158/1078-0432.CCR-10-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankic M, Pavlovic S, Chin Y, Brogi E, Padua D, Norton L, Massagué J, Benezra R. TGF-beta-Id1 Signaling Opposes Twist1 and Promotes Metastatic Colonization via a Mesenchymal-to-Epithelial Transition. Cell Rep. 2013;5:1228–1242. doi: 10.1016/j.celrep.2013.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS, Camphausen KA, Smith QR. Brain metastases as preventive and therapeutic targets. Nat Rev Cancer. 2011;11:352–363. doi: 10.1038/nrc3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens JK, Everson GT, Elliott CL, Kam I, Wachs M, Haney J, Bartlett ST, Franklin WA. Fatal transfer of malignant melanoma from multiorgan donor to four allograft recipients. Transplantation. 2000;70:232–236. [PubMed] [Google Scholar]
- Stockmann C, Doedens A, Weidemann A, Zhang N, Takeda N, Greenberg JI, Cheresh DA, Johnson RS. Deletion of vascular endothelial growth factor in myeloid cells accelerates tumorigenesis. Nature. 2008;456:814–818. doi: 10.1038/nature07445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss DC, Thomas JM. Transmission of donor melanoma by organ transplantation. Lancet Oncol. 2010;11:790–796. doi: 10.1016/S1470-2045(10)70024-3. [DOI] [PubMed] [Google Scholar]
- Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, Notch, and Hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Todaro M, Gaggianesi M, Catalano V, Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S, Cocorullo G, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell Stem Cell. 2014 doi: 10.1016/j.stem.2014.01.009. this issue. [DOI] [PubMed] [Google Scholar]
- Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell. 2012;22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Chang A, Benaich N, Reinhardt F, Weinberg RA. Activation of miR-31 function in already-established metastases elicits metastatic regression. Genes Dev. 2011;25:646–659. doi: 10.1101/gad.2004211. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XHF, Lee DJ, Chaft JE, Kris MG, Huse JT, Brogi E, Massagué J. Serpins promote cancer cell survival and vascular cooption in brain metastasis. Cell. 2014 doi: 10.1016/j.cell.2014.01.040. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S, Massagué J. Origins of metastatic traits. Cancer Cell. 2013;24:410–421. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanharanta S, Shu W, Brenet F, Hakimi AA, Heguy A, Viale A, Reuter VE, Hsieh JJ, Scandura JM, Massagué J. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat Med. 2013;19:50–56. doi: 10.1038/nm.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- Vermeulen L, Todaro M, de Sousa Mello F, Sprick MR, Kemper K, Perez Alea M, Richel DJ, Stassi G, Medema JP. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Holst A. Tenascin C in stem cell niches: redundant, permissive or instructive? Cells Tissues Organs. 2008;188:170–177. doi: 10.1159/000112848. [DOI] [PubMed] [Google Scholar]
- Wang X, Kruithof-de Julio M, Economides KD, Walker D, Yu H, Halili MV, Hu YP, Price SM, Abate-Shen C, Shen MM. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waning DL, Mohammad KS, Guise TA. Cancer-associated osteoclast differentiation takes a good look in the miRNA)ror. Cancer Cell. 2013;24:407–409. doi: 10.1016/j.ccr.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Le X, Zheng L, Wang L, Frey JA, Gao AC, Peng Z, Huang S, Xiong HQ, Abbruzzese JL, Xie K. Stat3 activation regulates the expression of vascular endothelial growth factor and human pancreatic cancer angiogenesis and metastasis. Oncogene. 2003;22:319–329. doi: 10.1038/sj.onc.1206122. [DOI] [PubMed] [Google Scholar]
- Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nat Rev Cancer. 2011;11:411–425. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CW, Lee A, Shientag L, Yu J, Dong Y, Kao G, Al-Mehdi AB, Bernhard EJ, Muschel RJ. Apoptosis: an early event in metastatic inefficiency. Cancer Res. 2001;61:333–338. [PubMed] [Google Scholar]
- Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- Xie TX, Huang FJ, Aldape KD, Kang SH, Liu M, Gershenwald JE, Xie K, Sawaya R, Huang S. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006;66:3188–3196. doi: 10.1158/0008-5472.CAN-05-2674. [DOI] [PubMed] [Google Scholar]
- Xu J, Lamouille S, Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Jones S, Bozic I, Antal T, Leary R, Fu B, Kamiyama M, Hruban RH, Eshleman JR, Nowak MA, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Kikuchi H, Ohta M, Kawabata T, Hiramatsu Y, Kondo K, Baba M, Kamiya K, Tanaka T, Kitagawa M, Konno H. TSU68 prevents liver metastasis of colon cancer xenografts by modulating the premetastatic niche. Cancer Res. 2008;68:9754–9762. doi: 10.1158/0008-5472.CAN-08-1748. [DOI] [PubMed] [Google Scholar]
- Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Toole BP, Stamenkovic I. Induction of apoptosis of metastatic mammary carcinoma cells in vivo by disruption of tumor cell surface CD44 function. J Exp Med. 1997;186:1985–1996. doi: 10.1084/jem.186.12.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Tan W, Wu X, Poustovoitov M, Strasner A, Li W, Borcherding N, Ghassemian M, Karin M. A NIK-IKKalpha module expands ErbB2-induced tumor-initiating cells by stimulating nuclear export of p27/Kip1. Cancer Cell. 2013a;23:647–659. doi: 10.1016/j.ccr.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Jin X, Malladi S, Zou Y, Wen YH, Brogi E, Smid M, Foekens JA, Massagué J. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell. 2013b;154:1060–1073. doi: 10.1016/j.cell.2013.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massagué J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra A, Lewis J, Degryse B, Stuhlmann H, Quigley JP. The inhibition of tumor cell intravasation and subsequent metastasis via regulation of in vivo tumor cell motility by the tetraspanin CD151. Cancer Cell. 2008;13:221–234. doi: 10.1016/j.ccr.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik A, Burkhardt AM, Homey B. Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol. 2011;11:597–606. doi: 10.1038/nri3049. [DOI] [PubMed] [Google Scholar]