Abstract
Human cytolytic T lymphocytes and NK cells can limit tumor growth and are being increasingly harnessed for tumor immunotherapy. One way cytolytic lymphocytes recognize tumor cells is by engagement of their activating receptor, NKG2D, by stress-antigens of the MICA/B and ULBP families. This study shows that surface upregulation of NKG2D ligands by human epithelial cells in response to ultraviolet irradiation, osmotic shock, oxidative stress, and growth factor provision, is attributable to activation of the EGF-receptor (EGFR). EGFR activation causes intracellular re-localisation of AUF1 proteins that ordinarily destabilise NKG2D ligand mRNAs by targeting an AU-rich element conserved within the 3′ ends of most human but not murine NKG2D ligand genes. Consistent with these findings, NKG2D ligand expression by primary human carcinomas positively correlated with EGFR expression that is commonly hyper-activated in such tumours, and was reduced by clinical EGFR inhibitors. Thus, stress-induced activation of EGFR not only regulates cell growth but concomitantly regulates the cells’ immunological visibility. Thus, therapeutics designed to limit cancer cell growth should also be considered in terms of their impact on immunosurveillance.
Introduction
Although variation in human health is genetically determined, it is increasingly acknowledged to be massively affected by the “exposome” which refers to the totality of environmental challenges to which genomes equip individuals to respond (1). While the challenge of microbial exposure has received considerable attention, inanimate components of the exposome, collectively termed “stress”, are also important. In this regard, lymphoid stress-surveillance describes how lymphocytes, as opposed to myeloid cells, respond rapidly and polyclonally to endogenous molecules whose expression is substantially altered by cell and/or tissue dysregulation (2). One molecular manifestation of this is provided by MHC Class I-related antigens of the MICA/MICB and ULBP families in humans, and by the murine Rae1, H60 and Mult1 genes. By engaging NKG2D, an activating receptor on NK cells and subsets of T cells, these ligands provoke immune effector responses including cytolysis and cytokine production, either by primary activation of responding lymphocytes, or by co-stimulation of cells receiving signals through the T cell antigen receptor (TCR) (3-5). By either means, NKG2D ligand up-regulation can be a major source of immunogenicity of dysregulated cells, complementing the activities of microbe-associated molecules such as Toll-like receptor (TLR) ligands. It therefore becomes important to understand the mechanisms by which NKG2D ligands are regulated, since their expression can be a key factor in determining whether or not cells become visible to the immune system. Were NKG2D ligand expression not to be regulated appropriately, normal cells might become targets of immune attack, potentially provoking autoinflammatory diseases such as psoriasis, to which NKG2D ligands are conspicuously linked by genome-wide association studies (6, 7).
NKG2D regulation and lymphoid stress-surveillance are of particular relevance to cancer immunotherapy. Many human tumours express very high levels of MICA, MICB, and one or more ULBPs (8), the significance of which is implied by the many immuno-evasive mechanisms adopted by tumours as well as by viruses to suppress NKG2D-mediated lymphocyte activation (9-11). The common association of NKG2D ligand expression with cancer cells is consistent with evidence that murine Rae1 is upregulated in response to DNA damage (12). Nonetheless, human NKG2D ligands showed much lower levels of responsiveness to DNA damage (12). Moreover, MICA upregulation is frequently associated with scenarios such as osmotic and oxidative stress, virus infection, and increased cellular proliferation that cannot collectively be explained by DNA damage (13-15). Hence, the regulation of human NKG2D ligand expression merited more thorough investigation.
Another fascinating and unresolved aspect of NKG2D-dependent lymphoid stress-surveillance is the multiplicity of ligands (e.g. human MICA, MICB, ULBP1-6) (16), which appear functionally non-redundant despite their all engaging the same receptor (17). One hypothesis is that multiple ligands permit multiple means of regulation, enabling the host to respond to myriad components of the exposome and collectively confounding immuno-evasive strategies (18). Consistent with this, murine Rae1 genes but not Mult1 or H60 genes were expressed by primary fibroblasts upon explant to culture (19). In sum, there may be forms of regulation that apply to all NKG2D ligands and those that apply to only one or few.
The stress-responsiveness of MICA was first implied by the identification of a heat shock response element in the MICA promoter (20). Thus, many studies have focussed on NKG2D ligand gene transcription, including recent work showing regulation of murine Rae1 by the E2F transcription factor (19, 21). This notwithstanding, the profound impact of NKG2D ligands on cellular recognition by T lymphocytes and NK cells makes it probable that they are regulated at multiple levels. Consistent with this, some human NKG2D ligand mRNAs could be regulated under specific circumstances via micro-RNA (miR) docking sites in their 3′ untranslated regions (22, 23). Furthermore, disease-associated allelic variation in MICA was shown to profoundly affect the post-transcriptional stability of MICA mRNA with directly proportionate affects on surface protein expression (5).
Given these various considerations, this study re-assessed human NKG2D ligand regulation in epithelial cells. The surprising result was that the regulation of MICA by a broad spectrum of challenges could be more readily attributed to activation of the Epidermal Growth Factor (EGF) receptor than to DNA damage. This mode of regulation applied generally to human NKG2D ligand genes, albeit with slight variation in mechanism, but did not apply to most murine NKG2D ligand genes. Because the EGFR pathway is one of the most frequently dysregulated and commonly targeted pathways in human cancer (24), this study directly links tumour cell surveillance by T cells and NK cells to a major cell-autonomous component of cancer progression that is also a prime target of cancer therapy.
Results
UVB induces human NKG2D ligand upregulation
Because the murine NKG2D ligand, Rae1, is regulated by DNA damage, MICA expression was measured following UVB treatment of human keratinocytes, the cell-type from which MICA cDNA was originally cloned (25, 26). Although immortalised, the human HaCat keratinocyte cell line maintains striking contact-dependent inhibition of growth at confluence and expresses little MICA mRNA, thus permitting the gene’s upregulation by environmental agents to be studied. However, because MICA expression can be perturbed by changes in cell growth conditions, all experiments were meticulously controlled (see Materials and Methods).
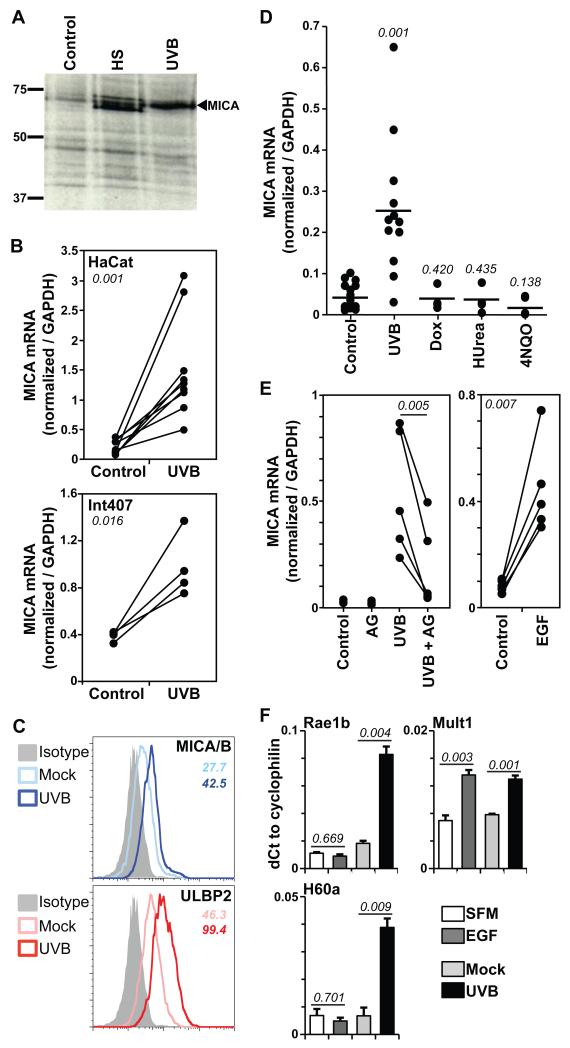
Acute exposure to 60mJ/cm2 of UVB (equivalent to twice the minimal erythematous dose) taking approximately 1-2 minutes, reproducibly upregulated total MICA protein expression to levels comparable to those induced by acute heat shock, the prototypic means of MICA upregulation (Figure 1A). Quantitative RT-PCR was used to measure MICA RNA relative to GAPDH RNA that was previously demonstrated to be extremely stable in response to UVB, including in HaCat cells (27-30). In each of nine independent experiments, MICA RNA was significantly upregulated, in several cases by >10-fold (comparable to the levels of protein induction shown in Fig 1A), and in other cases by 3-5-fold (Figure 1B). This response was not limited to HaCat cells, since MICA RNA was also significantly upregulated by UVB in four independent experiments using Int407, a foetal human intestinal epithelial cell line that also shows strict growth control (Figure 1B). It might be argued that UVB irradiation provoked MICA mRNA up-regulation via transient heat shock. However, this was not compatible with the observation that whereas MICA mRNA levels induced by heat shock peaked within 2h and returned almost to normal by 24h, those induced by UVB did not peak until after 24h (Figures S1A, S1B). UVB-induced NKG2D ligand upregulation extended to cultured primary human keratinocytes where increases in cell surface protein were detected using either an antibody specific for both MICA and MICB or one specific for ULBP2 (Figure 1C).
Figure 1. UVB upregulates NKG2D ligand expression via EGFR.
(A) Confluent HaCat cells were untreated (control) or exposed to UVB (60mJ/cm2) or heat shock (HS, 90mins 42°C), labelled with 35S-methionine/cysteine immediately after HS and 24h post-UVB, and lysed. MICA immunoprecipitates were analysed relative to known size markers (kD) by SDS-PAGE and autoradiography. (B) Confluent HaCat (top) or Int407 (bottom) cells were untreated or exposed to UVB. RNA was extracted 24h later and MICA quantified relative to GAPDH by real-time qPCR. Data were compiled from 9 (HaCat) and 4 (Int407) independent experiments. Numbers in italic indicate p values (paired t-test). (C) Primary human keratinocytes were untreated (Mock) or UVB-irradiated (60mJ/cm2) and MICA/B and ULBP2 expression monitored by flow cytometry 24h post-treatment. Numbers in italic indicate respective MFI values. (D) Confluent HaCat cells were exposed to UVB or treated with doxorubicin (Dox, 1μg/mL), hydroxyurea (HUrea, 1mM) or 4NQO (1μg/mL). MICA expression was monitored as in B. Each dot represents the analysis of an individual sample. Numbers in italic indicate p values relative to control (unpaired t-test). (E) Confluent HaCat cells were untreated (control) or treated with the EGFR inhibitor AG1478 (AG) alone or 30mins prior to UVB irradiation (left); or serum-deprived overnight and incubated +/− EGF (500ng/mL, right), and mRNA levels quantified as in B. Results from 5 independent experiments are shown. Numbers in italic indicate p values (paired t-test). (F) Confluent primary murine keratinocytes in serum free medium (SFM) were untreated or stimulated with EGF (100ng/mL) for 24h (left). As a positive control for Rae1b and H60a upregulation, cells were UVB irradiated (right). Expression was monitored by real-time qPCR. Data were normalized to cyclophilin and expressed as the mean of three treatments +/− SD. Numbers in italic indicate p values (unpaired t-test).
To examine the impact of other genotoxic stresses on MICA mRNA, HaCat cells were treated with doxorubicin, which intercalates into DNA (31); hydroxyurea that stalls or causes collapse of DNA replication forks following inhibition of ribonucleotide reductase (32); and 4-nitroquinoline-1-oxide (4NQO), a carcinogen that – akin to UV-light – provokes the formation of bulky purine adducts that are targeted by the nucleotide excision repair pathway, and which additionally induces DNA single strand breaks and alkali-labile sites (33). Most unexpectedly none of these significantly upregulated MICA mRNA levels (Figure 1D), although the agents were effective as judged by the upregulation of p21 RNA (Figure S1C) that is induced by the DNA damage-dependent activation of p53 (34). Thus MICA mRNA expression was upregulated in keratinocytes and intestinal epithelial cells by UVB but not obviously by a mechanism relating to DNA damage.
UVB upregulates NKG2D ligand RNAs via the EGFR
UVB irradiation can induce signalling through the EGFR, the major growth regulatory pathway for epithelial cells (35). To assess the potential contribution of this pathway to the regulation of MICA mRNA, cells were exposed to UVB but with or without AG1478 that inhibits several receptors with tyrosine kinase activity including EGFR. In each of five independent experiments, AG1478 significantly inhibited UVB-induced up-regulation of MICA mRNA (Figure 1E, left panel). Moreover, in five independent experiments in which confluent HaCat cells were rested in serum-free medium and then treated with EGF, MICA mRNA was sharply upregulated (Figure 1E, right panel), with kinetics and dose-dependence very similar to an established EGF-regulated gene, the urokinase-like plasminogen activator receptor, UPAR (36, 37) (Figure S1D). Cell surface expression of MICA/B and ULBP2 proteins was likewise induced by EGF with similar kinetics, with the most overt increase evident between 12h and 24h post-treatment (Fig S1E).
Consistent with the cells’ strict contact-inhibition, neither UVB nor EGF alone induced cell cycling in confluent HaCat cells (Figure S1F), thus distinguishing this regulation of MICA mRNA from the upregulated transcription of the MICA and ULBP2 genes induced by increased cell division (19). The experiments were then repeated in primary murine keratinocytes. Predictably, NKG2D ligand RNAs were upregulated by UVB irradiation: Rae1b and H60a by >4-5-fold, and Mult1 by ~2-fold. Strikingly, however, Mult1 was the only NKG2D ligand gene upregulated by EGF [again by ~2-fold] (Figure 1F). Thus, the mechanism of NKG2D ligand regulation by EGF/UVB most likely reflects a property that is shared by the Mult1, MICA/B and ULBP2 genes, but not by the Rae1b and H60a genes (see below).
EGFR promotes NKG2D-dependent cell killing
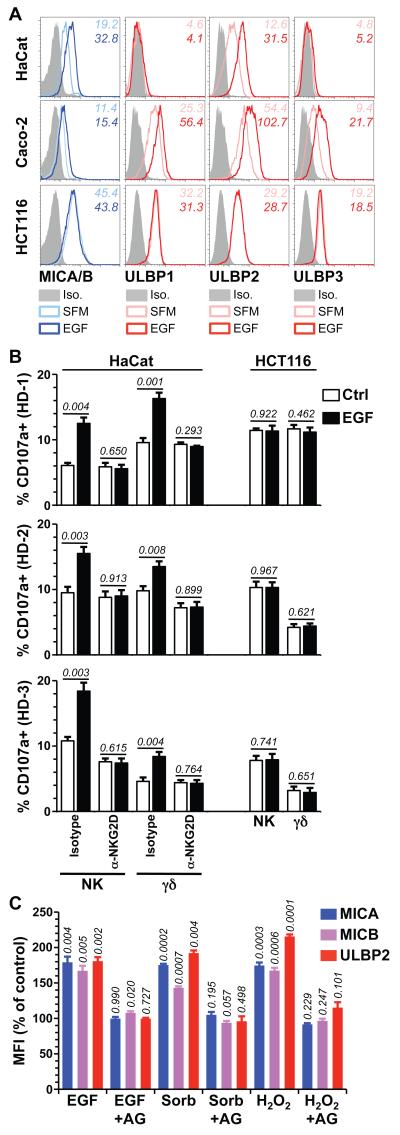
EGF treatment of confluent HaCat cells increased surface expression of ULBP2 and of MICA/B detected either with a widely-used antibody that detects both proteins, or with mono-specific antibodies for MICA and MICB, respectively (Figures 2A, S2A), but it did not induce expression of ULBP1 and ULBP3 which were not expressed pre-treatment (Figure 2A). Either EGF does not upregulate NKG2D ligands de novo, or it specifically does not regulate ULBP1 and 3. This was clarified by showing that EGF upregulated ULBP1, 2, and 3, and to a lesser extent MICA/B proteins on the surface of Caco-2 cells, an immortalised human intestinal epithelial cell line that like HaCat shows strong contact-dependent growth inhibition at confluence (Figure 2A). Thus, over the course of 12-24h EGF upregulated the surface display of several human NKG2D ligands in two distinct epithelial cell types but was seemingly more effective at increasing the level of expressed ligands rather than inducing ligands de novo. No EGF-mediated NKG2D ligand upregulation was shown by HCT116 cells that carry a KRAS mutation preventing surface EGFR expression (Figure 2A) (38), whereas the ligands were upregulated on both HaCat and HCT116 cells by phorbol-12-myristate-13-acetate (PMA) that activates signalling components downstream of EGFR (Figure S2B).
Figure 2. Cell surface expression of NKG2D ligands is upregulated by EGF and cellular stresses.
(A) Confluent HaCat, Caco-2 and HCT116 cells were serum-deprived overnight and stimulated with 500 ng/mL EGF (dark blue and red histograms) or left in serum-free medium (SFM) (light blue and red histograms) for 24h and cell surface expression of the indicated NKG2D ligands monitored by flow cytometry. Data are representative of at least 3 independent experiments (iso = isotype control staining). (B) HaCat and HCT116 cells were treated as in A (see Figure S2D for assesement of NKG2D ligand expression) and then co-incubated for 5h at 37°C with PBMCfrom 3 healthy donors (HD1-3) in the presence of PE-conjugated anti-CD107a antibody and a blocking anti-NKG2D or isotype control antibody (10μg/mL). Cells were washed and stained with APC-anti-CD3 and FITC-anti-pan TCRγδ, or with APC anti-CD3 and FITC anti-CD56 to analyse responses of γδ T and NK cells, respectively (see Figure S3 for gating controls). Data are means of triplicate stimulations +/− SD. Numbers in italic indicate p values (unpaired t-test). (C) HaCat cells serum-deprived overnight were treated with EGF (500ng/mL), Sorbitol (Sorb, 0.5M) or hydrogen peroxide (H2O2, 0.6mM) with or without a 1h pre-treatment with AG1478 (AG, 10μM). Surface MICA, MICB and ULBP2 expression levels were monitored 24h later by FACS. Data normalized to untreated cells (ctrl) and expressed as means of three treatments +/− SD. Numbers in italics are p values (unpaired t-test).
The implication of EGFR in the response of human NKG2D ligands to UVB and EGF raised the question of whether this major tyrosine kinase receptor pathway that regulates cell growth also contributed to the levels of ligand expression in actively growing cells. Supporting this, dividing, sub-confluent HaCat cells treated with AG1478 showed dose-dependent reductions in cell surface MICA/B and ULBP2 expression (Figure S2C), whereas no such reductions were seen in HCT116 cells in which the EGFR is impaired and does not regulate cell growth (Figure S2C).
Although the increases in NKG2D ligand surface expression were modest, they were well within the range reported for biologically relevant miRNA-mediated regulation of MICA and MICB (22) and typical of the levels of miRNA-mediated gene regulation more generally. Moreover, NKG2D+ γδ T cells and NK cells can be sensitive to only small changes in MICA expression, albeit with donor-to-donor variation (5). Therefore, to examine whether EGF regulates this critical function of NKG2D ligands, HaCat and HCT116 cells were treated with increasing concentrations of EGF; surface expression of MICA/B and ULBP2 measured on aliquots of cells (Figure S2D); parallel aliquots of cells co-cultured with primary PBMC from three independent healthy donors; and the induced killing potential of NK cells and γδ T cells within the PBMC measured by flow cytometry of CD107a that comes to the cell surface as cells de-granulate cytolytic lysosomes (5). Notwithstanding predictable donor-specific response variation, EGF-treated HaCat cells consistently provoked greater de-granulation-dependent CD107a expression by NK cells and γδ T cells than did control cultures (Figure 2B, Figure S3). Indeed, in some cases, ~20% of primary NK or γδ T cells were functionally activated. These effects were NKG2D-dependent (Figure 2B) and not attributable to relief of NK inhibition by potential EGF-induced reductions in HLA-expression, since levels of HLA-A/B/C and -E were unaltered by EGF (Figure S2E). Indeed, the lack of any impact on HLA refutes the hypothesis that EGF treatment provokes generalised immune dysregulation. Of note, the lack of an EGF-response pathway in HCT116 cells fully correlated with a failure to stimulate NK and γδ T cell de-granulation (Figure 2B; Figure S3). In sum, EGFR-mediated regulation markedly increased cells’ NKG2D-dependent susceptibility to cellular immune surveillance.
To investigate the generality of this regulatory pathway in relation to cell stress, cell surface MICA, MICB and ULBP2 expression were measured on HaCat cells treated with EGF; with sorbitol, that induces osmotic shock; and with hydrogen peroxide that induces oxidative stress. Of note, the EGFR can be activated by sorbitol and by oxidative stress, as well as by UV irradiation (39, 40). MICA, MICB and ULBP2 were each upregulated in an AG1478-sensitive mechanism (Figures 2C, S4). Thus, activation of EGFR and/or a closely related AG1478-sensitive receptor tyrosine kinase (RTK) mediates the upregulated surface expression of human NKG2D ligands in response to multiple components of the exposome.
EGF stabilizes MICA/B RNAs via the 3′ UTR
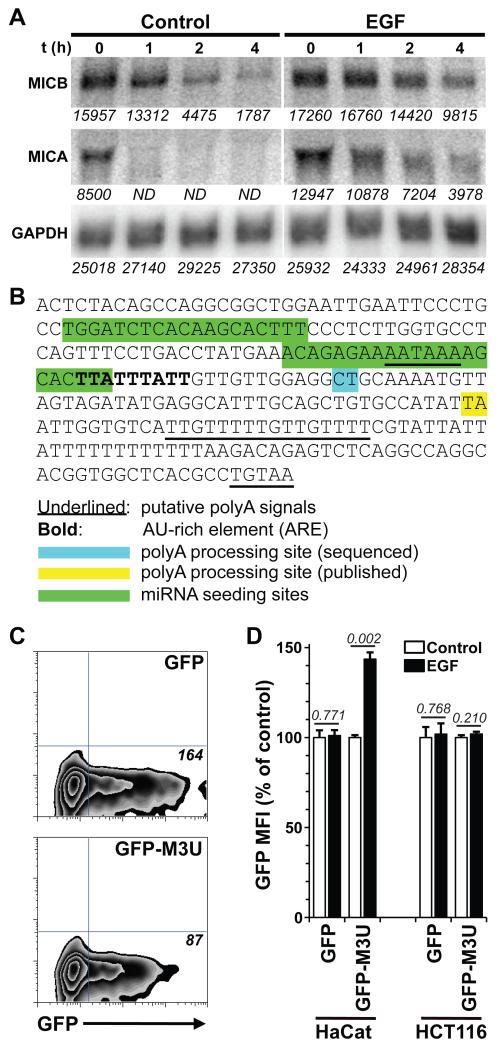
The sustained induction of MICA mRNA by UVB and EGF, rather than its induction de novo might reflect accumulation of stabilized mRNA. To test this, confluent HaCat cells were treated with actinomycin D to arrest transcriptional initiation 24h after treatment with either EGF or control medium. Northern analysis of RNAs sampled at hourly time-points thereafter permitted quantitation relative to GAPDH. This experiment showed that MICA and MICB mRNAs had very low half-lives in confluent cells (t1/2-MICA, ≤30 mins; t1/2-MICB, ~1.5h), consistent with the biological importance of limiting NKG2D ligand expression in growth-controlled cells. However, both mRNA species were substantially stabilized by EGF-treatment (t1/2-MICA, >2h; t1/2-MICB, ≥4h) (Figures 3A, S5A).
Figure 3. MICA mRNA stability is regulated by its 3′UTR.
(A) HaCat cells were serum-deprived and incubated overnight with or without EGF (500ng/mL). Actinomycin D was added and RNA extracted at the indicated time points [t (h)] for northern analysis. The blot was probed for MICA and MICB, and re-probed for GAPDH. Representative of 3 experiments. Densitometry analysis was performed with ImageJ and raw values are indicated in italics for each sample (ND, not detectable). See Figure S5A for the evaluation of mRNA half-lives (B) The sequence of the full 3′UTR of MICA was deduced from cloning and sequencing and from the genomic sequence (accession number NC_000006.11) to include poly-A signals (underlined), relative to which the poly-A processing site identified by re-sequencing (blue) shows a different location than the previously identified site (yellow) (see also Figure S6A). Also shown are microRNA seeding sites (green) and a canonical ARE (bold). (C) HaCat cells were transfected with the pMax-FP-Green plasmid (GFP) or a modified version including the 3′UTR of MICA (GFP-M3U). GFP expression was monitored by flow cytometry 24h post-transfection. Plots are representative of 3 independent transfections. Numbers in italic indicate MFI. (D) HaCat and HCT116 cells were transfected as in B. Medium was supplemented 24h post-transfection with G418 (0.5mg/mL). After 14 days, confluent cells were serum-deprived overnight and further treated (or not) with EGF for 24h before GFP expression was analyzed by flow cytometry. Data were normalized to untreated cells for each transfectant and are means of triplicate EGF treatments +/− SD. Numbers in italic indicate p values (unpaired t-test).
RNA instability is commonly regulated via the 3′-untranslated region (3′UTR), in relation to which seeding sites for micro-RNAs (miRs) mapped within the 3′UTRs of MICA/MICB mRNAs (green annotation in Figure 3B) were shown to reduce MICA/MICB expression (22, 23). However, the 3′ end also harbours a conspicuous AU-rich element (ARE) of the kind that can destabilize >5% of all cellular transcripts (41, 42). By re-sequencing and re-annotation of the 3′UTR of the MICA gene relative to published and archived sequences, the ARE could be oriented 6 nucleotides 3′ downstream to a canonical AATAAA polyA signal (bold, Fig 3B) relative to which a new poly-A tail addition site was identified (blue, Fig 3B) that is 26 nucleotides downstream, consistent with the reported mammalian mRNA range of 5-30 nucleotides (43, 44). This contrasts with the previously reported poly-A addition site (yellow, Fig 3B), which would have been at an highly atypical distance of 71 nucleotides downstream of the AATAAA signal (Figure 3B).
To test the potential impact on gene expression of the MICA 3′UTR region, it was cloned (starting immediately after the translational stop codon) downstream of the GFP coding region, creating a chimeric allele termed GFP-M3U. HaCat cells transiently transfected with GFP-M3U displayed reproducibly lower GFP expression than those transfected with unmodified GFP, consistent with GFP being down-regulated by the MICA 3′UTR (Figure 3C). Moreover, essentially the same result was shown by other human cell lines, with in each case there being fewer cells expressing high GFP levels (Figure S5B).
HaCat and HCT116 cells transfected with GFP and GFP-M3U constructs were then established as stable lines by drug selection over a 2-week period during which the expression of GFP in GFP-M3U transfectants continued to decline significantly, relative to unmodified GFP transfectants, as judged both by mean fluorescence intensity (MFI) and percent-cells positive (Figure S5C). However the MFI of GFP expression was significantly upregulated in EGF-treated GFP-M3U HaCat transfectants relative to mock-treated cells, whereas it was not upregulated in EGF-treated GFP HaCat transfectants. GFP was also not upregulated in HCT116 cells transfected with either the GFP or GFP-M3U constructs (Figure 3D), consistent with the refractoriness of HCT116 cells to EGF. Collectively, these data establish that the MICA 3′UTR destabilises gene expression, but that this can be relieved by EGF activation. Of note, AREs are conserved in all human NKG2D ligand genes (including several MICA alleles) for which reliable 3′UTR sequences are available, and in Mult1, the only murine NKG2D ligand tested that was regulated by EGF (Figure S6), but are not conserved in Rae1b or H60a genes.
An AU-rich element regulates NKG2D ligands
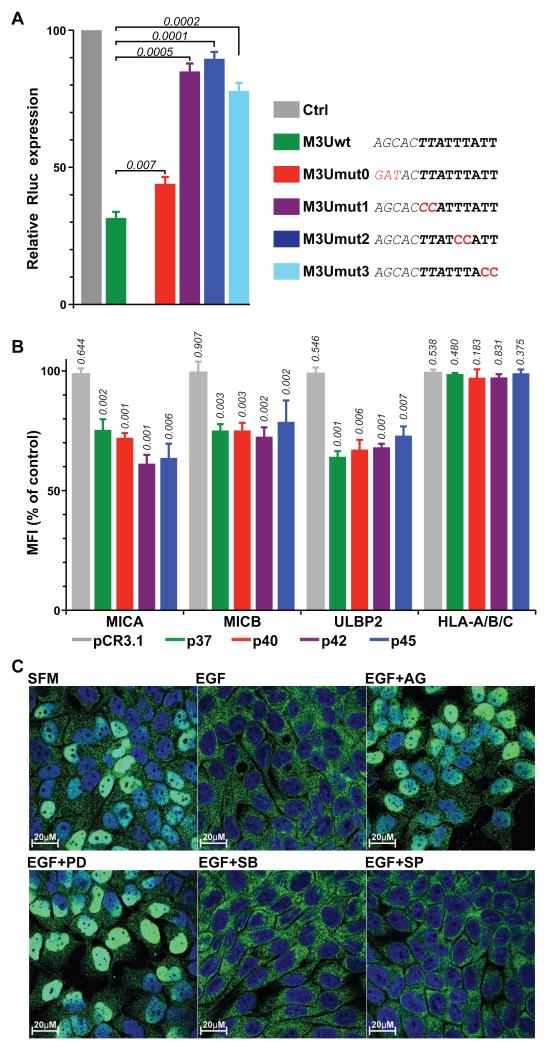
To confirm that MICA mRNA instability was attributable to the ARE, the chimeric reporter constructs were subject to several mutations that did not change the length of the constructs. Individual doublet mutations (M3U-mut1, mut2, and mut3) within the ARE significantly rescued the low expression levels of GFP-M3U expression (purple, blue, and cyan columns, Fig S7A), whereas mutation of the overlapping miR seeding site (M3U-mut0) had a lesser effect (red column). However, as an independent measure of mRNA destabilization, further chimeras were generated by fusing WT and mutant versions of the MICA 3′UTR to Renilla luciferase (Rluc) that encodes a less stable protein than GFP, thereby providing a more direct read-out of the destabilizing effect of the 3′UTR. In these studies, the activity of a co-introduced, unmanipulated Firefly luciferase served to normalize Rluc expression across different transfectants. Again, relative to non-chimeric expression, luciferase levels were greatly reduced by the 3′UTR (green column) (Figure 4A). Again, these were marginally rescued by mutation of the overlapping miR seeding site (M3U-mut0, red column), but were almost completely restored to normal by independent mutations of the ARE site (purple, blue, and cyan columns) (Fig 4A). Collectively, the results obtained with chimaeric GFP and luciferase constructs demonstrated the capacity of the MICA 3′UTR ARE to destabilize different RNAs at steady-state, with relief of this regulation being provided by EGFR activation.
Figure 4. The ARE and AUF1 destabilize MICA mRNA.
(A) HaCat cells were transfected with plasmids encoding Renilla luciferase (Rluc, pGL4.74, Ctrl) or modified versions including the wild-type (wt) or mutated (M3Umut0 to M3Umut3) 3′UTR of MICA and co-transfected with a plasmid encoding Firefly luciferase (Fluc) as an internal control. At 24h post-transfection, luciferase activity was measured using a Dual-Luciferase reporter assay system, and luminescence values for Rluc normalized to the internal Fluc values and to the wt Rluc control. Data are the mean of three independent transfections +/− SD. Numbers in italic indicate p values (unpaired t-test). Relevant sequences are indicated, including the miRNA seeding site (italic), ARE (bold) and mutations generated (red). (B) HaCat cells were mock-transfected, or transfected with pCR3.1 plasmids either empty or encoding each AUF1 isoform as indicated. Cell-surface expression of MICA, MICB and ULBP2 was analyzed by flow cytometry at 36h post-transfection. Data are normalized to mock-transfected cells expressed as the mean of three transfections +/− SD. Numbers in italic indicate p values (unpaired t-test). (C) HaCat cells grown on coverslips were treated as indicated, fixed and permeabilized, stained for endogenous AUF1, and analysed by confocal microscopy. Images are representative of 3 independent experiments and show overlays of AUF1 (green) and DAPI staining (blue) (see Figure S9B for individual channels).
In considering how EGFR might promote mRNA stabilization, it was reported that several regulators of mRNA stability that bind to AU-rich elements are regulated by MAP kinases (MAPK) (45). Therefore, confluent HaCat cells were stimulated with EGF with or without increasing doses of inhibitors of MEK, p38, and JNK. MEK inhibition by PD184352 recapitulated the inhibitory effects of AG1478 in a dose-dependent fashion; conversely JNK inhibition (by SP600125) had no effect, and p38 inhibition (by SB203580) impaired EGF-responsive upregulation of ULBP2 but not of MICA/B (Figure S7B). Thus, the EGFR-regulated cell surface display of NKG2D ligands shows selective dependence on MAP kinases.
AUF1 is one of the most widely implicated factors that destabilize mRNAs by binding to AU-rich elements (46). However, the four main isoforms vary in their effects on different genes (47), and it was therefore important to investigate whether any or all AUF1 isoforms might regulate NKG2D ligands at steady-state. Thus, cell surface expression was measured on HaCat cells transfected with constructs encoding each of the four AUF1 isoforms (p37, p40, p42, p45), respectively. Relative to mock-transfected cells, or cells transfected with the empty vector (pCR3.1), cells transfected with each of the four isoforms showed significant reductions in cell surface MICA and MICB and ULBP2 (Figures 4B, S8).
To investigate whether EGFR-mediated rescue of NKG2D ligand expression might reflect regulation of AUF1 by EGFR, serum-starved cells were treated with EGF with or without signaling inhibitors, and AUF1 protein assessed. There was no measurable impact on total or phosphorylated protein levels (Figure S9A). However, cell fractionation experiments had previously shown that generalized MAPK activation could provoke nuclear AUF1 re-localization to the cytoplasm (48), which is noteworthy given that deletion of the AUF1-p37 nuclear localization signal showed that nuclear import is required for AUF1-mediated regulation of RNA turnover (49). Strikingly, EGF treatment provoked near-complete exclusion of AUF1 from the nucleus (Figures 4C, S9B). Of note, this was prevented by AG1478 and PD184352 that both inhibited surface upregulation of MICA/B and ULBP2, as described above, but not by either SP600125 or SB203580, neither of which inhibited EGFR-regulation of MICA/B (Figure S9B).
EGFR and NKG2D ligand expression correlate in human cancer
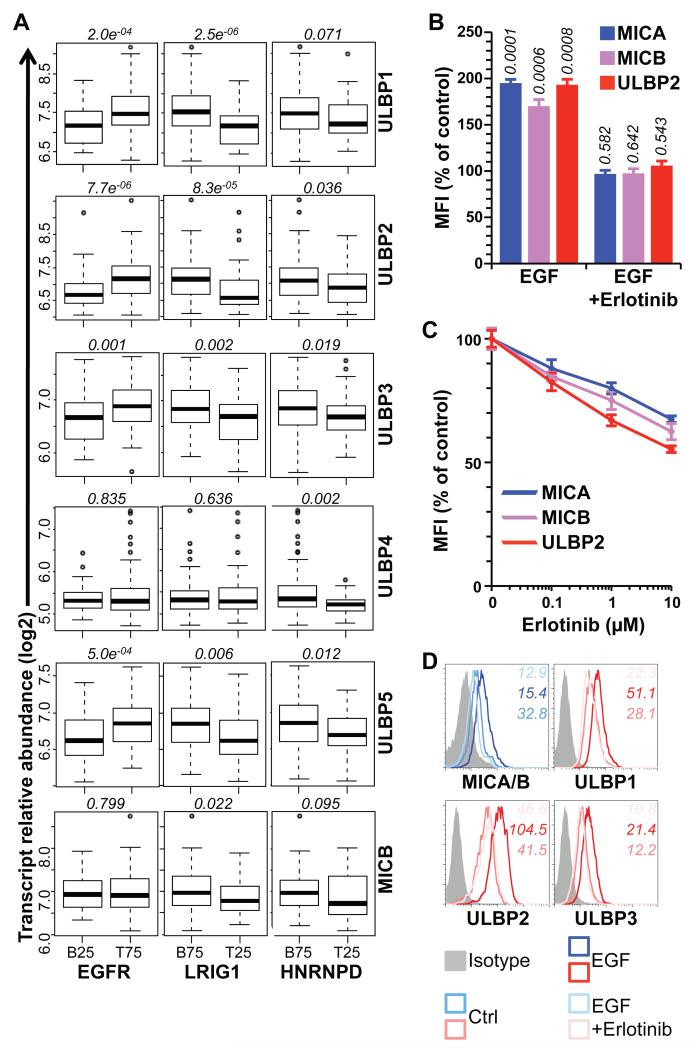
Were EGFR-mediated regulation of human NKG2D ligand mRNAs in vitro to reflect events in vivo, overt changes in EGFR activity should be reflected in NKG2D ligand expression. In this regard, many human carcinomas display specific dysregulation of the EGFR (24, 50-52). Thus, gene expression profiles of a collection of 172 fresh-frozen primary breast carcinomas (53) were examined for six NKG2D ligand RNAs (note: probes for MICA were not present on the array platform used) in relation to EGFR, the estrogen receptor (ESR1), and HER2, that respectively represent the major growth regulatory pathways for human breast carcinomas (54-56). Strikingly, tumors graded in the lowest quartile (B25) for their expression of EGFR showed significantly lower expression of four of six NKG2D ligand RNAs assayed (ULBP1, 2, 3, and 5) (Figure 5A), none of which showed any correlation with ESR1 or HER2 expression (Figure S10A). Independently corroborating the correlation of NKG2D ligand expression and the EGFR pathway, the expression of all NKG2D ligand mRNAs except ULBP4 negatively correlated with LRIG1 RNA, that encodes a negative regulator of the EGFR (57) (Figure 5A). Furthermore, all NKG2D ligand mRNAs showed negative correlations with HNRNPD RNA encoding AUF1, albeit that the correlations with MICB and ULBP1 failed to reach statistical significance (Figure 5A).
Figure 5. The EGFR-mediated upregulation of NKG2D ligand upregulation has clinical implications.
(A) Expression of NKG2D ligands in primary breast cancers with the bottom 25% (B25) EGFR or top 25% (T25) LRIG1 and HNRNPD gene expression compared to the remaining samples. Numbers in italic indicate p values (Wilcoxon rank sum test). (B) HaCat cells were grown to confluence, serum-deprived for 24h, treated or not with EGF (500ng/mL) for 24h with or without a 1h pre-treatment with Erlotinib (10μM), and stained for MICA, MICB and ULBP2. Data normalized to untreated cells (control) and expressed as the mean of three treatments +/− SD. Numbers in italic indicate p values (unpaired t-test). (C) Actively growing subconfluent HaCat cells grown in complete medium were treated or not with increasing doses of Erlotinib for 24h and stained for MICA, MICB and ULBP2. Data were normalized to untreated cells and are means of three treatments +/− SD. (D) Differentiated Caco-2 monolayers grown on collagen-coated transwell inserts until establishing electrically-resistant epithelial sheets were serum-deprived overnight, treated (or not) with EGF (500ng/mL) for 24h with or without a 1h pre-treatment with Erlotinib (10μM), and stained for MICA/B and ULBP2. Numbers in italic indicate MFI.
Because MICA was missing from the array, its expression, together with that of MICB, was investigated in a data-set of 26 triple-negative breast cancer cell lines (58): both genes showed positive correlation to EGFR expression (Figure S10B). In sum, although primary human tumours and cell lines are inevitably complex and heterogeneous, this complexity was super-imposed upon by an expression pattern fully consistent with the regulation of the majority of NKG2D ligand RNAs by the opposing effects of EGFR activation and AUF1.
The EGFR pathway is also a major therapeutic target in cancer. Illustrating the potential impact of such modalities on NKG2D ligand expression, Erlotinib (a clinically employed EGFR inhibitor) prevented the EGF-mediated upregulation of different NKG2D ligands on confluent HaCat cells (Figures 5B, S10C). Likewise, Erlotinib reduced NKG2D ligand expression on actively growing HaCat cells (Figures 5C, S10D) and phenocopied the effects of AG1478 and PD184352 on both sub-confluent and confluent Caco-2 cells (Fig S11). Analogous dose-dependent effects in sub-confluent and confluent HaCat cells were also achieved by Cetuximab, another clinical inhibitor of EGFR (Figure S12).
Finally, the regulation of NKG2D ligand by EGFR activation and its response to erlotinib were examined in differentiated, non-proliferative human intestinal epithelial cells. To achieve this, Caco-2 cells (which are commonly used to model a fully differentiated human intestinal epithelium) were grown in collagen-coated transwells in which they form electrically resistant monolayers (59). Over a three-week period, electrical resistance was regularly measured across a Caco-2 cell sheet seeded in this way, and when it had reached a stable plateau of 500 Ohms the cells were transferred to serum-free medium, with or without EGF. MICA/B and ULBP1-3 surface expression were up-regulated by EGF (Figure 5D), demonstrating that this form of regulation can occur in a fully confluent, functional monolayer, as might be the case at the earliest stages of human epithelial cell transformation in vivo. These EGF-mediated increases were substantially inhibited by Erlotinib (Figure 5D).
Discussion
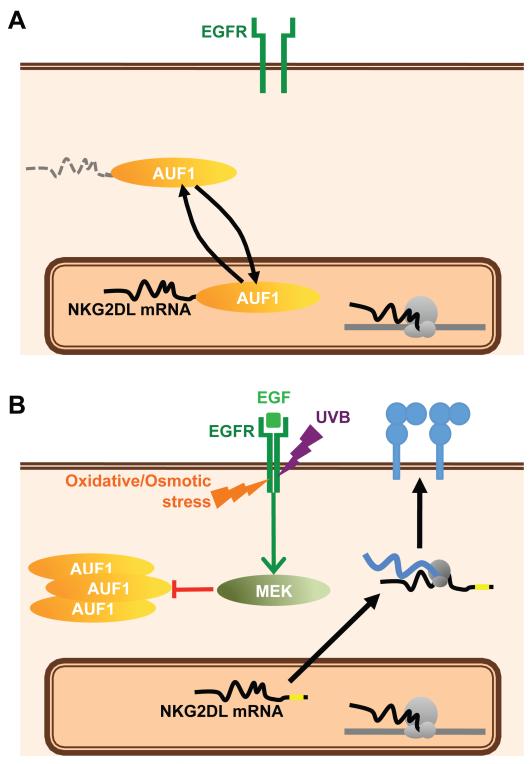
The NKG2D axis is increasingly viewed as a key component of host interactions with the environment. In response to microbial and non-microbial components of the exposome, NKG2D ligands can promote immunogenicity in the innate phase of immunity and can activate effector/memory cells in the adaptive phase. Given these profound potentials, it was predicted that NKG2D ligand expression would be tightly regulated. In that regard, this study has identified a pathway by which cell surface ligand expression is restricted via post-transcriptional regulation, in large part mediated by a consensus ARE-element that can be destabilized by AUF1 proteins (Figure 6A). In cells experiencing environmental perturbation by at least four components of the exposome - UVB irradiation, growth factors, osmotic stress, and oxidative stress - this restriction was relieved by signaling via the EGFR that among other things induced cellular re-localisation of AUF1 (Figure 6B).
Figure 6. The EGFR pathway induces NKG2D ligand upregulation via the abrogation of AUF1-mediated mRNA destabilization.
(A) Under normal conditions, NKG2D ligand mRNAs are constitutively targeted by AUF1 for degradation via their AU-Rich Elements. (B) Activation of the EGFR, either by its ligand or by any of several physico-chemical stresses leads activates the MEK pathway and the exclusion of AUF1 from the nucleus, allowing NKG2D ligand mRNAs to be stabilized and translated and the proteins to be expressed at the cell surface, where they may engage NKG2D on cytolytic lymphocytes.
These findings are consistent with evidence that diverse environmental stress provokes the oxidative inactivation of active sites of phosphatases that constitutively suppress EGFR activity (35). Some quantitative variation in the levels of induction across experiments (e.g. see Figures 1B, 1D and 1E) most likely reflects the fine balance in any cell preparation between the activity of those phosphatases; the factors mediating RNA degradation, including AUF1; and the levels of MAPK activity induced by various components of the exposome. This variation notwithstanding, the increases in surface ligand expression consistently induced by UVB and other EGFR activators were sufficient to render cells targets for NK and γδ T cell-mediated immune surveillance.
All human NKG2D ligand transcripts for which relevant sequence data are available contain AREs, and given that surface expression of several NKG2D ligands was regulated in parallel with that of MICA and MICB, this mode of NKG2D ligand regulation is probably a general one in humans. Indeed, no differential responses of surface MICA and MICB protein were noted to any of the agents tested, although this does not preclude additional and differential means of regulation under different circumstances. The convergence of many different forms of stress on a common node of NKG2D ligand activation should provoke efficient and robust immune surveillance of many potentially threatening components of the exposome.
At the same time, the fine details of the regulation of different ligands can vary (21). Thus, p38 inhibition shows a differential impact on ULBP2 expression compared to MICA/B, with our ongoing studies implicating HuR in the regulation of ULBP2 but not MICA/B RNAs. Such differential regulation is most evident in the mouse where regulation via EGFR applied only to Mult1, the one mouse gene clearly harbouring canonical 3′ ARE sequences. It is evident from the comparison of heat-shock and UVB regulation of MICA (Fig 1A; Fig S1A,B) that NKG2D ligands are regulated in multiple ways. Therefore it is possible that the DNA damage pathway and primary transcriptional control play a greater role in the regulation of murine Rae1 and H60 than they do in the regulation of most human NKG2D ligands. A species-specific dichotomy in the major pathways of regulation need not be surprising since the main ligands for NKG2D in mice and humans are not strictly orthologs, as is also the case for NK inhibitory receptors. Nonetheless, it is clearly appropriate to seek further clarification over the mechanisms by which human NKG2D ligands may be regulated by DNA damage. In the meantime, this study strengthens the implication of human NKG2D ligands in human cancer biology since the EGFR pathway is one of the most frequently dysregulated in tumors, particularly during the transformation of epithelial cells that are among the most established targets of lymphoid stress surveillance (24).
This study has begun to describe a pathway of human NKG2D ligand regulation, but there are as yet many gaps to fill. For example, how does AUF1 ordinarily destabilise NKG2D ligand RNAs and how does MAPK signaling promote its re-localisation? Moreover, future studies should examine whether disease associated allelic variation in NKG2D ligands might affect the sensitivity to this form of gene regulation. The dissection of gene regulation described here is very difficult to undertake in primary cells and given the lack of conservation of this form of regulation of NKG2D ligands in the mouse, it is equally challenging to design appropriate animal models. In this study, differentiated Caco-2 cells and primary human keratinocytes were used to validate key observations, but this does not detract from the need to cross-reference to other independent data sets as much as is possible. This was achieved here by the gene expression analysis of over 170 primary cancers that supported the hypothesis that EFGR regulates human NKG2D ligands in vivo.
Additionally, his study has not as yet integrated different means of NKG2D ligand regulation. For example, do conditions that upregulate MICA/B or ULBP promoter activity also stabilize the respective RNAs or do these steps require separate regulation, perhaps as a checkpoint averting the precocious expression of NKG2D ligands by normal cells? Related to this, this study has not begun to ask which mechanism of regulation might be most relevant in particular circumstances? Nonetheless, the gene expression profiling of primary human breast cancers showed a positive correlation of most NKG2D ligands with EGFR expression and a negative correlation with both LRIG1, an EGFR inhibitor, and HNRNPD (AUF1), that was shown here to destabilize NKG2D ligand RNAs. These correlations are particularly striking given the complexity that will inevitably underlie the composition of cancer transcriptomes (60). Indeed, the correlation might argue that the impact of the EGFR pathway may be a dominant means of NKG2D ligand regulation in vivo. Only ULBP4 showed no significant correlations with either EGFR or LRIG1, perhaps reflecting a ligand-specific difference since ULBP4, by contrast to other ligands, showed a significant positive correlation with HER2 that is related to but distinct from EGFR (Figure S6A). Thus, the possibility that ULBP4 is regulated independently should be investigated. Likewise, this study has been limited to the regulation of NKG2D ligands by the EGFR pathway, whereas the likelihood exists that some or all NKG2D ligand genes are also regulated by MAPK-dependent RNA stabilization downstream of other RTKs, such as the platelet-derived growth factor receptor (PDGFR) that is likewise implicated in pathologic cellular dysregulation.
Importantly, the evidence provided here that EGFR activation can modulate NKG2D ligand expression in confluent cells independent of cell proliferation offers a mechanism of immune surveillance that may be relevant to the early stages of cancer when the EGFR pathway and/or EGF levels become dysregulated within a small focus of transformed cells amid a differentiated epithelium. Conversely, this study does not suggest that normal epithelial cells responding to physiologic levels of EGF as part of their normal growth cycle will necessarily upregulate NKG2D ligands and thereby activate immune responses unnecessarily. A similar consideration applied to the E2F-mediated induction of NKG2D ligands in proliferating cells (19). Rather, it is more likely that acquired “immunological visibility” is a general property of epithelial cells that are either responding to supra-physiologic EGFR activation provoked by high levels of various “stressors”, or are experiencing abortive cell proliferation cues, unable to balance their differentiated state with chronic EGFR/MAP-kinase activation. Indeed, this may reflect the status of differentiated cells following infection with a number of viruses, several of which express molecules that suppress NKG2D-mediated immune-surveillance (61-64).
Post-transcriptional regulation is increasingly acknowledged to play a major role in the regulation of gene expression, particularly in response to environmental change. This has been brought into focus by the identification of many non-coding RNAs that regulate coding mRNAs in trans. However, ARE-mediated regulation remains a potent force, particularly in immune responses where it also limits the expression of cytokines. A survey recently identified seventeen categories of p37 AUF1-regulated genes, among which immunity and defence genes were the ones with the highest statistical significance (65). Of note, in being co-regulated by both EGF and AUF1, MICA segregates with a subset of profoundly important biological functions, including CXCL5 and IL-11 (65-68) - two genes that, like NKG2D ligands, are expressed by epithelial cells and have immuno-stimulatory potential.
Thus, one can view the data as further evidence that EGFR activation is de facto an immunological regulator. Consistent with this, EGFR can be trans-activated following LPS treatment (69), while two recently published papers described a large number of immunological mediators acutely regulated, up or down, by clinical inhibitors of the EGFR pathway, including Erlotinib (70, 71). Importantly, this impact on immune effectors could be related to skin pathologies that commonly accompany therapeutic use of EGFR inhibitors and which are a major problem in that they deter patient compliance. More specifically, the common association of EGFR inhibitor usage with susceptibility to Staphylococcus aureus and virus infection (72) would be consistent with defects in lymphoid stress surveillance caused by EGFR inhibition and the consequent impairment of NKG2D ligand upregulation, as predicted by this study. Indeed, there is widespread implication of the NKG2D pathway in anti-viral defenses and EGFR is reportedly triggered by several Staphylococcus aureus toxins (73-75) and by cytomegalovirus (CMV) infection (76), highlighting the potential host-benefit of the EGFR being directly linked to lymphoid stress surveillance.
Consistent with the mechanism of ligand regulation elucidated in this study, and based on the demonstrated lack of response of MHC Class I expression to EGFR activation, one may conclude that the EGFR-mediated regulation of NKG2D ligands is somewhat specific. Nonetheless, it must be noted that other authors have claimed that NKG2D ligand expression can be up-regulated by application of Erlotinib to human cells including Caco-2 cells (77, 78). Because this seemingly contrasts with this study, the experiments described here were repeated in multiple variations, but always with the same result; namely, the suppression of any of several NKG2D ligands by Erlotinib and by other EGFR inhibitors, consistent with the detailed molecular mechanism deduced. Possibly cells in other studies had become so stressed by inhibitor treatment that secondary mechanisms began to affect NKG2D ligand expression.
Importantly however, this study adds to several others that highlight that the capacity of clinical inhibitors of the EGFR pathway to regulate immune surveillance may militate the effects of those therapeutics, particularly given the capacity of a derepressed immune system to be of therapeutic benefit to patients with solid and hematological tumors. This fuels the view that EFGR, RTK, and MAPK pathways inhibitors that are used in caner should be stratified according to their effects on immune surveillance. Moreover, the identification of specific factors and mechanisms regulating human NKG2D ligands, as has been achieved here, might facilitate the targeting of EGFR by strategies that do not impair NK cell and T cell recognition of tumors. The current study also argues for a clinical trial to test whether therapeutics targeting RTKs might best be supplemented with immune enhancement, rather than anti-inflammatories, as a means of promoting cancer therapy and reducing adverse events such as defects in skin barrier protection.
Materials and Methods
Study Design
The core of the investigative approach was the comparison of NKG2D ligand RNA and protein expression under defined circumstances. Because NKG2D ligand expression can be perturbed by changes in cell growth conditions, all experiments were meticulously controlled. Thus, the cell lines primarily employed (HaCat and Caco2) cease growth at confluence owing to contact inhibition, and were subject to “stress” or growth factor addition only after reaching confluence 48h beforehand. All control and experimental cell aliquots were treated comparably, for example the cells’ temporary removal from incubators for experimental or mock treatment, and their subsequent re-incubation in medium in which they had been previously growing and that had been stored pro tem at 37°C. All experiments were repeated independently, sometimes by different investigators (P.V; C.W; A.Turner) over a period of time of over 5 years, and were internally controlled, such that all conclusions reached could be based on the analysis of single experiments repeated several times, rather than relying on comparisons across experiments. The sample sizes and experimental repetitions were sufficient to permit rigorous statistical analysis as described in the Figure legends and text, including correction for multiple parameter analysis for the comparison of gene expression profiles. The specificity of reagents, particularly antibodies, was verified.
Cells and culture conditions
Caco2, HCT116, Int407 and HeLa cells were from the ATCC; HaCat (immortalized human keratinocytes). Polarized Caco2 monolayers were established as described (59). Primary human keratinocytes were isolated as described (79) from human skin samples obtained as discarded material after cutaneous surgery (St Thomas’ Hospital, London) under ethical approval of the Guy’s and St Thomas’ NHS Foundation Trust Research Ethics Committee (06/Q0704/18), adhering to the principles of the Declaration of Helsinki. All cells were cultured in DMEM + 10% foetal calf serum (FCS), penicillin (50units/mL), streptomycin (50μg/mL) and glutamine (2mM).
Primary murine keratinocytes were obtained from shaved mouse skin digested in a trypsin-GNK solution for 2h to dissociate epidermis from dermis. The former was further digested for 15min in trypsin-GNK and DNAse, filtered through a 70μm cell strainer, and washed 2× with RPMI. Cells were plated at 106 cells/mL in Keratinocyte Serum-Free Medium (SFM) supplemented with EGF in wells pre-coated with 50μg/mL rat-tail collagen type I (BD).
PBMC were isolated by Ficoll gradient centrifugation from blood obtained from healthy volunteers after informed consent, according to the Declaration of Helsinki.
Antibodies and reagents
A647 anti-MICA/B, APC anti-CD3, FITC anti-CD56, APC anti-HLA-E, PE anti-HLA-A/B/C, blocking anti-NKG2D and all isotype control were from Biolegend. PE anti-MICA, -MICB, ULBP1, -ULBP2 and -ULBP3 were from R&D systems. FITC anti-pan γδTCR was from Beckman Coulter. PE anti-CD107a was from BD. Cetuximab (Erbitux) was from Merck. Recombinant human EGF was from Calbiochem. All cell culture media and reagents, Alexa 488 goat anti-rabbit IgG, and Prolongold were from Invitrogen. Chemical inhibitors and rabbit polyclonal anti-AUF1 and anti-phosphoAUF1 were from Sigma-Aldrich. The Dual Luciferase reporter system was from Promega.
Metabolic Labeling and Immunoprecipitation
Medium was replaced with methionine- and cysteine-free RPMI and 5% dialysed FCS for 1h. 150μCi of 35S labeled methionine/cysteine (Amersham) were added and cells cultured for 6h at 37°C, 5% CO2. Cells were washed 3× with PBS and lysed with 1% NP40, 1mM PMSF buffer containing 50μl/mL protease inhibitor cocktail (lysis buffer). Lysates were pre-cleared for 2h at 4°C with Protein G Sepharose beads. Immunoprecipitation was carried out overnight at 4°C with 2μg MICA monoclonal antibody coupled to Protein G Sepharose beads. Beads were then washed 2× with lysis buffer and once with PBS. Sodium dodecyl sulphate-polyacrylamide gel electrophoresis sample buffer was added; the samples boiled for 5 minutes; and run on a 10% polyacrylamide gel. The gels were dried in a Bio-Rad gel drier at 80°C for 1h and exposed to film.
UV irradiation
Cells were irradiated under a single Westinghouse FS20 tube (8cm from the source). The dose was calibrated using a phototherapy radiometer (IL442A) and head SEE 1240. Medium was removed from cells and stored at 37°C, monolayers were rinsed with pre-warmed PBS and irradiation was carried out on cells covered with a meniscus of PBS. Following irradiation, PBS was removed and the cells’ original medium that had been maintained pro tem at 37°C was added back to the cells. Control cells were treated in identical fashion but not irradiated.
RNA isolation and first strand cDNA synthesis
RNA was isolated using TRIzol reagent (Invitrogen) from cell monolayers lysed directly in their flasks, followed by extraction with chloroform and precipitation with isopropanol. 5μg of RNA were reverse transcribed for 1h in the presence of 500μM dNTPs, 25nM oligo(dT)12-18 primer, 10μM DTT and 200 units Superscript II Reverse Transcriptase (Invitrogen).
qPCR
Quantitative real-time PCR was carried out using the TaqMan system for cDNAs generated from human cell lines. Primers and probes for MICA and MICB were designed using the Primer-Express software (Perkin-Elmer). GAPDH primers and probe have been reported (80). Amplicons were designed to cover an exon-exon boundary. Reporter and quencher dyes were 6-carboxyfluorescein (FAM) and 6-carboxytetramethylrhodamine (TAMRA), respectively. For monitoring murine NKG2D ligand expression, the QuantiTect SYBR Green PCR Kit was used (Qiagen). All qPCR primers are listed in Table S1A.
Plasmids, RT-PCR, cloning and generation of MICA-3′UTR mutants
pGL4.74 (encoding Renilla luciferase) was from Promega; pMax-FP-Green-N was from Lonza. All molecular biology reagents were from New England Biolabs. PCR were performed in 50μL with Phusion Taq DNA polymerase according to manufacturer’s instructions. All primers are listed in Table S1B. AUF1 cDNA was cloned from HaCat cells. The amplicon product (generated in the presence of 10% DMSO as the first exon contains >80% GC) was directly digested with EcoRI and XhoI restriction enzymes, followed by ligation into the pCR3.1 plasmid (Invitrogen). All four isoforms (p37, p40, p42, p45) were obtained and their sequence verified. GFP reporter constructs were generated by overlapping PCR. GFP was cloned from the pMax-FP-Green-N plasmid (primers GFP forward and reverse) and the 3′UTR of MICA including all three polyA signals was amplified from genomic DNA extracted from HeLa cells (primers GFP-M3U forward and reverse). 2μL of both PCR products were used as a template for the overlapping PCR using primers, GFP forward and GFP-M3U reverse. The resulting PCR product was cloned into pMax-FP-Green-N using HindIII and AlfII restriction enzymes and the sequence verified. Mutants were generated from this template by overlapping PCR using primers GFP forward, GFP-M3U reverse, and the M3U-mut0 to M3U-mut3 primer pairs. Renilla luciferase reporter constructs were generated in the same way, using the pGL4.74 plasmid and primers RLuc, Rluc-M3U forward and reverse, and HindIII and BamHI restriction enzymes. All transfections were carried out using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions.
Flow Cytometry
Cells were collected with PBS 10mM EDTA and stained in 50μL PBS 5% FCS (FACS buffer) with the indicated antibodies (all used at a 1/100 final dilution) for 30mins at 4°C. Cells were washed 2× with FACS buffer, and data acquired on a FACScalibur flow cytometer (Becton Dickinson) and analyzed with FlowJo (Tree Star).
CD107a assay
Activation of effector cells by measuring the cell surface translocation of the CD107a marker was performed as described previously (81), except that brefeldin A was omitted. Briefly, target cells and PBMCs were mixed 1:1 in the presence of the PE anti-CD107a antibody (1/30 final dilution) and incubated for 5h at 37°C and 5% CO2. Cells were washed with FACS buffer and stained with combinations of APC anti-CD3 and FITC anti-CD56 (NK cells) or APC–anti-CD3 and FITC anti-pan γδTCR (γδ T cells), acquired and analyzed as described above.
mRNA stability experiments
Confluent HaCat cells were serum-deprived overnight and treated (or not) with EGF (500ng/mL). 5μg/mL Actinomycin D (Act.D, Sigma) was then added to replicate flasks, which were subsequently harvested for mRNA extraction at defined time points. mRNA was quantified by spectrophotometry and 10μg of each sample separated on a 1.2% formalin-agarose gel and transferred overnight onto nylon membranes (Hybond N+, Amersham) by capillary blotting in 10× SSC. Blots were UV-crosslinked before hybridization. MICA, MICB and GAPDH probes were prepared by PCR and labeled with α32P dCTP using Megaprime labeling kit (Amersham) following manufacturer’s instructions. Probes were denatured at 95°C for 5min and hybridization was carried out in Expresshyb solution (Clontech) following manufacturer’s instructions. Blots were incubated with probe in roller bottles for 1h at 68°C, washed with 2× SSC, 0.5% SDS at room temperature, then with 0.1× SSC, 0.1% SDS at 50°C, and exposed to a storage phosphor screen and acquired using the Storm PhosphorImager (Amersham). Bands were quantitated using ImageJ software.
Confocal Microscopy
HaCat cells were grown to confluence on 13mm coverslips placed in 24 well plates and treated as described. Cells were then washed 2× with PBS, fixed for 20mins at room temperature with BD CellFix buffer, washed 2× with PBS and permeabilised with PBS 0.1% Triton X-100 for 30mins. Cells were then incubated with primary and secondary antibodies in PBS 0.02% Triton X-100 for 1h each, washed 2× with PBS and once in ultrapure water, and then mounted on glass slides using 10μL Prolongold. Samples were analysed using a TCS SP2 AOBS confocal microscope (equipped with an HCX PL APO CS [confocal scanning], 63.0×/1.4 oil objective; Leica).
Analysis of gene expression in breast cancer samples and cell lines
The Human Exon 1.0 ST Affymetrix Array was used to generate the gene expression data of 172 primary breast carcinomas as described (53). Briefly, gene expression data were normalized by using the quantile normalization method of Aroma Affymetrix package (82) of the R software project (http://www.r-project.org/), followed by combat normalisation to adjust for batch (83). The difference in gene expression of several NKG2D ligands was determined between tumours with the lowest 25% EGFR, ESR1 or HER2 or highest 25% LRIG1 and HNRNPD expression versus the remaining samples. Publicly available gene expression data of 26 triple negative breast cancer cell lines was obtained from and pre-processed as described (58). Probe sets with the highest variability across the dataset were used as a representative for each gene (MICA, MICB, and EGFR). The association between their gene expression levels was determined using Pearson’s Correlation.
Statistical analysis
Statistical significance for all experimental data was determined by paired or unpaired (as indicated for each experiment) 2-tailed t-tests, using the Excel software (Microsoft). Correlations between NKG2D ligands and EGFR, ESR2, HER2, LRIG1 and HNRNPD expression in primary breast cancer samples and triple negative breast cancer cell lines were analysed by Wilcoxon rank sum test and Pearson’s Correlation, respectively. The appropriateness and correct implementation of the statistical tests was independently verified.
Supplementary Material
Acknowledgements
We are grateful to Professor Antony Young (Department of Photobiology, St John’s Institute for Dermatology (SJID) London, UK, for help during the early phases of this research, and to many colleagues for discussions. HaCat cells were a kind gift from Dr. M. Allen (SJID). The monoclonal antibody to MICA (hu MIC-A-M673) for immunoprecipitation was a kind gift of Dr. D. Cosman (Amgen Corporation). Primary human materials were provided through the National Institutes of Health Research Biomedical Research Centre of Guy’s and St Thomas’ Hospital and King’s College London. Statistical tests were independently verified by Dr. L. Abeler-Dörner.
Funding: We thank CRUK (A.H and P.V), a Wellcome Trust Programme Grant (A.H); the European Community’s Seventh Framework Programme FP7/2007-2013 under grant agreement #PIEF-GA-2009-255285 (P.V); the UK Medical Research Council (C.S); and National Institute for Health Research (NIHR) Biomedical Research Centre at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. (A.G and A. Tutt).
Footnotes
List of Supplementary Materials
Figure S1. Induction of MICA by UVB and EGF is not due to heat-shock, DNA damage or cell proliferation.
Figure S2. NKG2D ligands are upregulated at the cell surface following EGF treatment.
Figure S3. Cell surface increase in NKG2D ligand expression by EGF is detected by cytotoxic lymphocytes.
Figure S4. NKG2D ligand induction by various stress components of the exposome is EGFR-dependent.
Figure S5. NKG2D ligand expression is regulated post-transcriptionally.
Figure S6. NKG2D ligand mRNAs contain ARE sequences in their 3′UTRs.
Figure S7. The ARE sequence and the MEK pathway regulate NKG2D ligand expression.
Figure S8. AUF1 regulates NKG2D ligand expression.
Figure S9. The EGFR/MEK pathway regulates AUF1 localization.
Figure S10. NKG2D ligand expression correlates with EGFR expression levels and is abrogated by Erlotinib.
Figure S11. EGF-induced upregulation of cell surface NKG2D ligand expression by confluent differentiated Caco-2 cells is abrogated by EGFR and MEK inhibitors.
Figure S12. Cetuximab inhibits NKG2D ligand expression.
Table S1. Primers used in this study.
Competing interests: The authors declare that they have no competing interests.
References
- 1.Wild CP. Complementing the genome with an “exposome”: the outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev. 2005;14:1847–1850. doi: 10.1158/1055-9965.EPI-05-0456. [DOI] [PubMed] [Google Scholar]
- 2.Hayday AC. Gammadelta T cells and the lymphoid stress-surveillance response. Immunity. 2009;31:184–196. doi: 10.1016/j.immuni.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Nedellec S, Sabourin C, Bonneville M, Scotet E. NKG2D costimulates human V gamma 9V delta 2 T cell antitumor cytotoxicity through protein kinase C theta-dependent modulation of early TCR-induced calcium and transduction signals. J Immunol. 2010;185:55–63. doi: 10.4049/jimmunol.1000373. [DOI] [PubMed] [Google Scholar]
- 4.Rincon-Orozco B, et al. Activation of V gamma 9V delta 2 T cells by NKG2D. J Immunol. 2005;175:2144–2151. doi: 10.4049/jimmunol.175.4.2144. [DOI] [PubMed] [Google Scholar]
- 5.Shafi S, et al. An NKG2D-mediated human lymphoid stress surveillance response with high interindividual variation. Sci Transl Med. 2011;3:113ra124. doi: 10.1126/scitranslmed.3002922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petukhova L, et al. Genome-wide association study in alopecia areata implicates both innate and adaptive immunity. Nature. 2010;466:113–117. doi: 10.1038/nature09114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsoi LC, et al. Identification of 15 new psoriasis susceptibility loci highlights the role of innate immunity. Nat Genet. 2012;44:1341–1348. doi: 10.1038/ng.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nausch N, Cerwenka A. NKG2D ligands in tumor immunity. Oncogene. 2008;27:5944–5958. doi: 10.1038/onc.2008.272. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Messina L, et al. Differential mechanisms of shedding of the glycosylphosphatidylinositol (GPI)-anchored NKG2D ligands. J Biol Chem. 2010;285:8543–8551. doi: 10.1074/jbc.M109.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldhauer I, et al. Tumor-associated MICA is shed by ADAM proteases. Cancer Res. 2008;68:6368–6376. doi: 10.1158/0008-5472.CAN-07-6768. [DOI] [PubMed] [Google Scholar]
- 11.Wilkinson GW, et al. Modulation of natural killer cells by human cytomegalovirus. J Clin Virol. 2008;41:206–212. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molinero LL, et al. Activation-induced expression of MICA on T lymphocytes involves engagement of CD3 and CD28. J Leukoc Biol. 2002;71:791–797. [PubMed] [Google Scholar]
- 14.Yamamoto K, et al. Oxidative stress increases MICA and MICB gene expression in the human colon carcinoma cell line (CaCo-2) Biochim Biophys Acta. 2001;1526:10–12. doi: 10.1016/s0304-4165(01)00099-x. [DOI] [PubMed] [Google Scholar]
- 15.Zou Y, Bresnahan W, Taylor RT, Stastny P. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J Immunol. 2005;174:3098–3104. doi: 10.4049/jimmunol.174.5.3098. [DOI] [PubMed] [Google Scholar]
- 16.Bahram S, Inoko H, Shiina T, Radosavljevic M. MIC and other NKG2D ligands: from none to too many. Curr Opin Immunol. 2005;17:505–509. doi: 10.1016/j.coi.2005.07.016. [DOI] [PubMed] [Google Scholar]
- 17.Lanca T, et al. The MHC class Ib protein ULBP1 is a nonredundant determinant of leukemia/lymphoma susceptibility to gammadelta T-cell cytotoxicity. Blood. 2010;115:2407–2411. doi: 10.1182/blood-2009-08-237123. [DOI] [PubMed] [Google Scholar]
- 18.Eagle RA, Trowsdale J. Promiscuity and the single receptor: NKG2D. Nat Rev Immunol. 2007;7:737–744. doi: 10.1038/nri2144. [DOI] [PubMed] [Google Scholar]
- 19.Jung H, et al. RAE-1 ligands for the NKG2D receptor are regulated by E2F transcription factors, which control cell cycle entry. J Exp Med. 2012;209:2409–2422. doi: 10.1084/jem.20120565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groh V, et al. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci U S A. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raulet DH, et al. Regulation of Ligands for the NKG2D Activating Receptor. Annu Rev Immunol. 2013 doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stern-Ginossar N, et al. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- 23.Yadav D, et al. Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-gamma-induced microRNA. J Immunol. 2009;182:39–43. doi: 10.4049/jimmunol.182.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, et al. ErbB receptors: from oncogenes to targeted cancer therapies. J Clin Invest. 2007;117:2051–2058. doi: 10.1172/JCI32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahram S, Bresnahan M, Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci U S A. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boukamp P, et al. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106:761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aoki-Yoshida A, Aoki R, Takayama Y. Protective effect of pyruvate against UVB-induced damage in HaCaT human keratinocytes. J Biosci Bioeng. 2013;115:442–448. doi: 10.1016/j.jbiosc.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Fernau NS, et al. Role of HuR and p38MAPK in ultraviolet B-induced post-transcriptional regulation of COX-2 expression in the human keratinocyte cell line HaCaT. J Biol Chem. 2010;285:3896–3904. doi: 10.1074/jbc.M109.081430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang HC, Chang TM, Chang YJ, Wen HY. UVB irradiation regulates ERK1/2- and p53-dependent thrombomodulin expression in human keratinocytes. PLoS One. 2013;8:e67632. doi: 10.1371/journal.pone.0067632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, et al. Selection of reference genes for gene expression studies in ultraviolet B-irradiated human skin fibroblasts using quantitative real-time PCR. BMC Mol Biol. 2011;12:8. doi: 10.1186/1471-2199-12-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Momparler RL, Karon M, Siegel SE, Avila F. Effect of adriamycin on DNA, RNA, and protein synthesis in cell-free systems and intact cells. Cancer Res. 1976;36:2891–2895. [PubMed] [Google Scholar]
- 32.Frenkel EP, Skinner WN, Smiley JD. Studies on a Metabolic Defect Induced by Hydroxyurea (Nsc-32065) Cancer Chemother Rep. 1964;40:19–22. [PubMed] [Google Scholar]
- 33.Matsushima T, Kobuna I, Sugimura T. In vivo interaction of 4-nitroquinoline-1-oxide and its derivatives with DNA. Nature. 1967;216:508. doi: 10.1038/216508a0. [DOI] [PubMed] [Google Scholar]
- 34.el-Deiry WS, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 35.Zheng ZS, Chen RZ, Prystowsky JH. UVB radiation induces phosphorylation of the epidermal growth factor receptor, decreases EGF binding and blocks EGF induction of ornithine decarboxylase gene expression in SV40-transformed human keratinocytes. Exp Dermatol. 1993;2:257–265. doi: 10.1111/j.1600-0625.1993.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 36.Lund LR, et al. Transcriptional and post-transcriptional regulation of the receptor for urokinase-type plasminogen activator by cytokines and tumour promoters in the human lung carcinoma cell line A549. Biochem J. 1995;310(Pt 1):345–352. doi: 10.1042/bj3100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marschall C, et al. UVB increases urokinase-type plasminogen activator receptor (uPAR) expression. J Invest Dermatol. 1999;113:69–76. doi: 10.1046/j.1523-1747.1999.00631.x. [DOI] [PubMed] [Google Scholar]
- 38.van Houdt WJ, et al. Oncogenic KRAS desensitizes colorectal tumor cells to epidermal growth factor receptor inhibition and activation. Neoplasia. 2010;12:443–452. doi: 10.1593/neo.92088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng H, et al. Stress kinase p38 mediates EGFR transactivation by hyperosmolar concentrations of sorbitol. J Cell Physiol. 2002;192:234–243. doi: 10.1002/jcp.10134. [DOI] [PubMed] [Google Scholar]
- 40.Knebel A, Rahmsdorf HJ, Ullrich A, Herrlich P. Dephosphorylation of receptor tyrosine kinases as target of regulation by radiation, oxidants or alkylating agents. EMBO J. 1996;15:5314–5325. [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 42.Chen CY, Shyu AB. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 43.Proudfoot NJ. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tian B, Hu J, Zhang H, Lutz CS. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33:201–212. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clark A, Dean J, Tudor C, Saklatvala J. Post-transcriptional gene regulation by MAP kinases via AU-rich elements. Front Biosci. 2009;14:847–871. doi: 10.2741/3282. [DOI] [PubMed] [Google Scholar]
- 46.White EJ, Brewer G, Wilson GM. Post-transcriptional control of gene expression by AUF1: mechanisms, physiological targets, and regulation. Biochim Biophys Acta. 2013;1829:680–688. doi: 10.1016/j.bbagrm.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zucconi BE, Wilson GM. Modulation of neoplastic gene regulatory pathways by the RNA-binding factor AUF1. Front Biosci (Landmark Ed) 2011;16:2307–2325. doi: 10.2741/3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.David PS, Tanveer R, Port JD. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. RNA. 2007;13:1453–1468. doi: 10.1261/rna.501707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen CY, Xu N, Zhu W, Shyu AB. Functional dissection of hnRNP D suggests that nuclear import is required before hnRNP D can modulate mRNA turnover in the cytoplasm. RNA. 2004;10:669–680. doi: 10.1261/rna.5269304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aigner A, et al. Marked increase of the growth factors pleiotrophin and fibroblast growth factor-2 in serum of testicular cancer patients. Ann Oncol. 2003;14:1525–1529. doi: 10.1093/annonc/mdg416. [DOI] [PubMed] [Google Scholar]
- 51.Balcan E, et al. Serum levels of epidermal growth factor, transforming growth factor, and c-erbB2 in ovarian cancer. Int J Gynecol Cancer. 2012;22:1138–1142. doi: 10.1097/IGC.0b013e31825b7dcc. [DOI] [PubMed] [Google Scholar]
- 52.Kim BK, et al. The multiplex bead array approach to identifying serum biomarkers associated with breast cancer. Breast Cancer Res. 2009;11:R22. doi: 10.1186/bcr2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gazinska P, et al. Comparison of basal-like triple-negative breast cancer defined by morphology, immunohistochemistry and transcriptional profiles. Mod Pathol. 2013 doi: 10.1038/modpathol.2012.244. [DOI] [PubMed] [Google Scholar]
- 54.Jacot W, et al. The HER2 amplicon in breast cancer: Topoisomerase IIA and beyond. Biochim Biophys Acta. 2013;1836:146–157. doi: 10.1016/j.bbcan.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 55.Masuda H, et al. Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat. 2012;136:331–345. doi: 10.1007/s10549-012-2289-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renoir JM, Marsaud V, Lazennec G. Estrogen receptor signaling as a target for novel breast cancer therapeutics. Biochem Pharmacol. 2013;85:449–465. doi: 10.1016/j.bcp.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 57.Laederich MB, et al. The leucine-rich repeat protein LRIG1 is a negative regulator of ErbB family receptor tyrosine kinases. J Biol Chem. 2004;279:47050–47056. doi: 10.1074/jbc.M409703200. [DOI] [PubMed] [Google Scholar]
- 58.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ou G, et al. Contribution of intestinal epithelial cells to innate immunity of the human gut--studies on polarized monolayers of colon carcinoma cells. Scand J Immunol. 2009;69:150–161. doi: 10.1111/j.1365-3083.2008.02208.x. [DOI] [PubMed] [Google Scholar]
- 60.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chalupny NJ, Rein-Weston A, Dosch S, Cosman D. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun. 2006;346:175–181. doi: 10.1016/j.bbrc.2006.05.092. [DOI] [PubMed] [Google Scholar]
- 62.Cosman D, et al. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity. 2001;14:123–133. doi: 10.1016/s1074-7613(01)00095-4. [DOI] [PubMed] [Google Scholar]
- 63.Matusali G, et al. Soluble ligands for the NKG2D receptor are released during HIV-1 infection and impair NKG2D expression and cytotoxicity of NK cells. FASEB J. 2013;27:2440–2450. doi: 10.1096/fj.12-223057. [DOI] [PubMed] [Google Scholar]
- 64.Thomas M, et al. Down-regulation of NKG2D and NKp80 ligands by Kaposi’s sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2008;105:1656–1661. doi: 10.1073/pnas.0707883105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu X, et al. Combinatorial mRNA binding by AUF1 and Argonaute 2 controls decay of selected target mRNAs. Nucleic Acids Res. 2013;41:2644–2658. doi: 10.1093/nar/gks1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Furugaki K, et al. Erlotinib inhibits osteolytic bone invasion of human non-small-cell lung cancer cell line NCI-H292. Clin Exp Metastasis. 2011;28:649–659. doi: 10.1007/s10585-011-9398-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lal A, et al. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McMellen ME, et al. Epidermal growth factor receptor signaling modulates chemokine (CXC) ligand 5 expression and is associated with villus angiogenesis after small bowel resection. Surgery. 2010;148:364–370. doi: 10.1016/j.surg.2010.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hsu D, et al. Toll-like receptor 4 differentially regulates epidermal growth factor-related growth factors in response to intestinal mucosal injury. Lab Invest. 2010;90:1295–1305. doi: 10.1038/labinvest.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lichtenberger BM, et al. Epidermal EGFR Controls Cutaneous Host Defense and Prevents Inflammation. Sci Transl Med. 2013;5:199ra111. doi: 10.1126/scitranslmed.3005886. [DOI] [PubMed] [Google Scholar]
- 71.Mascia F, et al. Genetic Ablation of Epidermal EGFR Reveals the Dynamic Origin of Adverse Effects of Anti-EGFR Therapy. Sci Transl Med. 2013;5:199ra110. doi: 10.1126/scitranslmed.3005773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eilers RE, Jr., et al. Dermatologic infections in cancer patients treated with epidermal growth factor receptor inhibitor therapy. J Natl Cancer Inst. 2010;102:47–53. doi: 10.1093/jnci/djp439. [DOI] [PubMed] [Google Scholar]
- 73.Breshears LM, Schlievert PM, Peterson ML. A disintegrin and metalloproteinase 17 (ADAM17) and epidermal growth factor receptor (EGFR) signaling drive the epithelial response to Staphylococcus aureus toxic shock syndrome toxin-1 (TSST-1) J Biol Chem. 2012;287:32578–32587. doi: 10.1074/jbc.M112.352534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomez MI, Seaghdha MO, Prince AS. Staphylococcus aureus protein A activates TACE through EGFR-dependent signaling. EMBO J. 2007;26:701–709. doi: 10.1038/sj.emboj.7601554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haugwitz U, et al. Pore-forming Staphylococcus aureus alpha-toxin triggers epidermal growth factor receptor-dependent proliferation. Cell Microbiol. 2006;8:1591–1600. doi: 10.1111/j.1462-5822.2006.00733.x. [DOI] [PubMed] [Google Scholar]
- 76.Wang X, et al. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature. 2003;424:456–461. doi: 10.1038/nature01818. [DOI] [PubMed] [Google Scholar]
- 77.Bae JH, et al. Susceptibility to natural killer cell-mediated lysis of colon cancer cells is enhanced by treatment with epidermal growth factor receptor inhibitors through UL16-binding protein-1 induction. Cancer Sci. 2012;103:7–16. doi: 10.1111/j.1349-7006.2011.02109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He S, et al. Enhanced interaction between natural killer cells and lung cancer cells: involvement in gefitinib-mediated immunoregulation. J Transl Med. 2013;11:186. doi: 10.1186/1479-5876-11-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Modi BG, et al. Langerhans cells facilitate epithelial DNA damage and squamous cell carcinoma. Science. 2012;335:104–108. doi: 10.1126/science.1211600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barbany G, et al. Manifold-assisted reverse transcription-PCR with real-time detection for measurement of the BCR-ABL fusion transcript in chronic myeloid leukemia patients. Clin Chem. 2000;46:913–920. [PubMed] [Google Scholar]
- 81.Betts MR, et al. Sensitive and viable identification of antigen-specific CD8+ T cells by a flow cytometric assay for degranulation. J Immunol Methods. 2003;281:65–78. doi: 10.1016/s0022-1759(03)00265-5. [DOI] [PubMed] [Google Scholar]
- 82.Bengtsson H, Irizarry R, Carvalho B, Speed TP. Estimation and assessment of raw copy numbers at the single locus level. Bioinformatics. 2008;24:759–767. doi: 10.1093/bioinformatics/btn016. [DOI] [PubMed] [Google Scholar]
- 83.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.