Abstract
Background
Lipid synthesis and storage are accomplished by lipid droplets (LDs). The perilipin family of LD-associated proteins, comprising 5 members (PLIN1-PLIN5), has been well characterized in adipocytes but not in sebocytes, epithelial cells in which LD formation is a key feature of the cellular differentiation.
Methods
Perilipin expression in the sebaceous gland cell line SZ95 and in human sebaceous glands was studied by qRT-PCR, Western blots, and immunohistochemistry. Lipid accumulation was evaluated by Nile red staining and mass spectrometry.
Results
PLIN2 and PLIN3 are the most abundant perilipins in undifferentiated sebocytes. Induction of lipogenesis by linoleic acid (LA) resulted in increased transcript levels of all perilipins except for PLIN3 and in a time-dependent increase of PLIN2 protein. Nile red staining revealed that siRNA-mediated downregulation of PLIN2 significantly impaired basal and LA-induced lipid accumulation. Mass spectrometry revealed PLIN2 deficiency to cause a reduction in the amount of several specific lipid fractions, including di- and triacyl-glycerol esters, phosphatidylcholine lipids, and ceramides in sebocytes under basal conditions. In contrast, PLIN2 downregulation exerted a statistically significant inhibitory effect only on the accumulation of specific LA-induced triglycerides. PLIN2-deficient mice showed normal morphology of sebaceous glands. However, their sebaceous glands were significantly reduced in size and showed less cell proliferation.
Conclusions
PLIN2 is the major perilipin regulated during sebocyte differentiation in vitro. PLIN2 is also important for sebaceous lipid accumulation in vitro and regulates sebaceous gland size in vivo.
General significance
Our study provides the first systematic analysis of LD-associated proteins in sebocytes.
Keywords: Sebaceous glands, Lipid droplets, Perilipins
1. Introduction
Sebum is a holocrine secretion composed by lipids synthesized by sebocytes and the cellular debris of these cells, which are disrupted after terminal differentiation [1–3]. While several functions have been attributed to sebum, including a role in epidermal barrier and in hair follicle integrity, as well as antibacterial and antioxidant properties [4,5], its exact role remains uncertain. Unquestionable, however, is the key role played by increased or deregulated sebum production during the most common skin disease, acne [6,7].
During differentiation, sebocytes accumulate large amounts of lipids within cytoplasmic vesicles known as lipid droplets (LDs). Traditionally considered relatively inert storage organelles, LDs are now regarded as dynamic structures implicated in a remarkable number of biological processes. This new view is reflected in the large number of studies published in recent years covering lipid droplet genesis, composition, modification, and regulation [8–12]. LDs contain essentially neutral lipids and are limited by a phospholipid monolayer [13]. Several types of proteins are embedded in this membrane, including structural proteins, enzymes involved in lipid synthesis, lipases, and membrane-trafficking proteins. The perilipin family is a major group of LD-associated proteins with both structural and regulatory functions [14,15]. Perilipins are proteins present in a wide range of evolutionary distinct organisms sharing a highly conserved sequence organization and the ability to bind the surface of LDs. Formerly collectively known as the PAT-family in recognition of the first three members localized to LDs, perilipin, adipophilin, and TIP47, the five known mammalian perilipins are now denominated perilipins 1 to 5 (PLIN1 to PLIN5) [16].
Although LDs have been most intensively studied – for logical reasons – in adipocytes, it has often been stressed that they can be found in almost every cell type. Favored examples are mammary epithelial cells, steroidogenic cells of the adrenal cortex and gonads, and macrophages [17]. Although lipid accumulation is a key feature of sebocyte differentiation, the study of the composition and regulation of LD-associated proteins in these cells has been limited to a few reports [18,19]. There are, however, many facts suggesting that LD biology may differ significantly between adipocytes and sebocytes. For instance, the composition of sebaceous lipids differs quite strongly from that of adipocyte lipids. Also, while lipids are released by cell disruption in the sebaceous glands, lipids of adipocytes are mobilized in response to complex metabolic stimuli in a process termed lipolysis. The aim of this work is to study the expression and function of the perilipin family members in sebocytes as a first step to shed light on the biology of sebaceous LDs.
2. Materials and methods
2.1. Cell culture studies
SZ95 sebocytes [20] were cultured in Sebomed® medium (Biochrom, Berlin, Germany) supplemented with 10% fetal calf serum (PAA, Pasching, Austria) and 5 μg/l epidermal growth factor (EGF, Biochrom). For differentiation, cells were cultured in the medium indicated above including linoleic acid (LA; 10−4 M, Merck, Darmstadt, Germany) for the indicated periods of time and without EGF.
2.2. siRNA-mediated downregulation
Lipofectamine RNAiMAX (Invitrogen, Darmstadt, Germany) was used to transfect SZ95 sebocytes at ~40% confluence in 6-well plates with siRNAs for PLIN2 or with a negative control siRNA (Silencer Select, Ambion, Austin, TX, USA). 24 h after transfection, cells were induced to synthesize lipids by adding LA to the culture medium as described above. The cells were allowed to differentiate for 48 h, harvested with ice-cold PBS and stored as cell pellets at −80 °C for the lipid analysis or dissolved in protein lysis buffer for Western blot analysis.
2.3. Quantitative RT-PCR
Total RNA was isolated with TRIZOL reagent (Invitrogen) and 5 μg of RNA samples was reverse-transcribed in a final volume of 35 μl using Superscript™ II Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions. Quantitative RT-PCR was carried out in a LightCycler®480 (Roche, Mannheim, Germany) using the primers listed in Table 1 (0.5 μM), 1 μl cDNA, 0.2 μM probe (Universal ProbeLibrary Set, Roche), and the LightCycler® 480 Probes Master (Roche) in a final volume of 10 μl. Cycle conditions were 95 °C for 5 min for the first cycle, followed by 45 cycles of 95 °C for 10 s, 60 °C for 15 s, and 72 °C for 1 s. Transcript copy numbers were normalized to peptidylprolyl isomerase A (PPIA) mRNA copies. The ΔCt value of the sample was determined by subtracting the average Ct value of the target gene from the average Ct value of the PPIA gene. For each primer pair we performed no-template control and no-RT control assays, which produced negligible signals that were usually greater than 40 in Ct value. Experiments were performed in duplicates for each sample.
Table 1.
Sequences of primers and probes employed for the quantitative RT-PCR analysis.
| PLIN1 | Forward primer | 5′-acattaaagggaagaagttgaagc-′3 |
| Reverse primer | 5′-ttctcctgctcagggaggt-′3 | |
| Universal probe | #42 (cat. no. 04688015001) | |
| PLIN2 | Forward primer | 5′-tcagctccattctactgttcacc-′3 |
| Reverse primer | 5′-cctgaattttctgattggcact-′3 | |
| Universal probe | #72 (cat. no. 04688953001) | |
| PLIN3 | Forward primer | 5′-ggacaagttggaggagaacct-′3 |
| Reverse primer | 5′-acacaagctccttggtgtcc-′3 | |
| Universal probe | #66 (cat. no. 04688651001) | |
| PLIN4 | Forward primer | 5′-agttccaagccagggacac-′3 |
| Reverse primer | 5′-ctgctgggccttttcaatc-′3 | |
| Universal probe | #1 (cat. no. 04684974001) | |
| PLIN5 | Forward primer | 5′-gatcacttcctgcccatgac-′3 |
| Reverse primer | 5′-caccgaacccacttcagg-′3 | |
| Universal probe | #30 (cat. no. 04687639001) | |
| PPIA | Forward primer | 5′-cctaaagcatacgggtcctg-′3 |
| Reverse primer | 5′-tttcactttgccaaacacca-′3 | |
| Universal probe | #48 (cat. no. 04688082001) |
2.4. Western blot analysis
Protein was extracted using Laemmli-extraction-buffer, and the protein concentration was estimated via bicinchoninic acid protein assay. 30 μg of total protein was separated by 12% SDS-PAGE and transferred to PVDF membranes (Millipore, Schwalbach, Germany) by semidry blotting. Membranes were blocked in 5% w/v fat-free milk powder (Roth, Karlsruhe, Germany) for 1 h at room temperature. After washing in Tris-buffered saline solution with 1% Tween20 (Sigma, Taufkirchen, Germany), membranes were incubated over night at 4 °C in 5% w/v BSA (Sigma) with the appropriated primary antibody. Primary antibodies were guinea pig anti-PLIN2 (1:2000) (Progen, Heidelberg, Germany, #GP40), guinea pig anti-PLIN3 (1:2000) (Progen, #GP30), mouse anti-Keratin 7 (1:2500) (Santa Cruz, Heidelberg, Germany, #SC-70936) and rabbit anti-A Tubulin (1:5000) (Cell Signaling, Frankfurt, Germany, #2125). After washing, membranes were incubated in 5% w/v fat-free milk powder with horseradish peroxidase-labeled secondary antibodies: donkey anti-guinea pig (1:10.000; #43R-ID039; Fitzgerald, North Acton, MA), horse anti-mouse (1:2500; #7076, Cell Signaling) or donkey anti-rabbit (1:2000; NA934V, GE Healthcare, Munich, Germany). Signals were detected using an enhanced chemiluminescence detection reagent (GE Healthcare) and appropriated X-ray films (GE Healthcare).
2.5. Human skin samples, mouse tissue collection, histology, immunohistochemistry, and morphometry
Human skin biopsy samples obtained after informed consent of patients were kindly provided by C. Rose, M.D., from the Department of Dermatology, University of Lübeck, Lübeck, Germany. All animal studies have been approved by the author's institution Review Board (Baylor College of Medicine). Plin2-deficient mice have been described previously [21]. Tail and back skin were fixed in 4% formaldehyde (in PBS), dehydrated through a rising alcohol row and embedded in paraffin. For immunohistochemistry, 3 μm sections were boiled for antigen retrieval for 20 min in 10 mM sodium citrate buffer pH 6.0. Endogenous peroxidase was blocked with 3% H2O2 for 15 min. After washing in TBS, the samples were blocked with 5% rabbit serum for 30 min. Primary antibodies (Progen, Heidelberg, Germany), guinea pig anti-PLIN1 (#GP29), anti-PLIN2 (#GP40) anti-PLIN3 (#GP30), anti-PLIN4 (#GP34), and anti-PLIN5 (#GP 31), were diluted 1:250 and incubated over night at 4 °C. The secondary antibody was diluted 1:250 (rabbit anti-guinea pig, DAKO, #PO141), and 3,3′-diaminobenzidine (KEN-EN-TEC, Taastrup, Denmark) was used as chromogen.
For the quantitative evaluation of sebaceous gland area, three different sections from the back skin of 4-month-old control and Plin2−/− mice (n = 3 for each group) were H&E-stained. Pictures covering a total length of 39 mm of epidermis per animal were taken with a 200× magnification lens using a Leica DFC425C digital camera (Leica Microsystems, Wetzlar, Germany). The area of all visible sebaceous glands was recorded with LAS Software Version 3.8.0 (Leica Microsystems) and employed to calculate the mean gland area.
2.6. Lipid analysis
For the Nile red assay, SZ95 sebocytes were washed twice with PBS, stained for 10 min with DAPI (2.5 μ/ml, Sigma), washed once with PBS and then stained with AdipoRed (Lonza, Walkersville, MD, USA) according to the manufacturer's instructions. After 10 min, the released fluorescence was read on a fluorimeter (Infinite M1000 microplate reader, Tecan Group, Männedorf, Switzerland) with excitation at 485 nm and emission at 572 nm. The actual readout was presented in relative fluorescence units (RFU).
Lipid composition of sebocyte cells was investigated by analyzing crude lipid extracts with rapid resolution – reversed phase – high performance liquid chromatography coupled with electrospray ionization and time of flight mass spectrometry (RR–RP/HPLC–ESI–ToF/MS) as previously described [22]. Briefly, lipids were extracted from 2 × 106 SZ95 sebocytes for each sample with absolute ethanol containing 0.025% butylated hydroxytoluene (BHT, Sigma-Aldrich, St. Louis, MO, USA) to prevent autoxidation. N-lauroyl-d-erythro-sphingosylphosphorylcholine (SM 12:0, Avanti Polar Lipids INC., Alabaster, AL, USA), deuterated cholesterol-2,2,3,4,4,6-d6 (d6CH), and glyceryl-d5-trihexadecanoate (d5TG 48:0) (CDN isotopes INC., Pointe-Claire, Quebec, Canada) were added as the internal standards (ISTDs) at a final concentration of 5, 50 and 10 μM, respectively, to control the recovery of lipids, analytical performance and to calculate the relative abundance of detected lipids. The concentrated ethanol phase was extracted with ethyl acetate. Profiles of intact cell lipids were acquired in positive ion mode. Three experiments performed in duplicate were analyzed. Accurate mass spectra were acquired in the m/z mass range 120–1200. For the identification of neutral and polar lipids, the HPLC system was connected to a triple quadrupole (QqQ) mass detector by means of the ESI interface operating in positive ion mode. MS/MS experiments performed with the QqQ MS detector were set to acquire neutral loss (NL) scans of 141 and 297 Da to detect phosphatidylethanolamines and LA-containing glycerolipids, respectively; precursor ion scans of 184 Da to detect phosphatidylcholines and sphingomyelins, whereas the precursor ion scan of 264 Da allowed to individuate ceramides and related compounds [23,24]. Mass Hunter Workstation software (see below) was used to control data acquisition and qualitative analysis for both ToF and QqQ MS acquisition.
The molecular feature extraction algorithm was used to extract individual molecular species by their accurate mass detected with the ToF MS. Lists of molecular features, which were the detected species characterized by accurate mass, isotopic pattern and absolute abundance, were produced from each analyzed sample and converted into exchange files then processed with Mass Profiler Professional (MPP) 12 (see below). Molecular features shared by the analyzed samples were aligned by their retention time (RT) in the chromatographic run, and accurate mass axis in order to compare their expression across the different experimental conditions. Compounds detected in the different samples and presenting consistent RT (shift below 6% of the RT) and accurate mass (mass error below 6 ppm) were assigned as the same molecular species. Relative abundance of individual features was obtained by normalizing their peak area by the area of the ISTDs. Compound identification and annotation were performed on the basis of in house databases, built with previously collected MS data [22] and using the METLIN Personal Metabolite Database by means of the IDbrowser tool, and the Molecular Formula Generator algorithm. The Human Metabolome Database (http://www.hmdb.ca/) and LIPIDMAPS (http://www.lipidmaps.org/) were used to confirm and extend the identification. The annotation of free fatty acids (FAs), diglycerides (DGs), triglycerides (TGs), and glycerophospholipids included the number of carbon atoms and of double bonds of the side FAs. Other lipids were annotated consistently with the names reported on LIPIDMAPS. Equipments, data and statistical analysis packages were from Agilent Technologies, CA, USA.
2.7. Data pretreatment and statistical analysis
Pretreatment of analytical data consisted of RT and m/z alignment and normalization by ISTDs, baselining towards blank runs to remove background noise, and logarithmic transformation of normalized abundance. Statistical evaluation of the aligned and normalized HPLC-ToF/MS data was performed using univariate analysis, including Student t-test and one-way analysis of variance (ANOVA) using the MPP 12 package.
Ct values of gene targets of qRT-PCR analysis were shown relative to Ct values of the PPIA cDNA in Fig. 1A. In Fig. 1B all values were related to the mean value of the undifferentiated cells and compared by Student's t-test (GraphPad Prism version 5.0 for Windows, GraphPad Software, San Diego, CA, USA). Data are presented as means ± SD or box-plots with median. Group differences were considered to be statistically significant if p < 0.05.
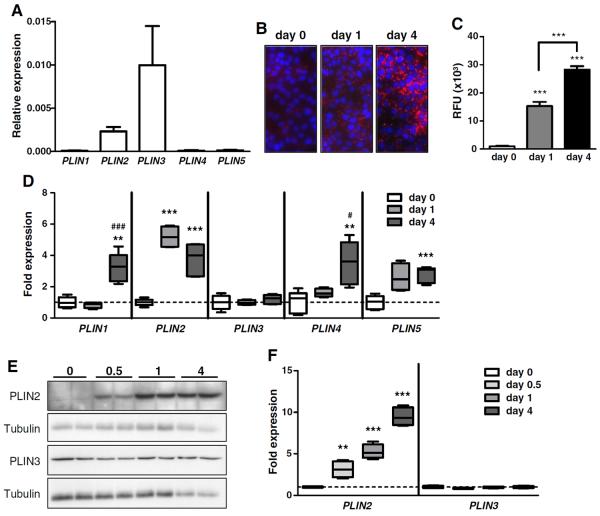
Fig. 1.
Abundance and regulation of perilipin members in SZ95 sebocytes. A, Analysis of perilipin transcript levels by quantitative RT-PCR in undifferentiated cells (n = 5 samples/group). B, Images of DAPI and Nile Red-stained SZ95 sebocytes before (day 0) and at days 1 and 4 after the addition of LA to the culture medium. C, Measurement of the emitted fluorescence of cells shown in B (n = 8 replicates/time point). D, Expression of perilipins at days 1 and 4 of LA-induced differentiation relative to undifferentiated cells (day 0). N = 5 replicates/group. E, Western blot analysis of PLIN2 and PLIN3 in undifferentiated cells (day 0) and at days 0.5, 1, and 4 after the induction of differentiation. The blots were stripped and employed for the detection of tubulin as a loading control. F, Quantification of PLIN2 and PLIN3 protein levels during the differentiation of SZ95 cells by densitometric analysis (n = 4 replicates/time point). Data were analyzed by Student's t-test. Asterisks indicate significant differences between day 1 or day 4 and undifferentiated cells (*p < 0.05, ** p < 0.01, *** p < 0.001). In D, # indicates significant differences between day 1 and day 4.
3. Results
3.1. The levels of PLIN2 increase during differentiation
Quantitative RT-PCR revealed that PLIN3 was the most abundant perilipin in undifferentiated SZ95 sebocytes (Fig. 1A). PLIN2 levels were about three times less abundant than those of PLIN3, while the remaining perilipins were practically undetectable (Fig. 1A). To evaluate relative changes in perilipin expression during differentiation, we treated SZ95 sebocytes with the essential fatty acid linoleic acid (LA), for 1 or 4 days. Staining with Nile red (Fig. 1B) and quantification of the released fluorescence in a fluorimeter (Fig. 1C) confirmed a progressive increase in lipid accumulation. As evaluated by qRT-PCR, PLIN2 transcripts were already significantly increased at day 1 and remained increased at day 4, while PLIN1, PLIN4, and PLIN5 levels only became significantly increased as compared to undifferentiated cells at day 4 (Fig. 1D). In contrast, PLIN3 transcripts did not change during differentiation (Fig. 1D). Western blot analysis revealed a time-dependent, progressive increase in PLIN2 levels with differentiation, while PLIN3 levels remained unchanged (Fig. 1E, F). The remaining perilipins were not detected in Western blots regardless of the differentiation stage (data not shown).
3.2. PLIN2 is strongly expressed in human sebaceous glands
To evaluate to which extent the expression of individual perilipins in cultured sebocytes reflects their relative abundances in human sebaceous glands we carried out immunohistochemistry in human skin. PLIN2 was the perilipin with the strongest expression in human sebaceous glands, where it localized properly to the surface of LDs (Fig. 2B, F). Weaker expression was detected for PLIN3 (Fig. 2C), PLIN4 (Fig. 2D), and PLIN5 (Fig. 2E), while PLIN1 was practically not detected (Fig. 2A). Negative control experiments, carried out by omitting the primary antibody, produced no specific signals (data not shown).
Fig. 2.
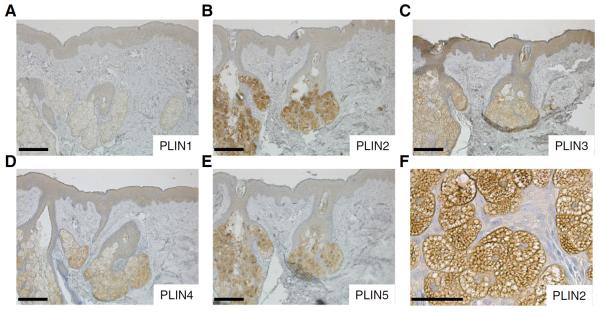
Detection of PLIN1-PLIN5 in human skin by immunohistochemistry. Size bars indicate 200 μm (A–E) or 50 μm (F).
3.3. PLIN2 downregulation impairs sebaceous lipogenesis in SZ95cells
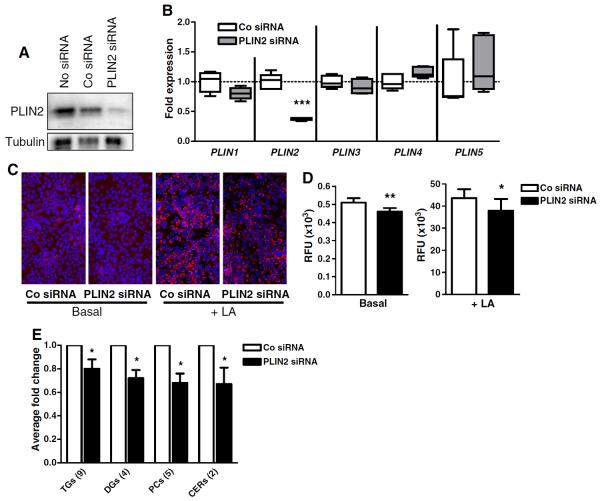
Next, we employed siRNA-mediated downregulation to evaluate the importance of PLIN2 during sebaceous lipogenesis. Since PLIN2 was the only perilipin showing a clear induction at both the mRNA and protein levels (Fig. 1D–F), we restricted our studies to this family member. Transfection of SZ95 sebocytes with PLIN2 siRNAs resulted in almost complete depletion of the corresponding PLIN, in contrast to untreated cells or cells transfected with a negative control siRNA (Fig. 3A). Quantitative RT-PCR revealed a ~70% reduction in PLIN2 transcript levels and unchanged transcript levels for the remaining perilipins (Fig. 3B), confirming the specificity of the employed siRNA. Nile red staining suggested a lower lipid accumulation following PLIN2 downregulation under basal conditions and after LA-induced lipid synthesis (Fig. 3C). Quantitative measurement of lipid accumulation using a fluorimeter confirmed this observation: PLIN2 downregulation resulted in significantly less lipid synthesis after 48 h of culture under both basal and LA-induced conditions (Fig. 3D).
Fig. 3.
Downregulation of PLIN2 impairs sebaceous lipogenesis. A, Western blot analysis showing the downregulation of PLIN2 protein following the transfection of SZ95 sebocytes with specific siRNAs. B, Quantitative RT-PCR revealed a ~70% reduction in PLIN2 transcript levels and unchanged transcript levels for the remaining PLINs (n = 5 replicates/group). C, Nile red and DAPI staining demonstrating reduced lipid accumulation in SZ95 cells with siRNA-mediated downregulation of PLIN2 after 48 h under basal conditions or LA treatment. D, Measurement of released fluorescence in a fluorimeter. The results (n = 8 replicates/group) are representative for two independent experiments and are presented as relative fluorescence units (RFU). E, Average fold change (FC) of the annotated lipids significantly less abundant in PLIN2 silenced cells under basal conditions grouped in their respective lipid class (TGs, triglycerides; DGs, diglycerides; PCs, glycerophosphatidylcholines, and CERs, ceramides). The figures in brackets indicate the number of the individual species characterized in each group and detailed in Table 2. *: p < 0.05; **: p < 0.01.
To investigate in greater detail the impact of PLIN2 downregulation on the lipid composition of SZ95 sebocytes, we employed a method based on rapid resolution reversed phase high performance liquid chromatography coupled to electrospray–time of flight mass spectrometry (RR-RP/HPLC-ESI-ToF/MS), which allows for the simultaneous detection of several hundreds of neutral lipid species in their intact form. Supplemental Fig. 1 shows total ion chromatograms (TICs) of lipid extracts under basal and LA-added conditions. TICs provided a clear evidence of the induction of lipid species in correspondence of the retention times of TGs in both control and PLIN2-downregulated cells stimulated with LA. Surprisingly, LA supply resulted in a comparable uptake of this substrate between control and PLIN2 siRNA-treated sebocytes (Supplemental Fig. 1), and siRNA-mediated PLIN2 downregulation had no visible effect on total TG accumulation after LA supplementation (data not shown). To assess the effects of LA on the individual TG species, molecular features were extracted with the MFE algorithm, and the relative abundances of the detected LA-containing TGs were summed and compared between experimental conditions. PLIN2 downregulation caused a modest (15.4%), yet significant (p < 0.05) effect on LA-containing TG accumulation after LA supplementation. This finding is at least in part consistent with the Nile red staining data (Fig. 3C–D), which showed a significant, ~14% lower lipid accumulation after LA treatment of PLIN2-depleted cells. By detailing the TG features we observed that, in contrast to trilinolein (TG 54:6; 18:2/18:2/18:2), which is more directly influenced by the availability of LA substrate and whose abundance was not influenced by PLIN2 loss, the abundance of some TGs such as TG 52:8, TG 52:6, TG 50:4, and TG 50:5, was significantly (p < 0.05) diminished when LA was administered to PLIN2-silenced cells (data not shown). These findings probably indicate that specific pathways of biotransformation of LA, or the availability of specific precursors, or the susceptibility to hydrolysis, may be under the influence of PLIN2. Nevertheless, it seems that LA supplementation provided such a powerful lipogenic stimulus that most of the negative effects resulting from the downregulation of a single perilipin family member were largely overridden.
Therefore, we next focused on species modified by downregulating PLIN2 under basal conditions. To address the complexity of the lipid signature consequent to PLIN2 downregulation, we investigated modifications of the abundance of the detected species with untargeted lipidomic strategies. ANOVA followed by Tukey HSD test showed that the downregulation of PLIN2 resulted in a statistically significant (p < 0.05) modification of the expression of 102 components in the lipid extracts. We could annotate several members of the TG lipid family and their precursors, such as diglycerides (DGs) among the species downmodulated in PLIN2 siRNA cells. Additionally, members of the phosphatidylcholines (PCs) and ceramides (CERs) were annotated on the basis of the experimental accurate mass and MS/MS data and of online resources (see Materials and methods). Fig. 3E shows the fold change of the annotated lipids significantly affected by PLIN2 downregulation grouped in their respective lipid class, while Table 2 reports the annotations, the HPLC–MS data, and the elemental composition of the individual species among the molecular features modified at a significant extent by PLIN2 downregulation.
Table 2.
HPLC–MS data and regulation of the molecular entities annotated among those found significantly changed following down regulation of PLIN2.
| Annotation | Elem. formula | Mass | Ion formula | m/z | RT | Co siRNA |
PLIN2 siRNA |
PLIN2 siRNA vs Co siRNA |
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| average RA | SD | average RA | SD | FC | p Value | ||||||
| TG 52:5 | C55 H96 O6 | 852,7207 | C55 H100 N O6 | 870,7545 | 21,157 | 0.032 | 0.008 | 0.027 | 0.006 | 0.83 | 0.026 |
| TG 52:3 | C55 H100 O6 | 856,7521 | C55 H104 N O6 | 874,7864 | 21,611 | 0.332 | 0.081 | 0.280 | 0.055 | 0.84 | 0.035 |
| TG 54:7 | C57 H96 O6 | 876,7207 | C57 H100 N O6 | 894,7551 | 21,071 | 0.046 | 0.007 | 0.038 | 0.008 | 0.82 | 0.016 |
| TG 54:4 | C57 H102 O6 | 882,7676 | C57 H106 N O6 | 900,8021 | 21,687 | 0.132 | 0.029 | 0.106 | 0.027 | 0.81 | 0.033 |
| TG 54:3 | C57 H104 O6 | 884,7833 | C57 H108 N O6 | 902,8171 | 21,907 | 0.838 | 0.137 | 0.521 | 0.087 | 0.62 | 0.040 |
| TG 56:8 | C59 H98 O6 | 902,7363 | C59 H102 N O6 | 920,7707 | 21,261 | 0.042 | 0.004 | 0.031 | 0.006 | 0.74 | 0.012 |
| TG 56:7 | C59 H100 O6 | 904,7521 | C59 H104 N O6 | 922,7864 | 21,471 | 0.162 | 0.019 | 0.127 | 0.022 | 0.78 | 0.006 |
| TG 56:6 | C59 H102 O6 | 906,7676 | C59 H106 N O6 | 924,8021 | 21,601 | 0.108 | 0.021 | 0.092 | 0.014 | 0.86 | 0.002 |
| TG 56:4 | C59 H106 O6 | 910,7989 | C59 H110 N O6 | 928,8333 | 22,018 | 0.081 | 0.019 | 0.070 | 0.016 | 0.87 | 0.046 |
| DG 34:2 | C37 H68 O5 | 592,5067 | C37 H72 N O5 | 610,5405 | 18,228 | 0.035 | 0.007 | 0.025 | 0.005 | 0.71 | 0.021 |
| DG 34:1 | C37 H70 O5 | 594,5223 | C37 H74 N O5 | 512,5562 | 18,608 | 0.311 | 0.046 | 0.238 | 0.034 | 0.76 | 0.050 |
| DG 36:3 | C39 H70 O5 | 618,5223 | C39 H74 N O5 | 636,5562 | 18,393 | 0.016 | 0.005 | 0.012 | 0.004 | 0.77 | 0.039 |
| DG 36:2 | C39 H72 O5 | 620,5381 | C39 H76 N O5 | 638,5718 | 18,836 | 0.417 | 0.056 | 0.260 | 0.033 | 0.62 | 0.029 |
| PC 32:0 | C40 H80 N O8 P | 733,5622 | C40 H81 N O8 P | 734,5694 | 17,965 | 1.751 | 0.299 | 1.403 | 0.229 | 0.80 | 0.039 |
| PC 37:2 | C45 H86 N O8 P | 799,6091 | C45 H87 N O8 P | 800,6164 | 18,954 | 0.176 | 0.049 | 0.117 | 0.032 | 0.66 | 0.032 |
| PC 40:8 | C48 H80 N O8 P | 829,5622 | C48 H81 N O8 P | 830,5694 | 17,252 | 0.088 | 0.012 | 0.061 | 0.008 | 0.69 | 0.009 |
| PC 42:10 | C50 H80 N O8 P | 853,5622 | C50 H 81 N O8 P | 854,5694 | 17,168 | 0.102 | 0.016 | 0.061 | 0.010 | 0.60 | 0.002 |
| PC 42:9 | C50 H82 N O8 P | 855,5778 | C50 H 83 N O8 P | 856,5851 | 17,414 | 0.061 | 0.010 | 0.038 | 0.006 | 0.62 | 0.015 |
| Cer (d18:1/16:0) | C34 H67 N O3 | 537,5121 | C68 H134 N2 O6 Na | 1098,0141 | 16,992 | 0.019 | 0.001 | 0.011 | 0.001 | 0.57 | 0.042 |
| Cer (d18:1/24:0) | C42 H83 N O3 | 649,6373 | C42 H82 N O2 | 632,6341 | 19,329 | 0.013 | 0.002 | 0.010 | 0.001 | 0.77 | 0.019 |
FC = fold change, m/z = mass to charge ratio, RA = relative abundance, RT = retention time, SD = standard deviation.
In summary, these data show that the loss of PLIN2 significantly affects the accumulation of diverse lipid classes in SZ95 cells under basal conditions. In contrast, in LA-treated cells, the deficiency of PLIN2 exerted a negligible influence on the uptake of this substrate and had a small, yet significant, impact on its overall biotransformation into TGs.
3.4. PLIN2 regulates sebaceous gland size in mice
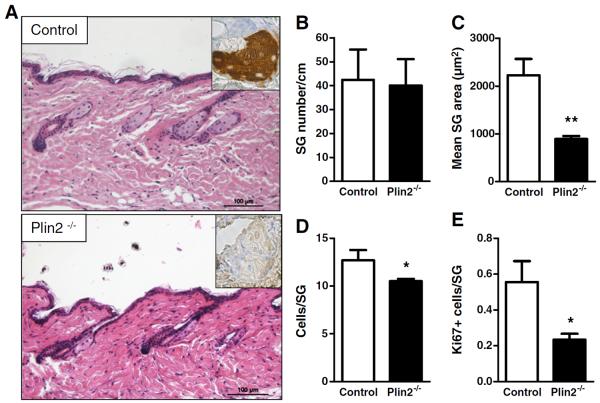
Perilipin 2 knockout mice (Plin2−/−) are known to have a significant reduction in hepatic triglycerides and to be resistant to diet-induced fatty liver, but their adipose differentiation is normal [21]. Also, no skin alterations have been reported so far. Evaluation of H&E-stained histological sections of the skin of Plin2−/− mice revealed the presence of normal-looking sebaceous glands (Fig. 4A), indicating that PLIN2 is dispensable for sebaceous gland formation. Quantitative analysis revealed that the number of sebaceous glands in Plin2−/− mice was not altered as compared to control mice (Fig. 4B). In contrast, the mean area of sebaceous glands in Plin2−/− mice was significantly reduced, reaching approximately half the size of the glands in control mice (Fig. 4C). Also, the sebaceous glands of PLIN2-deficient mice contained significantly less cells (Fig. 4D), and evaluation of cell proliferation by Ki67 staining revealed a significant reduction in the number of proliferating cells in PLIN2-deficient mice compared to control animals (Fig. 4E). Notably, thin layer chromatography (TLC) revealed no qualitative or quantitative changes in the epidermal lipids of Plin2−/− mice as compared to control animals (data not shown).
Fig. 4.
Reduced sebaceous gland size in Plin2−/− mice. A, H&E staining of the back skin of Plin2−/− mice (lower panel) and control littermates (upper panel). Inserts represent immunohistochemistry against PLIN 2 in sebaceous glands. Scale bars indicate 100 μm. Quantitative analysis showing unchanged number of sebaceous glands (B), reduced size of SGs (C), reduced cell number/SG (D), and reduced cell proliferation (E) in SGs of Plin2−/− as compared to control littermates. Data were analyzed by Student's t-test (n = 3 per group). *: p < 0.05; **: p < 0.01.
4. Discussion
Sebocyte differentiation involves cell dislodgement from the periphery to the core of the sebaceous gland with concomitant synthesis and accumulation of lipids within cytoplasmic structures known as LD. Once a certain size is attained, the cell disrupts and releases its contents into the gland, a process termed holocrine secretion. The synthesized lipids and cellular debris eventually reach the skin surface via the hair follicle canal, and represent the main component of epidermal sebum [1–3]. Although it is a key feature of these cells, the study of sebocyte lipid droplets has been so far largely neglected, and most studies have been carried out in other lipid-producing cells such as adipocytes and mammary gland epithelial cells. Here, we evaluated the expression and potential function of the perilipin family, the best studied group of LD-associated proteins, in cultured human sebocytes, human sebaceous glands, and in mice genetically lacking PLIN2.
Both quantitative RT-PCR and Western blot analysis revealed that PLIN2 and PLIN3 were the perilipins with the strongest expression in SZ95 sebocytes. PLIN2, also highly abundant in human sebaceous glands as shown by immunohistochemistry, was the only perilipin showing a time-dependent increase in expression during sebocyte differentiation. This finding is in accordance with the preferential expression of PLIN2 in human tissues with large amounts of lipids [25] and further justifies the frequent use of PLIN2 to identify sebocytes in genetically modified mouse lines with a sebaceous gland phenotype [26,27].
Collectively, TGs make up the largest fraction of the sebaceous mixture synthesized by the sebaceous gland. The mechanisms accounting for the synthesis of TGs from FAs have been poorly characterized in human sebaceous cells. In contrast, a larger body of evidence has been accumulated in adipocytes, demonstrating that, at least in part, the formation of DGs, which are the precursor of TGs, relies on the biotransformation of polar lipids, mostly glycerophospholipids. LA has been shown to be transformed into sebaceous lipids at a greater extent compared to other FA precursors [28]. While β-oxidation of LA accounts for the increment of non FA-containing lipids, such as squalene, most of the unmodified LA is esterified into TGs and polar lipids. Nile red staining and quantitative measurement of lipid accumulation using a fluorimeter revealed that siRNA-mediated downregulation of PLIN2 did not block the ability of SZ95 cells to produce lipid droplets and to synthesize LA-induced TGs, but it significantly reduced sebaceous lipid accumulation under basal conditions and after LA supplementation. There are at least two potential explanations for the rather mild reduction in lipid accumulation after PLIN2 downregulation in SZ95 cells. First, siRNA treatment did not completely deplete cells from PLIN2, and the residual levels of this perilipin may be sufficient to maintain most of its functions. Second, the loss of PLIN2 may induce a compensatory upregulation of other proteins. Evaluation of the expression of perilipin family members by qRT-PCR in PLIN2 siRNA-treated cells after 48 h of LA supplementation indeed revealed a significant, ~50% upregulation of PLIN4 transcripts, while PLIN1, PLIN3, and PLIN5 transcripts were unchanged and PLIN2 transcripts were, as expected, significantly reduced (data not shown). Considering the extremely low levels of PLIN4 in SZ95 cells (compare Fig. 1A), we consider it highly improbable that this increase in PLIN4 can compensate for the loss of PLIN2. However, we cannot exclude upregulation of and compensation by other unrelated LD-associated proteins.
In contrast to the Nile red method, mass spectrometry showed comparable amounts of LA and no significant changes in total TGs in control and PLIN2 siRNA conditions, demonstrating that PLIN2 influenced only weakly the uptake of LA and its transfer to TGs. Though, we detected statistically significant reductions in the amount of specific LA-derived TGs in PLIN2 siRNA-treated cells compared to control siRNA-treated ones. We postulate that the extremely strong lipogenic stimulus provided by LA supplementation overrides almost every negative influence the downregulation of a single perilipin family member may have. Alternatively, the latter findings may indicate that PLIN2 expression is less important for the final stages of TG synthesis. Besides, the hypothesis that PLIN2 can affect upstream pathways preceding the biotransformation of DGs into TGs may provide an explanation for the quantitative discrepancy between the Nile red and the mass spectrometry methods.
PLIN2 downregulation was associated with significant changes in the basal levels of TGs, DGs, PCs and CERs. This evidence suggests that the availability of precursors and alteration of membrane lipids might influence the levels of accumulated TGs [29]. Due to the complexity of the data, addressing the characterization of all the molecular features that were significantly modified could not be covered in the present study. Due to the lack of previous evidences, the untargeted approach used to address lipid composition of SZ95 sebocytes was suitable for the discovery of specific lipid changes associated with the altered PLIN2 expression. The present data provided first evidence that the downregulation of PLIN2 expression resulted in a moderate, yet significant, inhibition of TG formation accompanied by lower levels of members of DGs, which are the direct precursors in the biosynthetic pathways of TGs. The analysis of individual lipid species in an untargeted manner also highlighted changes in the abundance of members of the PC and sphingolipid classes. This observation indicated that PLIN2 may play a role in the composition of membrane lipids.
It has been suggested that a general function of perilipins is to restrict the access of lipases to LDs, thus preventing lipolysis [30]. Such a protective function has also been reported for PLIN2 by several groups (reviewed in [14]). Thus, while our data suggest that PLIN2 is important for normal lipogenesis in SZ95 cells, we cannot exclude that the reduced lipid accumulation is, at least in part, due to increased lipolysis.
Mice lacking PLIN2 are healthy and show normal adipose tissue differentiation and normal adipose lipolysis rate in both basal and stimulated conditions [21]. However, PLIN2-deficient mice have a significant reduction in hepatic TGs and are resistant to diet-induced fatty liver [21], and PLIN2-deficient females fail to form mature mammary gland alveoli and to nourish their first litters [31]. Here, we show that PLIN2 is dispensable for sebaceous gland development, as these glands are present and show a normal morphology in Plin2−/− mice. However, in the absence of PLIN2, the size of the sebaceous gland is significantly reduced, the glands contain fewer cells, and show reduced proliferation. Interestingly, no skin phenotype has been reported for mice lacking PLIN1 or PLIN5, which show, respectively, a significant reduction in adipose tissue mass [32,33], or a lack of LD specific to the heart [34]. The phenotype of mice lacking PLIN3 or PLIN4 has not been reported so far.
5. Conclusion
In conclusion, we studied for the first time the expression and potential function of the perilipin family, a group of LD-associated proteins, in cultured sebocytes and in sebaceous glands. PLIN2 emerged as the second most abundant perilipin in sebocytes, and as the major member regulated during the differentiation of these cells. In addition, the downregulation of PLIN2 in cultured sebocytes clearly impaired lipid accumulation. Finally, PLIN2-deficient mice showed significant reductions in sebaceous gland size, cell number, and cell proliferation. The reduced cell proliferation in sebaceous glands, allied to the changes in the abundance of membrane lipids such as PCs and sphingolipids in PLIN2 deficient SZ95 cells, suggests that PLIN2 may have unexplored roles in cellular functions beyond lipid droplet metabolism. However, there were no readily detectable changes in skin lipids compared to control mice. Collectively, these rather mild changes suggest a high degree of functional redundancy within the perilipin family or among LD-associated proteins in general. Such an attribute would lead to a high degree of robustness, allowing the LD to perform their essential function despite the loss of a single perilipin family member.
Supplementary Material
Acknowledgements
The authors would like to thanks C. Rose, M.D. (Department of Dermatology, University of Lübeck, Germany), for the human skin biopsy samples, Dr. Matteo Ludovici (Institute of Dermatology San Gallicano, Rome, Italy) for the preparation and LC/MS analysis of SZ95 sebocyte lipid extracts, and Stefanie Riesemann (Gene Center, Munich) for Western blot analysis. E.C., M.P., and M.L. were supported by the Italian Ministry of Health through the grant RF-2010-2316435. L.C. and B.C. were each supported by grants (HL-51586 and DK-084495, respectively) from the US National Institutes of Health.
This work was supported by a grant from the Cicatricial Alopecia Research Foundation to M.R.S.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagen.2013.05.016.
References
- [1].Jenkinson DM, Elder HY, Montgomery I, Moss VA. Comparative studies of the ultrastructure of the sebaceous gland. Tissue Cell. 1985;17:683. doi: 10.1016/0040-8166(85)90004-7. [DOI] [PubMed] [Google Scholar]
- [2].Thody AJ, Shuster S. Control and function of sebaceous glands. Physiol. Rev. 1989;69:383. doi: 10.1152/physrev.1989.69.2.383. [DOI] [PubMed] [Google Scholar]
- [3].Schneider MR, Paus R. Sebocytes, multifaceted epithelial cells: lipid production and holocrine secretion. Int. J. Biochem. Cell Biol. 2010;42:181. doi: 10.1016/j.biocel.2009.11.017. [DOI] [PubMed] [Google Scholar]
- [4].Smith KR, Thiboutot DM. Thematic review series: skin lipids. Sebaceous gland lipids: friend or foe? J. Lipid Res. 2008;49:271. doi: 10.1194/jlr.R700015-JLR200. [DOI] [PubMed] [Google Scholar]
- [5].Zouboulis CC, Baron JM, Bohm M, Kippenberger S, Kurzen H, Reichrath J, Thielitz A. Frontiers in sebaceous gland biology and pathology. Exp. Dermatol. 2008;17:542. doi: 10.1111/j.1600-0625.2008.00725.x. [DOI] [PubMed] [Google Scholar]
- [6].Webster GF. Acne vulgaris. BMJ. 2002;325:475. [PMC free article] [PubMed] [Google Scholar]
- [7].Kurokawa I, Danby FW, Ju Q, Wang X, Xiang LF, Xia L, Chen W, Nagy I, Picardo M, Suh DH, Ganceviciene R, Schagen S, Tsatsou F, Zouboulis CC. New developments in our understanding of acne pathogenesis and treatment. Exp. Dermatol. 2009 doi: 10.1111/j.1600-0625.2009.00890.x. [DOI] [PubMed] [Google Scholar]
- [8].Martin S, Parton RG. Lipid droplets: a unified view of a dynamic organelle. Nat. Rev. Mol. Cell Biol. 2006;7:373. doi: 10.1038/nrm1912. [DOI] [PubMed] [Google Scholar]
- [9].Fujimoto T, Ohsaki Y, Cheng J, Suzuki M, Shinohara Y. Lipid droplets: a classic organelle with new outfits. Histochem. Cell Biol. 2008;130:263. doi: 10.1007/s00418-008-0449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Walther TC, Farese RV., Jr. The life of lipid droplets. Biochim. Biophys. Acta. 2009;1791:459. doi: 10.1016/j.bbalip.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guo Y, Cordes KR, Farese RV, Jr., Walther TC. Lipid droplets at a glance. J. Cell Sci. 2009;122:749. doi: 10.1242/jcs.037630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Brasaemle DL, Wolins NE. Packaging of fat: an evolving model of lipid droplet assembly and expansion. J. Biol. Chem. 2012;287:2273. doi: 10.1074/jbc.R111.309088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T. The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 2002;277:44507. doi: 10.1074/jbc.M207712200. [DOI] [PubMed] [Google Scholar]
- [14].Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J. Lipid Res. 2007;48:2547. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- [15].Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. 2009;1791:419. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kimmel AR, Brasaemle DL, McAndrews-Hill M, Sztalryd C, Londos C. Adoption of PERILIPIN as a unifying nomenclature for the mammalian PAT-family of intra-cellular lipid storage droplet proteins. J. Lipid Res. 2010;51:468. doi: 10.1194/jlr.R000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Murphy DJ. The biogenesis and functions of lipid bodies in animals, plants and microorganisms. Prog. Lipid Res. 2001;40:325. doi: 10.1016/s0163-7827(01)00013-3. [DOI] [PubMed] [Google Scholar]
- [18].Akimoto N, Sato T, Iwata C, Koshizuka M, Shibata F, Nagai A, Sumida M, Ito A. Expression of perilipin A on the surface of lipid droplets increases along with the differentiation of hamster sebocytes in vivo and in vitro. J. Invest. Dermatol. 2005;124:1127. doi: 10.1111/j.0022-202X.2005.23718.x. [DOI] [PubMed] [Google Scholar]
- [19].Kaneta K, Honma M, Kishibe M, Iizuka H. Perilipin and adipophilin expression regulated by Rho-kinase during differentiation of hamster sebocyte. J. Dermatol. 2012;39:641. doi: 10.1111/j.1346-8138.2011.01359.x. [DOI] [PubMed] [Google Scholar]
- [20].Zouboulis CC, Seltmann H, Neitzel H, Orfanos CE. Establishment and characterization of an immortalized human sebaceous gland cell line (SZ95) J. Invest. Dermatol. 1999;113:1011. doi: 10.1046/j.1523-1747.1999.00771.x. [DOI] [PubMed] [Google Scholar]
- [21].Chang BH, Li L, Paul A, Taniguchi S, Nannegari V, Heird WC, Chan L. Protection against fatty liver but normal adipogenesis in mice lacking adipose differentiation-related protein. Mol. Cell. Biol. 2006;26:1063. doi: 10.1128/MCB.26.3.1063-1076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Camera E, Ludovici M, Galante M, Sinagra JL, Picardo M. Comprehensive analysis of the major lipid classes in sebum by rapid resolution high-performance liquid chromatography and electrospray mass spectrometry. J. Lipid Res. 2010;51:3377. doi: 10.1194/jlr.D008391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Taguchi R, Houjou T, Nakanishi H, Yamazaki T, Ishida M, Imagawa M, Shimizu T. Focused lipidomics by tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005;823:26. doi: 10.1016/j.jchromb.2005.06.005. [DOI] [PubMed] [Google Scholar]
- [24].Sullards MC. Analysis of sphingomyelin, glucosylceramide, ceramide, sphingosine, and sphingosine 1-phosphate by tandem mass spectrometry. Methods Enzymol. 2000;312:32. doi: 10.1016/s0076-6879(00)12898-8. [DOI] [PubMed] [Google Scholar]
- [25].Heid HW, Moll R, Schwetlick I, Rackwitz HR, Keenan TW. Adipophilin is a specific marker of lipid accumulation in diverse cell types and diseases. Cell Tissue Res. 1998;294:309. doi: 10.1007/s004410051181. [DOI] [PubMed] [Google Scholar]
- [26].Horsley V, O'Carroll D, Tooze R, Ohinata Y, Saitou M, Obukhanych T, Nussenzweig M, Tarakhovsky A, Fuchs E. Blimp1 defines a progenitor population that governs cellular input to the sebaceous gland. Cell. 2006;126:597. doi: 10.1016/j.cell.2006.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Dahlhoff M, Muller AK, Wolf E, Werner S, Schneider MR. Epigen transgenic mice develop enlarged sebaceous glands. J. Invest. Dermatol. 2010;130:623. doi: 10.1038/jid.2009.251. [DOI] [PubMed] [Google Scholar]
- [28].Pappas A, Anthonavage M, Gordon JS. Metabolic fate and selective utilization of major fatty acids in human sebaceous gland. J. Invest. Dermatol. 2002;118:164. doi: 10.1046/j.0022-202x.2001.01612.x. [DOI] [PubMed] [Google Scholar]
- [29].Harris CA, Haas JT, Streeper RS, Stone SJ, Kumari M, Yang K, Han X, Brownell N, Gross RW, Zechner R, Farese RV., Jr. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 2011;52:657. doi: 10.1194/jlr.M013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bell M, Wang H, Chen H, McLenithan JC, Gong DW, Yang RZ, Yu D, Fried SK, Quon MJ, Londos C, Sztalryd C. Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes. 2008;57:2037. doi: 10.2337/db07-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Russell TD, Schaack J, Orlicky DJ, Palmer C, Chang BH, Chan L, McManaman JL. Adipophilin regulates maturation of cytoplasmic lipid droplets and alveolae in differentiating mammary glands. J. Cell Sci. 2011;124:3247. doi: 10.1242/jcs.082974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Martinez-Botas J, Anderson JB, Tessier D, Lapillonne A, Chang BH, Quast MJ, Gorenstein D, Chen KH, Chan L. Absence of perilipin results in leanness and reverses obesity in Lepr(db/db) mice. Nat. Genet. 2000;26:474. doi: 10.1038/82630. [DOI] [PubMed] [Google Scholar]
- [33].Tansey JT, Sztalryd C, Gruia-Gray J, Roush DL, Zee JV, Gavrilova O, Reitman ML, Deng CX, Li C, Kimmel AR, Londos C. Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc. Natl. Acad. Sci. U.S.A. 2001;98:6494. doi: 10.1073/pnas.101042998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kuramoto K, Okamura T, Yamaguchi T, Nakamura TY, Wakabayashi S, Morinaga H, Nomura M, Yanase T, Otsu K, Usuda N, Matsumura S, Inoue K, Fushiki T, Kojima Y, Hashimoto T, Sakai F, Hirose F, Osumi T. Perilipin 5, a lipid droplet-binding protein, protects heart from oxidative burden by sequestering fatty acid from excessive oxidation. J. Biol. Chem. 2012;287:23852. doi: 10.1074/jbc.M111.328708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.