Abstract
The genus Orbivirus of the family Reoviridae includes a genetically diverse group of dsRNA arthropod-borne viruses that infect a wide variety of animal species. Here, we report the complete genome and phylogenetic analysis of a novel orbivirus (IAn-66411 or Sathuvachari virus, SVIV) isolated in 1963 from starlings (Brahminy myna) collected in Vellore, Tamil Nadu, India. Comparative genetic analysis of the SVIV polymerase (VP1 protein), core protein (VP3) and outer core protein (VP7) confirmed that SVIV is most closely related to the mosquito-borne orbiviruses, but that it is equally divergent from all known species. Therefore, SVIV should be tentatively considered as the prototype of a novel mosquito-associated Orbivirus species. These findings will aid in the development of molecular reagents that can identify genetically similar orbiviruses and help elucidate their geographical distribution, epidemiology, species tropism and possible disease association.
The genus Orbivirus of the family Reoviridae includes 22 virus species as well as 10 unclassified or unassigned viruses (Attoui et al., 2011). Orbiviruses are icosahedral, non-enveloped viruses with dsRNA genomes that encode seven distinct structural proteins (virion proteins, VP1–VP7) and four distinct non-structural proteins (NS1–NS4) (Attoui et al., 2011; Ratinier et al., 2011). Orbiviruses are global in distribution and infect a wide variety of vertebrate hosts, including wild and domestic ruminants and equids, rodents, bats, marsupials, birds, sloths and primates, including humans. Orbiviruses also replicate in and are transmitted by a variety of haematophagous arthropods (mosquitoes, ticks, phlebotomine sandflies and biting midges, depending on the virus).
Genetic characterization is crucial for the appropriate classification of unidentified viruses (Kapoor et al., 2008a, b). In turn, genomic sequence data can be used to develop molecular reagents for assessing the epidemiology and disease associations of novel viruses and their genetic relatives (Burbelo et al., 2012). We report here the complete genomic sequence of a novel orbivirus that was initially isolated in 1963 from a starling (Brahminy myna) collected in Vellore (Tamil Nadu, India). Based on comparative genetic analysis of all 10 genomic segments (GenBank accession nos KC432629–KC432638), IAn-66411 isolate (Sathuvachari virus, SVIV) is highly divergent from other known Orbivirus species, but is most closely related to the mosquito-borne virus species. A homology-based search against the NCBI non-redundant nucleotide sequence database yielded a partially characterized/unclassified orbivirus, JKT-8132, as the nearest genetic relative of SVIV. JKT-8132 (Tagtag virus, TGV) was isolated at the US Naval Medical Research Unit 2 (NAMRU-2), Jakarta (Indonesia), from a pool of Culex vishnui mosquitoes collected in Tagtag, Bali, Indonesia in 1980. On the basis of the phylogenetic analysis presented herein, we propose that SVIV and TGV viruses define a novel mosquito-transmitted species within the genus Orbivirus.
SVIV was initially isolated by intracerebral inoculation of newborn mice at the Virus Research Centre, Christian Medical College, Vellore, India (Carey et al., 1968a, b). TGV was first isolated in baby hamster kidney (BHK) and Aedes pseudoscutellaris (MOS-61) cells at NAMRU-2 (J. D. Converse, NAMRU-2, personal communication, 1982). Both viruses were subsequently sent to the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch for further characterization. Our initial attempts to culture SVIV from old lyophilized stocks by inoculation of newborn mice and culture in BHK and Vero cells were unsuccessful. However, subsequent attempts to grow the virus in C6/36 (Aedes albopictus) cells were successful. In contrast, TGV was viable and produced moderate cytopathic effect (CPE) in BHK and Vero E6 cells after 5 days of incubation at 37 °C. It also produced CPE in C6/36 cells after 4 days of incubation at 28 °C. Newborn mice inoculated intracerebrally with TGV became ill on the fourth and fifth days post-infection with symptoms of spasticity and incoordination. Antisera for serological tests were prepared in adult mice, using 10 % crude homogenates of TGV-infected newborn mouse brain in PBS as the immunogens. The immunization schedule consisted of four intraperitoneal injections of antigen mixed with Freund’s adjuvant, given at weekly intervals (Beaty et al., 1989). After the final immunization, mice were inoculated with sarcoma 180 cells, and the resulting immune ascitic fluids were collected. Complement fixation (CF) tests were performed by the microtitre technique, using 2 U of guinea pig complement and overnight incubation of the antigen and antibody at 4 °C (Knudson et al., 1984; Tesh et al., 1986). Antigens used in the CF tests were prepared from infected newborn mouse brain by the sucrose acetone extraction method and were inactivated with 0.05 % β-propiolactone (Sigma). No antigenic relationship could be shown between TGV and SVIV and other known orbiviruses in CF tests; however, transmission electron microscopy confirmed both viruses to have characteristic reovirus morphology (Fig. 1).
Fig. 1.
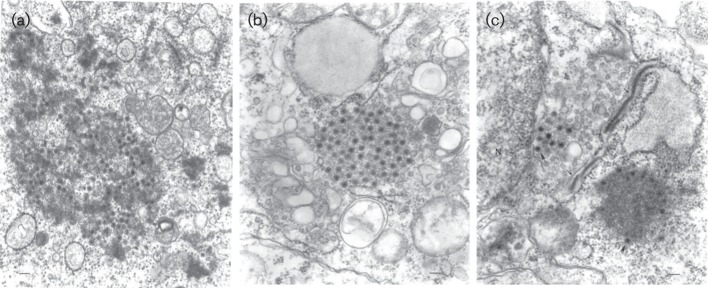
Electron micrographs of viruses SVIV (IAn-66411), TGV (JKT-8132) and TGV in BHK cells. Virions are shown by pointed arrows. (a) JKT-8132. Ultrastructure of reovirus fibrillar aggregate in the cytoplasm of a BHK cell with virus particles and cores. (b) IAn-66411 #5670. An aggregate of reovirus particles ~60 nm in diameter in the cytoplasm of a C6/36 cell, distance bar, 100 nm. (c) IAn-66411 #5672. A portion of a C6/36 cell infected with reovirus IAn-66411 showing viral protein aggregates with forming cores and virus particles ~60 nm in diameter (thick arrows) and microtubules (thin arrow) inside a cistern of granular endoplasmic reticulum which is expanded at one end. Bars, 100 nm.
For genetic characterization of SVIV, mouse brain suspensions, prepared by reconstituting old lyophilized virus stocks prepared in 1968. Virus nucleic acids were extracted from the filtrate and subjected to unbiased high-throughput sequencing (454 Roche) and applicable bioinformatics approaches (Kapoor et al., 2011; Victoria et al., 2009). Initial bioinformatics analysis indicated SVIV as a highly divergent orbivirus. Sequencing results were analysed further to acquire partial genomic sequences of all 10 segments of the virus. Assembled contigs (batch of sequences showing >95 % nt identity over >40 nt length) from the shotgun 454 sequencing reads were amplified by RT-PCR, which was followed by Sanger sequencing. Gaps between contigs were closed by designing PCR primers from the existing contigs spanning each gap. When needed, PCR products were cloned into pGEMT-easy vector and sequenced. 5′ Rapid amplification of cDNA ends (RACE) and 3′ RACE were used to acquire the terminal sequence of all 10 genomic segments (Kapoor et al., 2011).
The complete genome of SVIV comprises 18 834 nt pairs. SVIV segments 1 to 10 are 4015, 2860, 2393, 1997, 1789, 1644, 1205, 1188, 933 and 810 nt long and encodes VP1, VP3 (T2), VP2, VP4, NS1, VP5, NS2, VP7, VP6 and NS3 viral proteins, respectively. All these sequences were submitted to GenBank under the accession numbers: KC432629–KC432638. Genomic features and genetic relatedness of all 10 segments of SVIV is described in Table S1 (available in JGV Online). The orbiviruses share hexanucleotide termini that are partially conserved between the genomic segments of viruses within the same species and, to a lesser extent, between viruses of different species (Belaganahalli et al., 2011). The genomic segments of SVIV share six completely conserved nucleotides at their 5′ ends as well as four conserved nucleotides at their 3′ ends (5′-GGUUU/AA- virus gene- UACC-3′). Moreover, the first and last pair of nucleotides for each genome segment are inverted complements and identical to those reported for other known orbiviruses (Attoui et al., 2005, 2009; Belaganahalli et al., 2011, 2012). Previous studies have observed that the genomes of orbiviruses contain 5.03–5.695 % of non-coding region (NCR) in the mosquito-borne group, 4.47–4.9 % of NCR in the tick-borne group, and 3.5–4.1 % NCR in Culicoides-borne viruses (Belaganahalli et al., 2011). Analysis of the SVIV genome revealed 4.874 % NCR in its entire genome. It is noteworthy that SVIV has a lower percentage of NCRs relative to previously characterized mosquito-borne orbiviruses. The biological significance of lower percentage of NCR observed in SVIV is unknown. In this respect, SVIV more closely resembles the tick-borne agent Great Island virus (4.978 % NCR). Moreover, the T2- subcore protein (VP3) of SVIV also possesses more sequence similarity (46 %) with Great Island virus (Belhouchet et al., 2010) than with the mosquito-borne viruses (45 % NCR). In addition, SVIV shares <36 % identity in its VP5 protein with the Great Island virus, the same percentage observed with other mosquito-borne orbiviruses. The G+C content of SVIV is 42.3 mol%. Previous studies of other mosquito-borne viruses found a G+C content of 36.72 mol% in Peruvian horse sickness virus, 41.53 mol% in Umatilla virus and 41.55 mol% in Yunnan virus (Attoui et al., 2009). For tick-borne viruses, G+C content is between 51.93 mol% (St. Croix River virus) and 57.29 mol% (Great Island virus), while the midge-borne orbiviruses have an intermediate G+C content: 39.89 mol% in Chuzan virus to 45.89 mol% in equine encephalosis virus (Belaganahalli et al., 2011). Our analysis demonstrates that SVIV has a slightly higher G+C content than previously characterized mosquito viruses, and that this percentage is more similar to midge-borne viruses. In most orbivirus genome segments, the 5′ NCRs are shorter than the 3′ NCRs. In case of SVIV, all of the segments have shorter 5′ NCRs than 3′ NCRs, except for segment 7 (NS2) that has a 5′ NCR that is 57 bp and a 3′ NCR that is 47 bp. An earlier report found that segments 7 (NS2) and 9 (VP6) from Umatilla virus express 5′ NCRs longer than their 3′ NCRs. Also, segment 6 (VP5) of Yunnan orbivirus and segment 9 of Great Island virus contains longer 5′ NCRs relative to their 3′ NCRs (Belaganahalli et al., 2011). A unique feature of the orbivirus genome is the coding assignment of its different segments, i.e. each segment encodes protein(s) with their own putative function. The coding assignment varies between the strains/species, and the pattern can indicate which arthropod vector is preferred for the individual viruses (Attoui et al., 2005; Belaganahalli et al., 2011). Therefore, each segment of SVIV was analysed by blastx and/or conserved domain search (NCBI) to determine the type of protein that it encodes and its putative function. The SVIV coding assignments were compared to assignments previously published for other orbiviruses. The analysis revealed that segments 1, 9 and 10 of SVIV putatively encode polymerase, helicase and the viral release protein, respectively. SVIV segment 2 encodes VP3 (T2) and segment 3 is VP2 (OCP1), which is distinct from previously investigated orbiviruses.
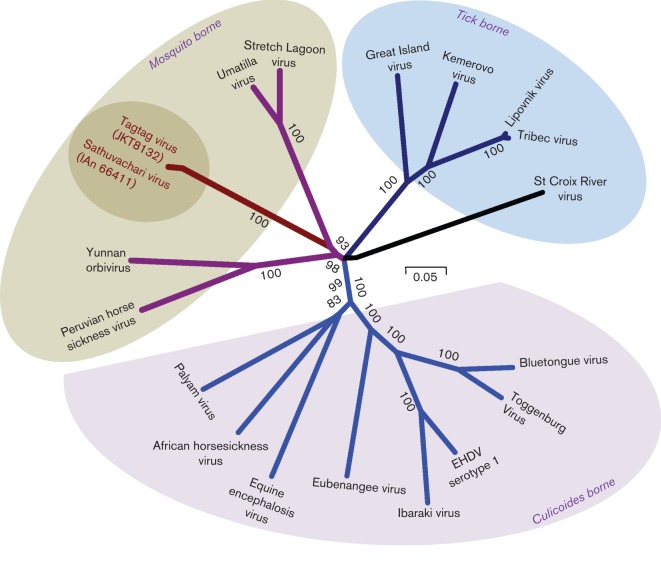
To determine the genetic relationship of SVIV with other viruses in the genus Orbivirus, we conducted phylogenetic analysis on all 10 virus genome segments (Figs 2 and 3). The type member for each Orbivirus species was included in the phylogenetic trees. The deduced amino acid sequences of SVIV virus were aligned with the homologous protein sequences of well-characterized orbiviruses using clustal w default parameters and BLOSUM protein weight matrix, as implemented in mega5 (Tamura et al., 2011). Protein alignments were used to calculate the Bayesian information criterion (BIC) for 48 unique protein substitution models, and the maximum-likelihood amino acid substitution model with the lowest BIC score was used to construct the phylogenetic tree (Tamura et al., 2011). The RNA-dependent RNA polymerase VP1 protein is the most evolutionarily conserved of all orbivirus proteins and has been used to classify novel orbiviruses into taxonomic groups (Attoui et al., 2009; Belaganahalli et al., 2011, 2012). Our phylogenetic analysis of the VP1 protein from SVIV and TGV suggest their close relationship with other mosquito-borne orbiviruses; thus, suggestive of their vector origin. However, the sequences from both novel viruses formed a separate branch within the group (Fig. 2). Confidence in phylogenetic analyses was assessed using the bootstrap method. We observed that the trees recapitulated previously reported classification of all well-characterized orbiviruses and that there was genetic clustering of viruses transmitted by a common arthropod vector (shown as different colour shades in Fig. 2) (Attoui et al., 2009; Belaganahalli et al., 2011, 2012). Almost the same tree topology was observed in phylogenetic analyses for all remaining nine segments (Fig. 3). Statistically, the VP1 protein of SVIV showed 10 % more amino acid identity with mosquito-borne viruses than with the tick- or midge-borne orbiviruses. The comparison of deduced amino acid sequences of all the segments of SVIV revealed that it is clearly distinct from other Orbivirus species investigated. SVIV showed <55 % identity in the polymerase protein, <46 % in T2 subcore protein, <18 % in outer capsid protein 1 (VP2) and <36 % in outer capsid protein 2 (VP5) with known orbiviruses (data not shown). We sequenced partial VP1, VP5, VP6, VP7, NS1 and NS2 genes of the TGV (GenBank accession nos KC439154–KC439159) using the same primers that were used for SVIV. The analysis showed that TGV had a maximum of 99 % (range of 94–99 %) identity with SVIV at the amino acid level. Our analysis confirms that SVIV and TGV viruses are genetically related variants of the same Orbivirus species.
Fig. 2.
Phylogenetic analyses of inferred amino acid sequences of the VP1 fragment of SVIV and TGV with other known orbiviruses; bootstrap values of >70 % are shown. The strains used for comparison with SVIV and TGV were retrieved from GenBank (accession numbers are YP_052968, ACY02806, YP_003240108, BAD89093, AFH41509, AEE98368, YP_002925132, YP_460038, YP_443925, YP_003896058, YP_052966, YP_052935, ADM88609, ADM88603, ADM88606, ACJ06234, YP_052942).
Fig. 3.
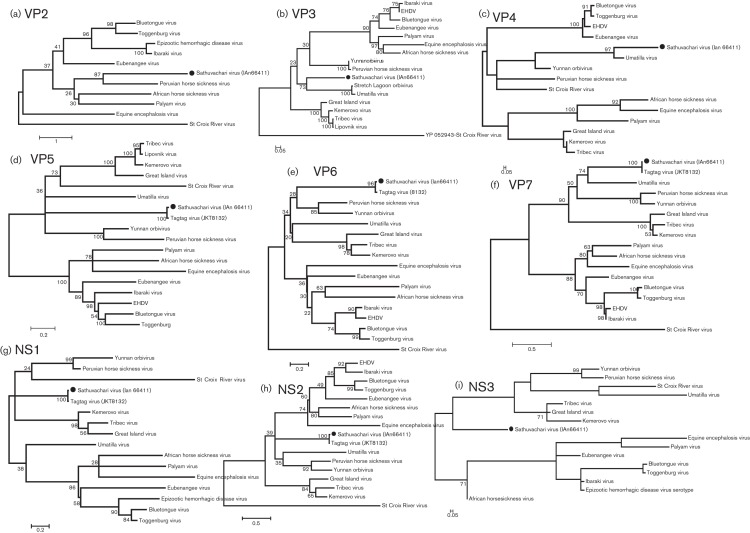
Phylogenetic analyses of inferred amino acid sequences of the nine genomic segments of SVIV. (a–i) The phylogenetic trees showing analysis of nine proteins encoded by segments VP2–7 and NS1–3, respectively. The GenBank accession numbers of strains that were used for this analysis are as follows: VP2, YP_052943, AFH41510, ADI79209, ACJ06245, AEY69029, YP_052931, CAN89166, YP_460040, ACJ06702, ADU57369; VP3, YP_002925133, YO_443926, YP_003896059, ADM88610, ADM88607, ADM88604, YP_052943, ACJ06236, AEE98369, AC053603, BAC67379, YP_052934, AEY69030, AAC40995, ACR58460, AFH41511; VP4, YP_460041, YP_443928, AEE98372, YP_052936, CAN89107, CAP04843, ACJ06237, YP_003896060, ADZ96231, ADZ96221, YP_052945, ACR58461, AFH41512, ACY02808; VP5, AEE98373, YP_003896063, YP_443930, ADZ96224, ADZ96234, ADM88605, YP_460042, YP_052946, YP_052932, CAE52975, YP_003240113, ACJ06239, ACJ06704, BAA93693, AFH41514, YP_052963; VP6, YP_460043, ADZ96227, YP_003896066, AEE98376, ADZ96237, ACO53605, AFH41517, CAN89173, YP_052937, ACJ65038, ACJ06250, CAN89112, YP_052950, ACJ06707, ACJ06242; VP7, AEE98375, YP_460044, YP_003896064, YP_443932, ADZ96226, ADZ96236, YP_052933, CAP04847, ACJ06241, P18259, YP_052949, ACJ06705, AFH41515, CAN89110, BAC20279; NS1, YP_443929, YP_460045, AEE98371, ADZ96222, ADZ96233, YP_003896061, ACH92681, YP_052938, AFH41513, CA085724, AAA91963, YP_052947, ACJ06238, ACJ06703; NS2, YP_460046, YP_443931, YP_003896065, ADZ96225, ADZ96235, CAP04848, YP_003240115, CAP12633, ACJ06240, YP_052948, ACJ 06706, AFH41516, YP_052939, AEE98374, BAC22192; NS3, AAB03411, AFH41518, ABU48536, ADZ96228, BAF40427, YP_443934, ACO53602, YP_003240117, AEP95960, YP_052951, YP_003896068, AEE98377, ADZ96238, YP_052940, ACJ06708.
Orbiviruses are known to infect multiple animal species (Attoui et al., 2011). Traditionally orbivirus isolates were classified based on their serological properties (Attoui et al., 2005; Belaganahalli et al., 2011, 2012; Palacios et al., 2011). The development of next generation sequencing technologies have repeatedly demonstrated their utility in identifying non-cultivable viruses and also in characterizing existing virus isolates that cannot be identified by more traditional laboratory methods (Victoria et al., 2008). Many studies that included all known orbiviruses have demonstrated that phylogenetic analysis and genetic relatedness can be used to classify uncharacterized orbiviruses (Belaganahalli et al., 2011, 2012; Palacios et al., 2011). Moreover, the orbiviruses transmitted by a common arthropod vector (i.e. mosquito, tick and midge) also show common ancestry or closer genetic relatedness. In the present study, these concepts and methods were adopted to completely characterize the genome of SVIV, which was isolated almost 50 years ago. Both SVIV and TGV were deposited in the WRCEVA collection as unknowns. However, their full characterization was not possible until full genome sequencing was done. Moreover, the sequence data generated for SVIV helped us to characterize another previously unclassified orbivirus, TGV http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=10892. The genetic information presented in this study should allow development of molecular reagents (PCR primers, serological assays etc.) that can now be used to define the prevalence, epidemiology, host range and possible disease association of SVIV and TGV.
Nonetheless, this is also an example of the continuing importance of primary virus isolation and deposition of unknown or novel viruses in permanent virus collections or repositories, so that such agents are available for study as new techniques become available or new pathogens appear (Arrigo et al., 2012). To date, nothing is known about the potential public health or veterinary importance of SVIV and TGV viruses, but their characterization now makes such studies feasible. Wider geographical sampling of vectors, animals and humans will provide a better description of the genetic diversity of this proposed new Orbivirus species. Serological assays will be needed to determine whether these viruses infect animals, including humans (Burbelo et al., 2011, 2012). The genetic characterization of a second novel virus (TGV) with a genetically divergent VP1 and other genes indicates that wider geographical sampling for related viruses will likely reveal other novel variants. The genetic diversity within this proposed species may also reflect a range of disease phenotypes upon their host. In conclusion, the sequence data of SVIV should provide sufficient information to develop specific molecular diagnostic assays that will allow confirmation of future outbreaks or cases of orbivirus infection and retrospective analysis of previously unconfirmed case; and it will also facilitate epidemiological studies.
Acknowledgements
This work was supported by the National Institutes of Health grants AI090196, AI081132, AI079231, AI57158 (North-east Biodefence Center-Lipkin), AI070411 and by the Defense Threat Reduction Agency. V. L. P., A. T. R. and R. B. T. were supported by NIH contract HHSN272201000040I/HHSN27200004/DO4.
Footnotes
A supplementary table is available with the online version of this paper.
References
- Arrigo N. C., Briese T., Calisher C. H., Drebot M. A., Hjelle B., LeDuc J. W., Powers A. M., Repik P. M., Roehrig J. T. & other authors (2012). Recommendations for publication of viral genetic data and sample access for novel viruses and strains. Am J Trop Med Hyg 86, 189–191 10.4269/ajtmh.2012.11-0523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attoui H., Mohd Jaafar F., Belhouchet M., Aldrovandi N., Tao S., Chen B., Liang G., Tesh R. B., de Micco P., de Lamballerie X. (2005). Yunnan orbivirus, a new orbivirus species isolated from Culex tritaeniorhynchus mosquitoes in China. J Gen Virol 86, 3409–3417 10.1099/vir.0.81258-0 [DOI] [PubMed] [Google Scholar]
- Attoui H., Mendez-Lopez M. R., Rao S., Hurtado-Alendes A., Lizaraso-Caparo F., Jaafar F. M., Samuel A. R., Belhouchet M., Pritchard L. I. & other authors (2009). Peruvian horse sickness virus and Yunnan orbivirus, isolated from vertebrates and mosquitoes in Peru and Australia. Virology 394, 298–310 10.1016/j.virol.2009.08.032 [DOI] [PubMed] [Google Scholar]
- Attoui H., Mertense P. P. C., Becnel J., Belaganahalli S., Bergoin M., Brussaard C. P., Chappell J. D., Ciarlet M., del Vas M. & other authors (2011). Virus taxonomy. In Ninth Report of the International Committee on Taxonomy of Viruses, 1 edn, p. 1338 Edited by King A. M. Q., Adams M. J., Carstens E. B., Lefkowitz E. J. Elsevier [Google Scholar]
- Beaty B., Calisher C. H., Shope R. E. (1989). Diagnostic Procedures for Viral, Rickettsial and Chlamydial Infections, 5th edn Edited by Schmidt N. J., Emmons R. W. Washington: American Public Health Association [Google Scholar]
- Belaganahalli M. N., Maan S., Maan N. S., Tesh R., Attoui H., Mertens P. P. (2011). Umatilla virus genome sequencing and phylogenetic analysis: identification of stretch lagoon orbivirus as a new member of the Umatilla virus species. PLoS ONE 6, e23605 10.1371/journal.pone.0023605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belaganahalli M. N., Maan S., Maan N. S., Nomikou K., Pritchard I., Lunt R., Kirkland P. D., Attoui H., Brownlie J., Mertens P. P. (2012). Full genome sequencing and genetic characterization of Eubenangee viruses identify Pata virus as a distinct species within the genus Orbivirus. PLoS ONE 7, e31911 10.1371/journal.pone.0031911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belhouchet M., Mohd Jaafar F., Tesh R., Grimes J., Maan S., Mertens P. P., Attoui H. (2010). Complete sequence of Great Island virus and comparison with the T2 and outer-capsid proteins of Kemerovo, Lipovnik and Tribec viruses (genus Orbivirus, family Reoviridae). J Gen Virol 91, 2985–2993 10.1099/vir.0.024760-0 [DOI] [PubMed] [Google Scholar]
- Burbelo P. D., Ching K. H., Esper F., Iadarola M. J., Delwart E., Lipkin W. I., Kapoor A. (2011). Serological studies confirm the novel astrovirus HMOAstV-C as a highly prevalent human infectious agent. PLoS ONE 6, e22576 10.1371/journal.pone.0022576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo P. D., Dubovi E. J., Simmonds P., Medina J. L., Henriquez J. A., Mishra N., Wagner J., Tokarz R., Cullen J. M. & other authors (2012). Serology-enabled discovery of genetically diverse hepaciviruses in a new host. J Virol 86, 6171–6178 10.1128/JVI.00250-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey D. E., Reuben R., Myers R. M. (1968a). Japanese encephalitis studies in Vellore, South India. I. Virus isolation from mosquitoes. Indian J Med Res 56, 1309–1318 [PubMed] [Google Scholar]
- Carey D. E., Rodrigues F. M., Myers R. M., Webb J. K. (1968b). Arthropod-borne viral infections in children in Vellore, South India, with particular reference to dengue and West Nile viruses. Indian Pediatr 5, 285–296 [PubMed] [Google Scholar]
- Kapoor A., Victoria J., Simmonds P., Slikas E., Chieochansin T., Naeem A., Shaukat S., Sharif S., Alam M. M. & other authors (2008a). A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc Natl Acad Sci U S A 105, 20482–20487 10.1073/pnas.0807979105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Victoria J., Simmonds P., Wang C., Shafer R. W., Nims R., Nielsen O., Delwart E. (2008b). A highly divergent picornavirus in a marine mammal. J Virol 82, 311–320 10.1128/JVI.01240-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor A., Simmonds P., Gerold G., Qaisar N., Jain K., Henriquez J. A., Firth C., Hirschberg D. L., Rice C. M. & other authors (2011). Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci U S A 108, 11608–11613 10.1073/pnas.1101794108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson D. L., Tesh R. B., Main A. J., St George T. D., Digoutte J. P. (1984). Characterization of the Palyam serogroup viruses (Reoviridae: Orbivirus). Intervirology 22, 41–49 10.1159/000149532 [DOI] [PubMed] [Google Scholar]
- Palacios G., Cowled C., Bussetti A. V., Savji N., Weir R., Wick I., Travassos da Rosa A., Calisher C. H., Tesh R. B. & other authors (2011). Rapid molecular strategy for orbivirus detection and characterization. J Clin Microbiol 49, 2314–2317 10.1128/JCM.00337-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratinier M., Caporale M., Golder M., Franzoni G., Allan K., Nunes S. F., Armezzani A., Bayoumy A., Rixon F. & other authors (2011). Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog 7, e1002477 10.1371/journal.ppat.1002477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28, 2731–2739 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesh R. B., Peleg J., Samina I., Margalit J., Bodkin D. K., Shope R. E., Knudson D. (1986). Biological and antigenic characterization of Netivot virus, an unusual new Orbivirus recovered from mosquitoes in Israel. Am J Trop Med Hyg 35, 418–428 [DOI] [PubMed] [Google Scholar]
- Victoria J. G., Kapoor A., Dupuis K., Schnurr D. P., Delwart E. L. (2008). Rapid identification of known and new RNA viruses from animal tissues. PLoS Pathog 4, e1000163 10.1371/journal.ppat.1000163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victoria J. G., Kapoor A., Li L., Blinkova O., Slikas B., Wang C., Naeem A., Zaidi S., Delwart E. (2009). Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J Virol 83, 4642–4651 10.1128/JVI.02301-08 [DOI] [PMC free article] [PubMed] [Google Scholar]